Abstract
Serratia marcescens is a model organism for the study of secondary metabolites. The biologically active pigment prodigiosin (2-methyl-3-pentyl-6-methoxyprodiginine), like many other secondary metabolites, is inhibited by growth in glucose-rich medium. Whereas previous studies indicated that this inhibitory effect was pH dependent and did not require cyclic AMP (cAMP), there is no information on the genes involved in mediating this phenomenon. Here we used transposon mutagenesis to identify genes involved in the inhibition of prodigiosin by glucose. Multiple genetic loci involved in quinoprotein glucose dehydrogenase (GDH) activity were found to be required for glucose inhibition of prodigiosin production, including pyrroloquinoline quinone and ubiquinone biosynthetic genes. Upon assessing whether the enzymatic products of GDH activity were involved in the inhibitory effect, we observed that d-glucono-1,5-lactone and d-gluconic acid, but not d-gluconate, were able to inhibit prodigiosin production. These data support a model in which the oxidation of d-glucose by quinoprotein GDH initiates a reduction in pH that inhibits prodigiosin production through transcriptional control of the prodigiosin biosynthetic operon, providing new insight into the genetic pathways that control prodigiosin production. Strains generated in this report may be useful in large-scale production of secondary metabolites.
INTRODUCTION
Prodigiosin is a highly biologically active molecule with no well-established physiological role (58). This membrane-associated red pigment is produced by some biotypes of Serratia marcescens (16), a Gram-negative bacterium found in the rhizosphere (22, 36), salt and fresh water, insect and mammal guts, and hospitals (16). Evidence suggests that prodigiosin functions in bacterial dispersal (5), competition against other organisms (14, 59), regulation of proton gradients, energy, and pH homeostasis (21, 41, 44), and determination of surface hydrophobicity (42, 43). Recent work has shown that prodigiosin has immunosuppressive and antitumor properties that may be of high clinical value (reviewed by Perez-Tomas and Vinas [38]). The regulatory pathways that control S. marcescens prodigiosin production are incompletely understood; however, more insight into another species of the genus Serratia, species ATCC 39006, has been acquired. Salmond and colleagues have identified a network of transcriptional regulators that control production of prodigiosin and other secondary metabolites in response to environmental and other stimuli (13, 17–19, 59). Improved knowledge of regulatory pathways may direct the design of strains with elevated prodigiosin production for use in industrial settings.
Prodigiosin production by S. marcescens and other Serratia species is regulated by a number of physiological and environmental factors, including, but not limited to, growth phase, temperature (56, 57), pH (50), oxygen concentration (24), and nutrient and mineral content of the medium (4, 15, 21, 49, 60).
d-Glucose is an excellent carbon source to promote growth of S. marcescens; however, as early as 1949, d-glucose was reported as a potent inhibitor of prodigiosin production (4). Because other useful secondary metabolites are inhibited by glucose (6, 11, 20, 26, 45), it would be advantageous to engineer strains for which glucose could support both growth and generation of secondary metabolites. The basis for the glucose inhibition of the prodigiosin phenotype (GIP) is incompletely understood, but GIP has been observed in several studies (4, 8, 15, 27). It is known that GIP occurs in a crp mutant strain and therefore does not require cyclic AMP (cAMP) receptor protein (CRP) (27). GIP was also associated with acidification of the growth medium (4, 50, 52). We reasoned that a genetic approach to find mutants able to generate prodigiosin in glucose-rich medium would provide novel insight into prodigiosin production pathways and perhaps provide useful strains for applied microbiology. The goal of this study was to use a genetic screen to identify the genes required for GIP.
While it is clear from previous studies that the inhibition of prodigiosin production by nonproliferating cells in glucose-rich medium stems from a reduction in pH (50, 52), the genes required for the pH change and to respond to the pH change are unknown. In addition, it is not clear whether low pH inhibits transcription of pigment biosynthesis genes or if it inhibits prodigiosin production at a posttranscriptional level. In this study, we characterize the glucose effect in growing cultures rather than nonproliferating cells, contrary to several previous studies, and use a genetic approach to provide insight into the mechanism by which glucose inhibits S. marcescens prodigiosin production. We report mutants that were defective in acidification of the medium and had mutations in either the quinoprotein glucose dehydrogenase gene (gdhS) or other genes contributing to GdhS activity. Using a transcriptional reporter, we observed that growth in glucose-rich medium inhibited transcription of the pigment biosynthesis operon and that this inhibition was dependent upon GdhS. Together these data support the model that the metabolism of d-glucose by quinoprotein glucose dehydrogenase (GDH) initiates a reduction in pH that inhibits prodigiosin production through transcriptional control of the prodigiosin biosynthetic operon.
MATERIALS AND METHODS
Strains, carbon sources, and growth conditions.
Bacterial strains used in this study are listed in Table 1. All bacteria were grown in lysogeny broth (LB; per liter, 10 g Bacto-tryptone, 5 g yeast extract, 5 g NaCl) alone or supplemented with different carbohydrates as listed in the text and on LB with agar (1.5% [wt/vol]). LB broth supplemented with d-glucose (110 mM) is referred to as LBG, and LB agar supplemented with d-glucose (220 mM) is referred to as LBG agar. The glucose concentrations used were determined in preliminary experiments to elicit complete inhibition of prodigiosin production, and a higher glucose concentration was required for the inhibitory effect with agar plates (data not shown). Carbohydrates used and their sources and product numbers are as follows: d-glucose (Fisher Scientific, BP350-1), l-glucose (Sigma, G5500), Sodium d-gluconate (Sigma, G9005), d-gluconic acid (Sigma, G1951), d-glucono-1,5-lactone (Sigma, G4750), methyl-α-d-glucopyranoside (Fluka, 66940), methyl α-d-pyrannoside (Sigma, M6882), d-galactose (Fisher Scientific, S80022), d-mannose (Sigma, M2069), l-arabinose (Sigma, A3252), lactose (Sigma, L8773), glycerol (Acros, 15892), and raffinose (Acros, 19567). Carbon sources were dissolved in LB broth and filter sterilized. Tris-HCl (Sigma, T3253) at 120 mM was used to buffer medium when noted; the Tris-HCl powder was added to LB or LBG medium that was adjusted to pH 8.0 and filter sterilized. Antibiotics were used at the following concentrations: tetracycline at 10 μg/ml, gentamicin at 10 μg/ml, and kanamycin at 50 μg/ml for Escherichia coli and 100 μg/ml for S. marcescens. Pyrroloquinoline quinone (PQQ) (Sigma, product number D7783) was added to LB liquid medium where noted to 3.03 μM. Biotyping of strains by carbon source utilization was performed following the protocol of Grimont and Grimont (16). All experiments were conducted at 30°C unless otherwise noted.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| Saccharomyces cerevisiae | ||
| InvSc1 | MATa/MATα leu2/leu2 trp1-289/trp1-289 ura3-52/ura3-52 his3-Δ1/his3-Δ1 | Invitrogen |
| E. coli | ||
| SM10 λpir | thi thr leu tonA lacY supE recA::RP4-2Tc::Mu pir | 34 |
| S17-1 λpir | thi pro hsdR hsdM+ ΔrecA RP4-2::TcMu-Km::Tn7 pir | 34 |
| S. marcescens | ||
| CMS376 | WT Presque Isle Cultures strain no. 3611 | 46, 47 |
| Nima | Pigmented laboratory strain | Pryce Haddix |
| CHASM | Pigmented environmental isolate | 27 |
| CMS613 | crp-1 (insertion mutation), null allele | 28 |
| CMS1083 | gdhS (rig-1) with restored crp gene | This study |
| CMS1687 | Δcrp has crp-Δ4 deletion mutation, null allele | 27 |
| CMS1758 | pigA::lacZ (pStvZ3 insertion at pigA) | 27 |
| CMS2526 | gdhS (rig-1) pigA::lacZ (pStvZ3 insertion at pigA) | This study |
| Plasmids | ||
| pMQ118 | nptII rpsL oriT oriR6K URA3 CEN6/ARSH4 | 47 |
| pMQ125 | aacC1 oriT orip15a URA3 CEN6/ARSH4 PBAD-lacZα | 47 |
| pMQ132 | oripBBR1 aacC1 oriT URA3 CEN6/ARSH4 | 47 |
| pMQ185 | pMQ125 + crp | This study |
| pStvZ3 | oriR6K lacZ nptII promoter probe; oriT URA3 CEN6/ARSH4 | 27 |
| pMQ268 | pStvZ3 + pigA internal fragment | 27 |
| pMQ335/pgdhS | pMQ132 + gdhS | This study |
| pMQ339/pgdhS | pMQ335 with aphA-3 replaced by aacC1 from pMQ132 | This study |
| pMQ346/pgcd | pMQ132 + gcd | This study |
| pMQ372/pubiB | pMQ132 + ubiB | This study |
Transposon mutagenesis and DNA manipulations.
Transposon mutations were generated as previously described (48) using the mariner transposon delivery vectors pBT20 (31) and pSC189 (7). Pools of mutants were plated on LB agar supplemented with 4% (wt/vol) d-glucose to inhibit prodigiosin production. After 48 to 72 h of incubation at 30°C, plates were screened for red colonies and no more than one pink/red colony was taken per plate to eliminate the possibility of isolating colonies with the same mutation. Two hundred fifty-eight separate mutagenesis pools were generated such that ∼180,000 mutant colonies were screened.
Transposon insertion locations were mapped using arbitrary PCR (37) or by rescue of the oriR6K-containing pSC189-derived plasmid as previously described (7).
Cloning and directed mutagenesis.
All genes were cloned using yeast in vivo recombination using previously described vectors (Table 1) (46, 47). DNA was amplified using Phusion high-fidelity DNA polymerase (New England BioLabs), and oligonucleotide primers were obtained from Integrated DNA Technologies. To clone the gdhS gene, the open reading frame (ORF) was amplified with primers 2235 and 2236 (all primers are listed in Table 2). The gcd gene was amplified from E. coli strain S17-1 using primers 2418 and 2419. Cloning vector pMQ132 (47) was used, and the constructs were verified by sequencing. The ubiB ORF was cloned in the same way as gdhS but using primers 2673 and 2676.
Table 2.
Oligonucleotide primers used in this study
| Primer no. | Primer sequencea |
|---|---|
| 2235 | tgtgagcggataacaatttcacacaggaaacagctATGCAAAATAAAGCGTCACTATCG |
| 2236 | attcgccattcaggctgcgcaactgttgggaaggTTACTTCTGATCGGGCAGGGCGTAAG |
| 2418 | cgccagggttttcccagtcacgacgttgtaaaacgacggTTACTTCACATCATCCGGCAG |
| 2419 | gaattgtgagcggataacaatttcacacaggaaacagATGGCAATTAACAATACAGGCTC |
| 1254 | ttatcagaccgcttctgcgttctgatttaatctgtatcaggaTTAACGGGTGCCGTAGAC |
| 1255 | aactctctactgtttctccatacccgtaggaggaaaaagaATGGTTCTCGGCAAACCGCA |
| 2673 | acgttgtaaaacgacggccagtgccaagcttgcatgcCCAGAGTAACGAATTAAATCAGG |
| 2676 | tggaattgtgagcggataacaatttcacacaggaaacagATGACCCCAGGCGAACTGCGT |
Parts of sequences in upper case target amplification by PCR. Parts of sequences in lowercase target recombination using yeast in vivo cloning.
The wild-type crp gene was restored in the rig-1 mutant as previously described (53). To complement crp in rig-13 and rig-A21, we cloned the full-length crp gene under the control of the E. coli PBAD promoter into a modified pMQ125 plasmid using primers 1254 and 1255. pMQ125 has a low- to medium-copy-number replicon, p15a, and is effective for complementation of mutations in S. marcescens (31, 48). The resulting plasmid, pMQ185, was verified by sequencing and introduced into rig-13, rig-A21, and Δcrp strains by conjugation. The Δcrp pigment and biofilm phenotypes were complemented, indicating that the construct was functional (data not shown). Arabinose was used at 0.2% (vol/vol) to induce expression of crp.
Growth, pH, d-gluconate, and prodigiosin measurements.
Prodigiosin was measured from liquid cultures as previously reported using cells from 1 ml of culture (27). The pH was determined from overnight cultures using Corning pH/ion analyzer model 350. Briefly, 1 ml of culture was placed in a microcentrifuge tube and centrifuged for 1 min at 16,100 × g, and the pH was recorded by inserting the probe directly into the supernatant.
The time course experiments were performed as follows: four overnight cultures from different single colonies of wild-type (WT) and gdhS strains were grown in 5 ml of LB broth and subcultured into either fresh LB medium or LBG to a final bacterial optical density at 600 nm (OD600) of 0.1. In order to prevent theoretical changes caused by reducing culture volume over time by the removal of sample aliquots, independent culture tubes were prepared for each time point. The cultures were placed at 30°C on a TC-7 tissue culture roller (New Brunswick) and rotated at high speed (speed setting 9). At 0-, 3-, 6-, 9-, 12-, and 24-h time points, culture turbidity (OD600) was measured to assess growth (Beckman DU-70 spectrophotometer, 1-cm path length), prodigiosin was measured from cells, pH of the culture medium was determined as described above, and samples of cells and supernatants were frozen for GDH and gluconic acid analysis. Each time course experiment was repeated at least twice with similar results.
The d-gluconic acid concentration was determined using an enzymatic d-gluconic acid detection kit (Megazyme, product K-GATE) as described by the manufacturer except that the volumes were scaled down for use in a microtiter plate, and a standard curve and linear regression were used to determine d-gluconic acid levels in the linear range of the assay. d-Gluconic acid was measured from supernatants from the time course described above; at least 6 independent replicates were measured per genotype and condition. d-Glucono-1,5-lactone was not measured, as it was only transiently present in the medium (data not shown).
Enzymatic assays.
Glucose dehydrogenase was measured using a chromogenic reaction based on the method of Matsushita et al. (33) but modified for use with 96-well plates. Bacteria were grown in 5-ml cultures of LB or LBG for 18 to 20 h. Aliquots (0.5 ml) were removed, and cells were pelleted. The supernatant was discarded, and the cell pellets were either frozen at −80°C or immediately processed. Pellets were washed twice with 0.5 ml of 135 mM potassium phosphate buffer (KPB; pH 6) and resuspended in 0.8 ml of KPB. Cells were lysed by sonication on ice, and the lysates were clarified by centrifugation at 16,100 × g for 10 min at 4°C. The total protein concentration was determined by Bradford analysis (Thermo-Fisher Scientific, product number 23238) using bovine serum albumin as a standard. Lysates were normalized to the same protein concentration (200 μg) using KPB and kept on ice. Twenty microliters of KPB, 40 μl of 165 mM carbon substrate (d-glucose, methyl-alpha-d-glucopyrannoside, and other listed carbohydrates), 20 μl each of two electron acceptors (2 mM 2,6-dichlorophenol-indophenol [Sigma, D-1878] and 13 mM phenazine methosulfate [Sigma, P-9625]), and 100 μl of either KPB (mock) or sample protein lysate (200 μg) were added to individual wells of a flat-bottomed 96-well plate. Then the 96-well plate was incubated at 30°C, and the change in absorbance at 600 nm (ΔA600) over 15 to 30 min was determined as a measure of GDH activity.
β-Galactosidase assays were determined as previously reported using 200 μg of protein lysate in each reaction (27).
Statistical analysis.
A one-way analysis of variance (ANOVA) with Tukey's posttest and Student's t test was performed as noted using GraphPad Prism 5 software with significance set to a P value of <0.05.
RESULTS
d-Glucose inhibition of prodigiosin phenotype in growing cultures.
The most complete previous study of GIP used a nonproliferating culture methodology and a different strain of S. marcescens (52). We verified that GIP was evident in our strain, CMS376 (biotype TCT), and culture conditions. In addition, we tested for GIP in other pigmented S. marcescens strains, laboratory strain Nima (biotype A2a), environmental isolate CHASM (biotype A2a), and four ocular clinical isolates (biotypes A2a and A6a). In every case, LB broth supplemented with glucose to 110 mM (LBG) completely inhibited red pigment production, indicating that GIP was common among pigmented strains tested (Fig. 1 and data not shown; see also Fig. S1 in the supplemental material). LB growth medium was chosen because it supports robust prodigiosin production and equivalent growth rates for the wild-type (WT) and crp mutant strains which were used in the genetic studies detailed below (27, 28).
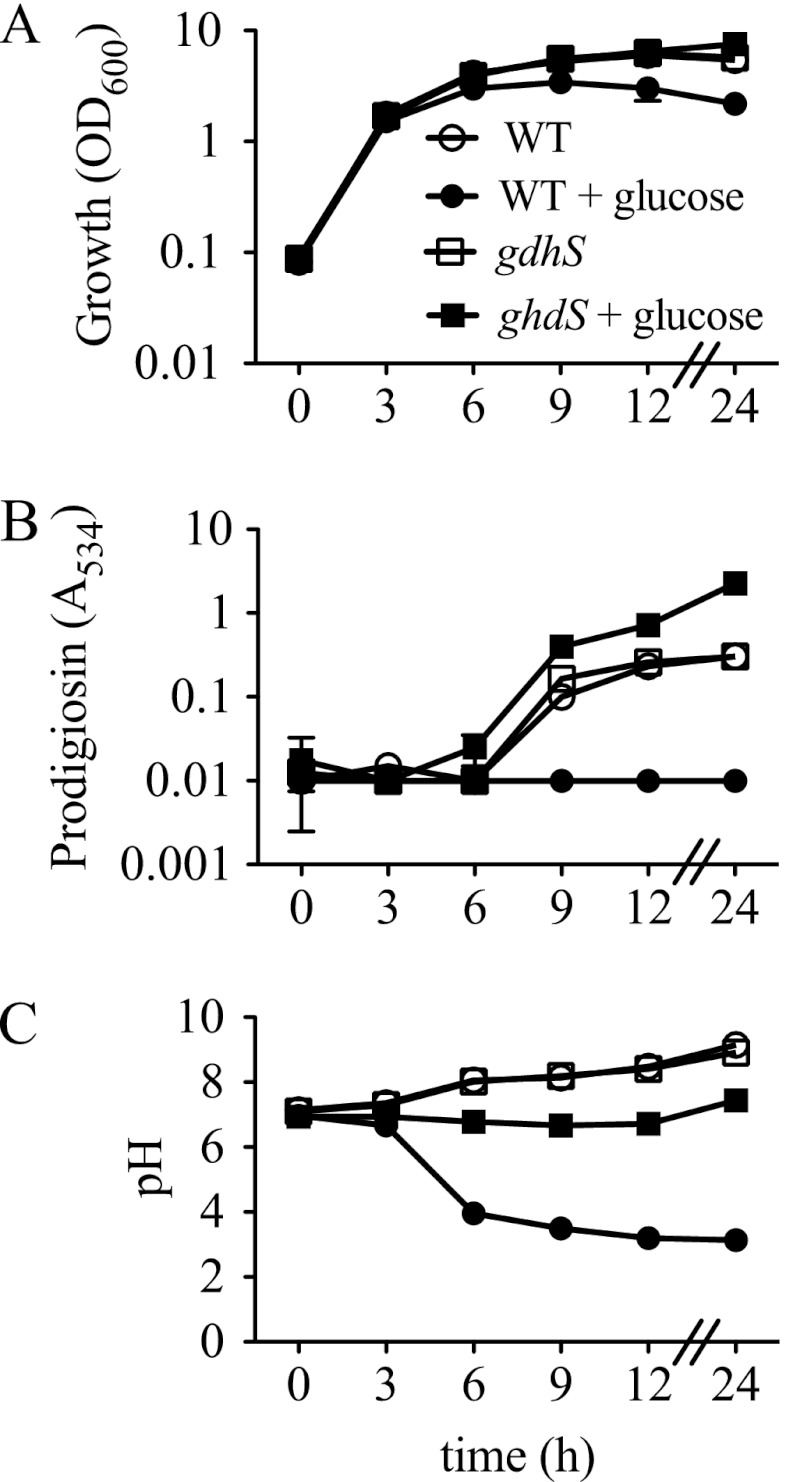
Fig 1.
Growth, prodigiosin production, and culture pH of WT (CMS376) and gdhS mutant (CMS1083) cells grown in LB and LBG. Genotype symbols are consistent for each panel. The gdhS strain is the rig-1 mutant with a wild-type crp gene. Cultures were grown in LB and in LBG (LB supplemented with 110 mM glucose). Culture turbidity (OD600 nm) (A), prodigiosin content (A534) (B), and culture pH (C) were measured. Time course experiments were performed with 4 independent biological replicates per time point per condition and repeated two times with consistent results. Mean values from one representative time course are shown. Error bars indicate one standard deviation.
A time course was performed to further characterize the effect of glucose on growth, prodigiosin production, and the pH of the culture medium under our experimental conditions (Fig. 1). Exogenous glucose did not inhibit the initial rate of growth but was associated with a more rapid achievement of stationary phase and a lower final culture density (Fig. 1A). As expected, prodigiosin production occurred in stationary phase in LB medium and was not detected in glucose-rich medium (Fig. 1B). Culture pH increased over time in LB medium but became highly acidified in LBG, with the maximal change occurring between 3 and 6 h (Fig. 1C).
Experiments tested whether GIP was d-glucose specific with wild-type strain CMS376. Unlike d-glucose, other sugars, including the nonmetabolizable glucose analogs (l-glucose, methyl-alpha-d-glucopyrannoside), the metabolizable sugar mannose, an aldose sugar, and galactose, an epimer of glucose (see Fig. S1A in the supplemental material), supported high levels of prodigiosin production. This result shows that prodigiosin is not directly inhibited by nonmetabolized glucose analogs, by a change in culture osmolarity, by phosphoenolpyruvate transferase system import, or by fermentation of a sugar. None of the tested sugars (at 2% [wt/vol]) except d-glucose conferred a reduction in pH below pH 6 (see Fig. S1B in the supplemental material). In the absence of bacteria, each of the tested carbohydrates lowered the pH of LB medium from 8.6 to 6.6 to 6.8. All subsequent references to glucose will mean d-glucose unless specifically stated.
To confirm that the prodigiosin production defect is a function of pH rather than the presence of glucose (50), we buffered the medium with Tris-HCl to pH 8.0. The Tris-HCl concentration was optimized to 120 mM, as it was determined to be the maximum concentration that did not impede S. marcescens growth (data not shown). Buffering the medium restored robust prodigiosin production to the WT strain grown in LBG and reduced the extent of the pH change (see Fig. S1C and D in the supplemental material).
Genetic screen to identify glucose-insensitive mutants.
Transposon mutagenesis was performed to gain insight into the mechanism by which glucose inhibits prodigiosin. The screen to find mutant colonies able to produce prodigiosin in the presence of glucose was performed in a crp mutant background, because GIP was found to be stronger in crp mutants, leading to fewer false-positive isolates (data not shown). Transposon-mutagenized cells were plated on LBG agar, and pigmented colonies were taken for further analysis. Candidate colonies that produced prodigiosin in LBG broth as well as on LBG plates were noted as red in glucose (rig) mutants. These fell into two classes based on colony and broth culture color: dark red and bright pink (Table 3).
Table 3.
Strain phenotypes in glucose-rich medium
| Straina | Culture colorb | Culture pHc | Mutation (insertion site)d |
|---|---|---|---|
| Δcrp | White | 3.54 ± 0.08 | NA |
| rig-1 | Dark red | 5.99 ± 0.09 | gdhS (972) |
| rig-2 | Dark red | 5.29 ± 0.14 | gdhS (−2) |
| rig-5 | Dark red | 6.23 ± 0.03 | gdhS (2181) |
| rig-10 | Dark red | 6.05 ± 0.03 | gdhS (123) |
| rig-11 | Dark red | 6.05 ± 0.02 | gdhS (2008) |
| rig-12 | Bright pink | 3.51 ± 0.05 | hexS (213) |
| rig-13 | Dark red | 6.16 ± 0.03 | pqqE (172) |
| rig-A2 | Dark red | 6.27 ± 0.17 | gdhS (121) |
| rig-A6 | Dark red | 6.31 ± 0.10 | gdhS (2340) |
| rig-A14 | Dark red | 6.30 ± 0.23 | gdhS (121) |
| rig-A21 | Bright pink | 3.92 ± 0.37 | ubiB (675) |
| rig-A50 | Dark red | 6.23 ± 0.04 | pqqE (172) |
| rig-A54 | Dark red | 6.30 ± 0.07 | pqqE (172) |
| rig-A55 | Dark red | 6.32 ± 0.10 | pqqE (172) |
| rig-A112 | Dark red | 6.01 ± 0.38 | pqqE (172) |
| rig-A185 | Dark red | 6.11 ± 0.11 | pqqE (172) |
| rig-A228 | Dark red | ND | gdhS (1936) |
| rig-A237 | Dark red | 6.17 ± 0.04 | gdhS (1930) |
| rig-A243 | Dark red | 6.33 ± 0.14 | pqqE (686) |
| rig-A258 | Bright pink | 3.91 ± 0.22 | ubiB (736) |
All strains are in a Δcrp mutant background except rig-1 and rig-2, which are in a crp-1 background.
Color by visual inspection after growth in LBG medium for 20 h.
pH as determined from the culture medium after incubation at 20 h in LBG medium (n ≥ 3 experiments per reading). Values are means and standard deviations. ND, not determined.
NA, not applicable. Numbers in parentheses indicate the insertion sites of the transposon with respect to the first base pair of the open reading frame.
gdhS is required for GIP.
Several mutants from the dark-red class had transposon insertions in a gene that codes for a quinoprotein glucose dehydrogenase enzyme (Table 3). This gene was previously cloned from another S. marcescens strain in a screen for genes able to confer the ability to solubilize mineral phosphate to an E. coli gcd mutant strain supplied with exogenous pyrroloquinoline quinone (PQQ), as E. coli does not make its own PQQ (30). The S. marcescens quinoprotein dehydrogenase gene was also previously amplified by PCR and named gdh (2). In this paper, it is referred to as gdhS to differentiate it from gdh genes from other organisms and the glycerol dehydrogenase gene of E. coli, which is also referred to as gdh. This gene is annotated as the SMA2373 open reading frame (ORF) in the sequenced S. marcescens Db11 genome and is predicted to be monocistronic based upon the orientation of adjacent ORFs (Wellcome Trust Sanger Institute).
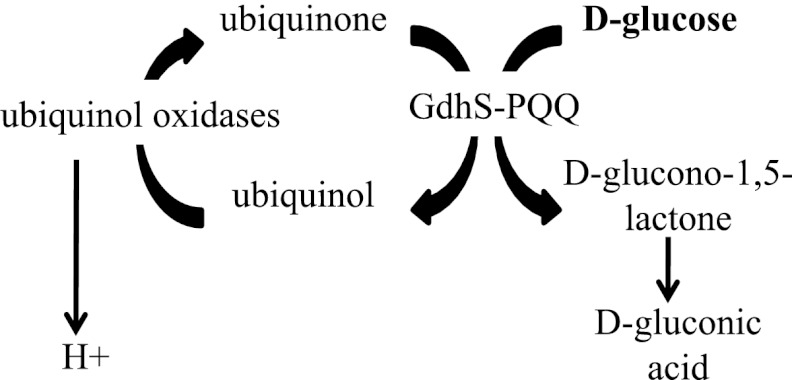
Quinoprotein glucose dehydrogenases (QGDHs; Enzyme Commission number 1.1.5.2) are found in many bacterial species and function to catalyze the extracytoplasmic conversion of d-glucose to d-glucono-1,5-lactone, the cyclic ester of d-gluconic acid, which is then rapidly converted to d-gluconic acid spontaneously and theoretically through the action of a gluconolactonase enzyme(s) (29) (Fig. 2).
Fig 2.
Model for the role of quinoprotein glucose dehydrogenases in extracytoplasmic glucose metabolism. d-Glucose is oxidized to d-glucono-1,5-lactone by GdhS in the periplasm, a process requiring both pyrroloquinoline quinone (PQQ) and ubiquinone (1, 32). Ubiquinone is reduced, generating ubiquinol, a molecule that is then converted to ubiquinone by one of several possible ubiquinol oxidases, creating a proton gradient (1, 32). d-Glucono-1,5-lactone is hydrolyzed to d-gluconic acid nonenzymatically and possibly through the action of a theoretical gluconolactonase enzyme. Production of d-glucono-1,5-lactone, its subsequent change to d-gluconic acid, and the establishment of a proton gradient are expected to decrease extracellular pH.
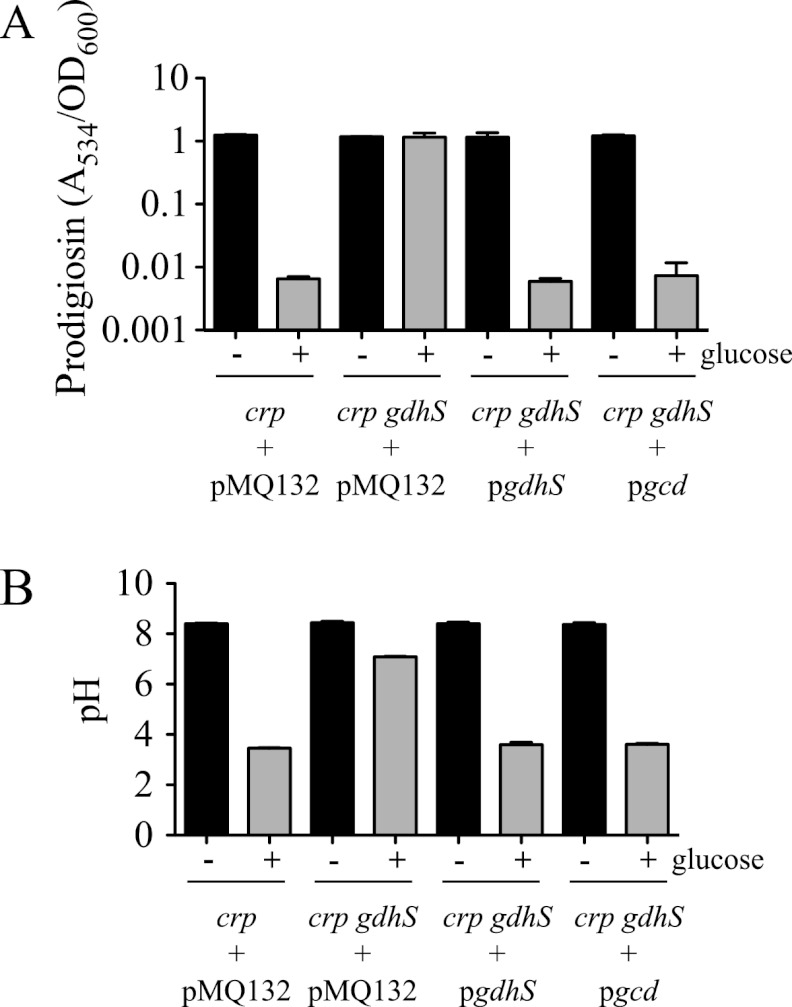
Multiple independent mutations in the gdhS gene (Table 3) support the idea that an insertional mutation in this gene confers a RIG phenotype and that it is unlikely that an unknown mutation elsewhere in the chromosome is responsible for the RIG phenotype. However, the RIG phenotype may be due to a polar effect on an adjacent gene(s). To control for this possibility, we performed complementation experiments. The gdhS ORF was cloned from S. marcescens and placed under transcriptional control of the E. coli Plac promoter on a medium-copy-number vector. This plasmid (pgdhS) was moved into the Δcrp gdhS (rig-10) double mutant and was able to restore GIP (Fig. 3A and B). The pgdhS plasmid was also moved into a modified rig-1 strain in which the intact crp gene had been restored so that it was isogenic to the WT except for the transposon mutation in gdhS (strain CMS1083). Together, these data indicate that a mutation of gdhS rather than a polar effect on transcription of adjacent genes caused the loss of GIP.
Fig 3.
Glucose dehydrogenase is necessary for glucose inhibition of prodigiosin (GIP). Complementation of the rig-10 mutant (Δcrp gdhS) glucose insensitivity phenotype using the wild-type S. marcescens gdhS gene or the E. coli gcd gene expressed from a medium-copy-number plasmid. The vector pMQ132 was used as a negative control. Cultures were incubated at 30°C for 20 h; “− glucose” indicates growth in LB (black bars); “+ glucose” indicates growth in LBG (gray bars). Prodigiosin levels (A534), normalized by culture turbidity (OD600 nm) (A), and culture pH (B) are shown. The charts show an average of three independent biological replicates per condition and genotype, and error bars indicate one standard deviation. The experiment was performed twice with consistent results. Strain CMS1687 with pMQ132 served as the crp mutant control.
To test whether the GIP-defective phenotype of the gdhS mutants is specific to glucose dehydrogenase activity, we cloned the gcd gene, which codes for a known glucose dehydrogenase that is ∼72% identical to GdhS at the amino acid level, from E. coli (9). The gcd gene was able to complement the gdhS mutant phenotype with respect to restoring GIP and acidification of the medium, further supporting the conclusion that glucose dehydrogenase activity is required for GIP (Fig. 3A and B).
Strain CMS1083 (Table 1) was used to test the impact of a gdhS mutation upon growth rate, prodigiosin production, and medium acidification. Growth curve analysis revealed that the gdhS mutant and the WT strain were indistinguishable in LB medium (Fig. 1A). However, unlike the WT strain, whose final culture turbidity was decreased in LBG, the gdhS mutant exhibited the same growth kinetics regardless of the presence of glucose (Fig. 1A). In the absence of glucose, both the gdhS mutant and WT strains generated prodigiosin. Strikingly, with the addition of glucose, the gdhS mutant strain reproducibly produced elevated prodigiosin levels, whereas the WT did not generate any detectable prodigiosin (Fig. 1B). The pH of the medium in gdhS mutant cultures grown in LB was identical to that of the WT culture pH (Fig. 1C). In LBG, the gdhS mutant exhibited little pH change and was found to be significantly more basic (P < 0.05) than the WT strain grown in glucose for 6 to 24 h (Fig. 1C). These data indicate that GdhS is required for medium acidification under tested conditions.
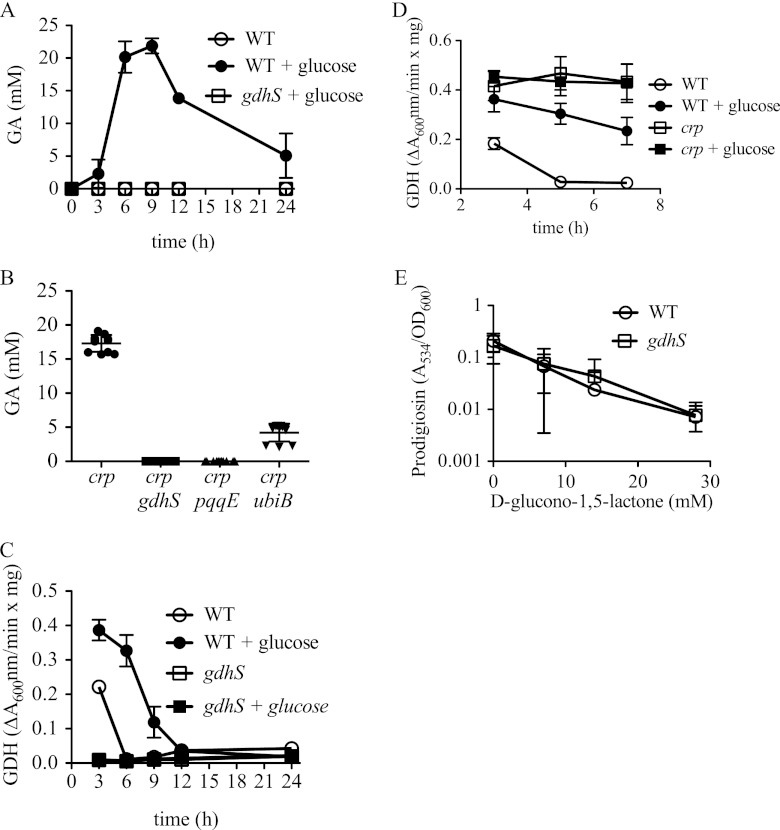
d-Gluconic acid, the indirect product of glucose dehydrogenase (GDH) activity, was measured from supernatants taken from the same time courses as in Fig. 1. No d-gluconic acid was measured from the WT culture grown in LB without glucose, whereas d-gluconic acid was evident starting at 3 h and peaking in concentration at 6 to 9 h in WT cultures grown in LBG (Fig. 4A). The major increase in d-gluconic acid concentration at 3 to 6 h was coincident with the steepest decrease in culture pH (Fig. 1C). It is known that S. marcescens can externally convert d-gluconic acid to 2-keto-d-gluconate and can internally convert d-gluconic acid to gluconate-6-phosphate, previously shown to account for the reduction in d-gluconic acid in S. marcescens cultures after hour 9, similar to that seen in Fig. 4A (3). Unlike the WT culture supernatants, no d-gluconic acid was measured from supernatants of the gdhS mutant grown in LB or LBG, verifying that measured d-gluconic acid was a product of GdhS activity (Fig. 4A and data not shown). Similar to the WT, d-gluconic acid was measured in a Δcrp mutant at 15 to 20 mM and was absent in the Δcrp gdhS (rig-10) double mutant (Fig. 4B). This result indicates that GdhS is the primary enzyme involved in oxidizing glucose under these experimental conditions.
Fig 4.
Gluconic acid production, GDH activity, and the effect of d-glucono-1,5-lactone on prodigiosin levels. “+ glucose” indicates growth in LBG; otherwise, strains were grown in LB. (A and B) Gluconic acid (GA) was measured from culture medium to assess GDH activity in different genetic backgrounds and culture conditions over a time course (A) or at 6 h of incubation at 30°C after being subcultured to an A600 of 0.1 (B). (C and D) Glucose dehydrogenase activity from crude cellular extracts measured over time. (E) Prodigiosin production measured from d-glucono-1,5-lactone-treated cultures, normalized by culture density and measured at 20 h. (A, C, and E) The charts show the averages of four biological replicates per time point and condition from a representative experiment that was repeated on at least one other occasion with consistent results. (B) The chart depicts the combined data from two experiments performed on different days, using a total of 8 biological replicates per genotype. Error bars indicate 1 standard deviation. WT, CMS376; gdhS, CMS1083; crp, CMS1687; crp gdhS, rig-10; crp pqqE, rig-13; crp ubiB, rig-A21.
GDH activity was measured from crude lysates of cells isolated from the same time courses as in Fig. 1; however, 0-h samples were excluded because of insufficient protein (Fig. 4C). The WT exhibited the highest levels of GDH activity at 3 h and elevated GDH levels when grown in LBG compared to those when grown in LB. The gdhS mutant exhibited no measurable GDH activity, suggesting that any hypothetical cytoplasmic GDH enzymes either play a relatively minor role in glucose oxidation or are not active under our assay conditions (Fig. 4C). Additionally, we noted that d-glucose, but not the glucose analog methyl-α-d-glucopyranoside, acted as a substrate for GDH (see Fig. S2 in the supplemental material), consistent with the inability of methyl-α-d-glucopyranoside to elicit acidification of culture medium (see Fig. S1B in the supplemental material). A Δcrp mutant exhibited higher GDH activity than did the WT in LB (Fig. 4D). Also different from the WT, GDH levels in the crp mutant remained stable over the tested time frame and were unchanged by glucose in the growth medium, suggesting that the CRP regulates GDH production or activity (Fig. 4D).
A small survey of substrate specificity was performed using lysates from the Δcrp mutant and the Δcrp gdhS (rig-10) double mutant, and glucose exhibited the most activity of the tested sugars, followed distantly by galactose and mannose (see Fig. S2A in the supplemental material). No activity was exhibited by the crp gdhS double mutant, indicating that GdhS was necessary for the measured activity with all positive reactions (see Fig. S2B in the supplemental material). Carbohydrates with no activity included the glucose analog methyl-α-d-glucopyranoside, the mannose analog methyl-α-d-mannopyranoside, the disaccharide lactose, the trisaccharide raffinose, and glycerol (see Fig. S2 in the supplemental material).
d-Glucono-1,5-lactone and d-gluconic acid, but not d-gluconate, confer a GIP-like phenotype to gdhS mutants.
As GdhS processing of glucose appears to be necessary for GIP, we tested whether the product of GDH enzymatic activity inhibited pigment production. GDH converts d-glucose to d-glucono-1,5-lactone, and in aqueous environments, d-glucono-1,5-lactone is rapidly hydrolyzed to d-gluconic acid (39). Therefore, we tested whether either d-glucono-1,5-lactone or d-gluconic acid would inhibit prodigiosin production. d-Glucono-1,5-lactone reduced the medium pH in the absence of bacteria. After 20 h of aeration, LB alone had a pH of 7.3, compared to 4.9 for LB supplemented with 14 mM d-glucono-1,5-lactone (pH 4.5 for 28 mM, pH 3.9 for 56 mM, and pH 3.5 for 112 mM). The maximum d-glucono-1,5-lactone concentration for which growth of CMS376 was not different from that in LB alone was 28 mM (data not shown); therefore, this was the maximum concentration used in our assays. d-Gluconic acid similarly caused a reduction in medium pH without bacteria (pH 7.5 for 0 mM, pH 5.3 for 25.5 mM, and pH 4.1 for 51 mM). d-Gluconate, the sodium salt form of d-gluconic acid did not reduce medium pH (data not shown; see Fig. S1B in the supplemental material).
Whereas d-gluconate did not inhibit WT prodigiosin production at the tested concentration of 2% (91.7 mM) (see Fig. S1A in the supplemental material), d-glucono-1,5-lactone strongly inhibited prodigiosin production by the WT in a dose-dependent manner (Fig. 4E). A similar dose-dependent inhibition of prodigiosin production was observed by the addition of d-gluconic acid (see Fig. S1E in the supplemental material) at concentrations similar to those measured in WT cultures grown in LBG.
If gdhS mutants do not have GIP because they cannot make d-glucono-1,5-lactone, then it follows that exogenous d-glucono-1,5-lactone should restore GIP (inhibit prodigiosin production) to a gdhS mutant. In support of this prediction, we observed that exogenous d-glucono-1,5-lactone, but not d-gluconate, inhibited gdhS mutant prodigiosin production (Fig. 4E; see also Fig. S1A in the supplemental material).
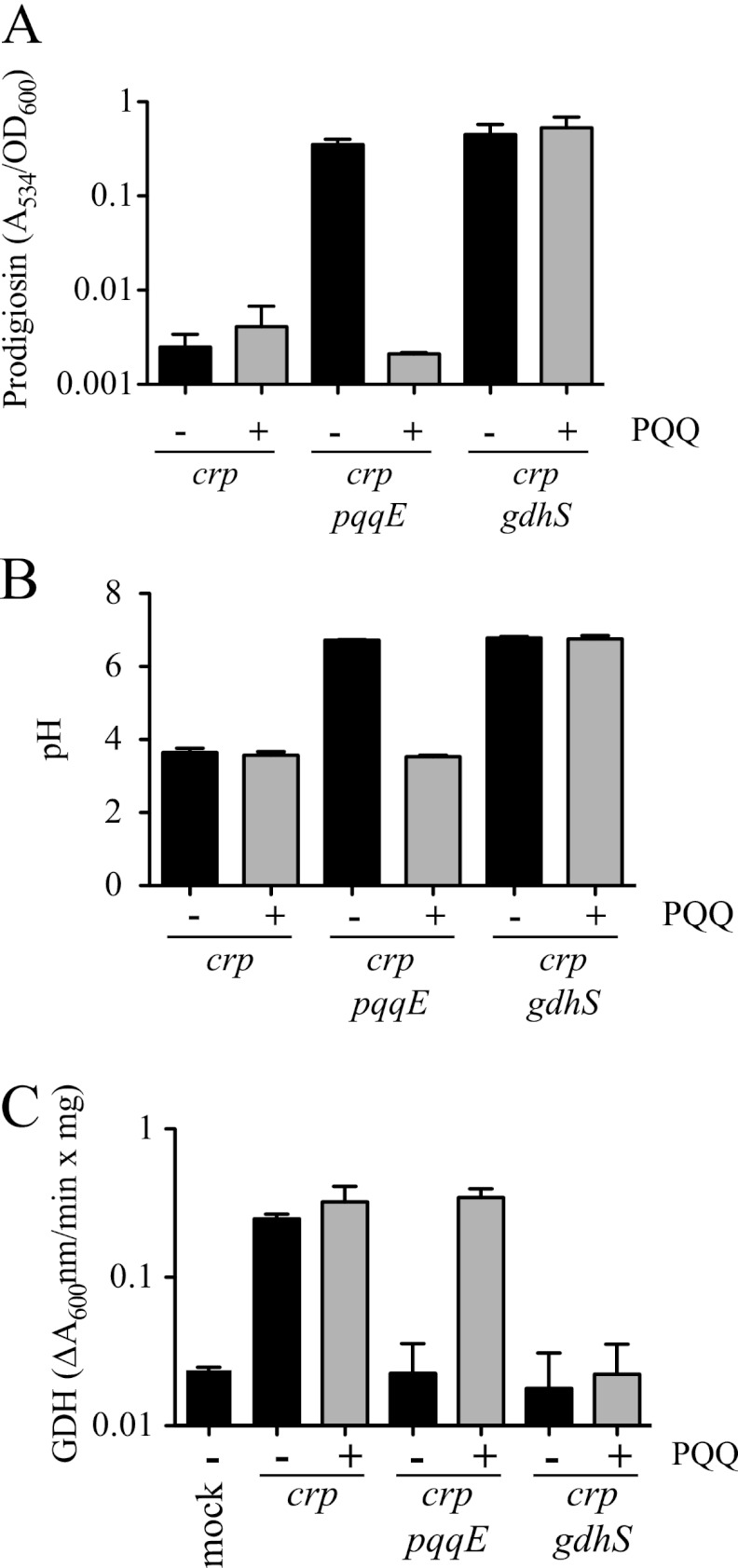
PQQ is required for QGDH activity and GIP.
Quinoprotein GDHs use PQQ as a cofactor, and its substrates are d-glucose and ubiquinone, a component of the electron transport chain (1, 32) (Fig. 2). S. marcescens GdhS has previously been shown to require PQQ for GDH activity when expressed in E. coli (30). As GdhS is necessary for GIP and GdhS requires PQQ, it is expected that mutations in the PQQ biosynthetic genes would impart a GIP-defective phenotype, similar to a gdhS mutant. Consistently, we found several rig strains with mutations in the pqqE gene (Table 3). There may be a mariner transposon insertion hot spot in pqqE, as six rig mutants isolated from different mutagenesis events over the course of several months were found in the same position in the pqqE gene. A seventh rig mutation was isolated with a transposon insertion at a different site in pqqE. Growth curve analysis showed no difference between the Δcrp mutant and a Δcrp pqqE (rig-13) double mutant in LB medium (data not shown).
When tested in LBG, crp pqqE double mutants were defective in GIP (Fig. 5A). The crp pqqE mutants also behaved identically to crp gdhS mutants with respect to growth medium acidification (Fig. 5B), d-gluconic acid generation (Fig. 4B), and GDH activity (Fig. 5C). To further verify the lack of PQQ as the reason for the RIG phenotype of pqqE mutants, we observed that GIP could be restored to crp pqqE mutants using exogenous PQQ (3.025 μM). The crp pqqE transposon mutant (rig-13) produced high levels of prodigiosin in LBG but made no pigment when PQQ was added to LBG (Fig. 5A). As a negative control, an isogenic Δcrp gdhS mutant (rig-10) was able to produce equivalently high levels of prodigiosin in LBG with and without PQQ (P = 0.35, Student's t test), showing that exogenous PQQ itself does not prevent generation of prodigiosin. As expected, the parental Δcrp mutant was unable to produce prodigiosin in LBG, with or without PQQ (Fig. 5A). In the same experiment, it was observed that exogenous PQQ restored glucose-dependent medium acidification and GDH enzyme activity to the crp pqqE mutant (rig-13) but not the crp gdhS mutant (rig-10), whereas the crp mutant acidified the medium regardless of the presence of PQQ (Fig. 5B).
Fig 5.
Pyrroloquinoline quinone (PQQ) is required for GIP. (A to C) Prodigiosin levels, normalized by culture turbidity, culture pH, and GDH activity of cellular extracts, were determined from cultures grown for 20 h in LBG with (+) or without (−) 3.03 μM PQQ. The crp mutant (Δcrp, CMS1687), crp pqqE mutant (rig-13), and crp gdhS mutant (rig-10) were used for this analysis. Panels A to C show the averages of 8 biological replicates per condition from two separate experiments. (C) Mock, no-protein control. Error bars indicate one standard deviation.
To ensure that the RIG phenotype of pqqE mutants was similar in a strain with a functional crp gene, we introduced a wild-type crp gene on a plasmid, pMQ185 (Table 1), into the crp pqqE double mutant. The rig-13 strain (crp pqqE) with pMQ185 was insensitive to glucose with respect to prodigiosin production and pH (prodigiosin/pH in LBG, 2.0/6.5 for rig-13 and pMQ185 and 0.01/3.55 for crp and pMQ185), indicating that a pqqE mutation alone is sufficient for the phenotype.
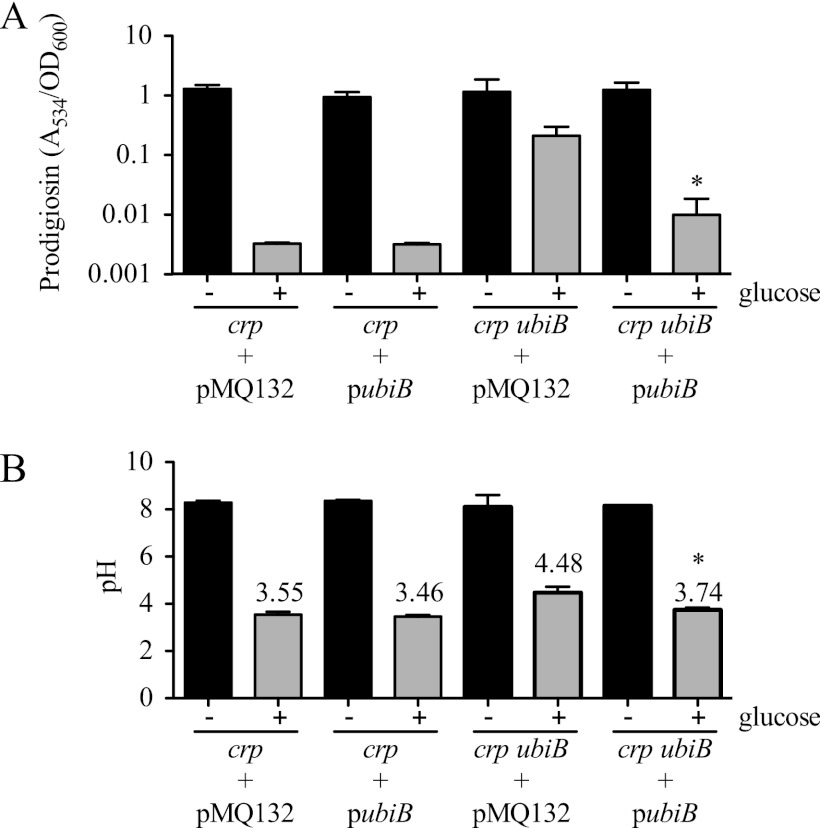
Mutations in the ubiB gene confer intermediate pigment phenotypes and gluconic acid production.
Ubiquinone (coenzyme Q), a substrate for quinoprotein GDH protein activity (Fig. 2), is reduced to ubiquinol by GDH (1). Two rig mutants (rig-A21 and rig-A258) had transposon insertions that mapped to a homolog of the ubiB gene (Table 3). The ubiB gene product is important but not absolutely required for ubiquinone biosynthesis (40). These mutations did not have a dark red phenotype when grown in glucose medium; rather, they exhibited intermediate prodigiosin production, resulting in a bright pink color (Fig. 6A and data not shown). The crp ubiB strains exhibited a reduced ability to acidify the medium (Fig. 6B and Table 3), though the acidification defect was not as severe as those of the crp gdhS and crp pqqE mutants. Furthermore, the crp ubiB mutants generated a reduced amount of gluconic acid (Fig. 4B). Lastly, the crp ubiB mutants exhibited a small-colony phenotype and a slightly reduced rate of growth compared to that of the Δcrp strain (data not shown).
Fig 6.
Complementation of the ubiB mutant phenotype. (A and B) Cultures of the Δcrp (CMS1687) and Δcrp ubiB (rig-A21) strains, bearing either an empty vector control (pMQ132) or the wild-type ubiB gene expressed from a medium-copy-number plasmid (pubiB), were grown in LB (− glucose) or LBG (+ glucose) for 20 h. Prodigiosin levels (A534), normalized by culture turbidity (OD600 nm) (A), and culture pH (B) are shown. The average pH values are shown for the rig-21 mutant. The experiments show the averages of 8 biological replicates per genotype from two experiments performed on different days. The error bars indicate one standard deviation. The asterisk indicates a significant difference between the strain with ubiB, pMQ132, and glucose and the strain with ubiB, pubiB, and glucose, as determined using a Student t test (P < 0.001).
To ensure that the ubiB mutation was responsible for the diminished GIP of the rig-A21 and rig-A258 mutants, we performed a complementation experiment. The ubiB gene was cloned under the control of the Plac promoter on a multicopy plasmid (pMQ132). The resulting plasmid (pubiB) and empty vector control (pMQ132) were moved into the Δcrp ubiB mutants rig-A21 (Δcrp ubiB) and rig-A258 (Δcrp ubiB). When grown in the absence of glucose, the pubiB plasmid did not cause any obvious phenotypic change with respect to pigment production in any tested strain background but did complement the reduced growth rate phenotype of the rig-A21 and rig-A258 mutants (data not shown). The Δcrp parental strain exhibited identical robust GIPs with both the vector alone and pubiB (Fig. 6A and B). The rig-A21 and rig-A258 mutants both exhibited a strong RIG phenotype with the vector alone (Fig. 6A and B), whereas the pubiB plasmid restored GIP (Fig. 6A and B), that is, a significant reduction in prodigiosin and a modest change in medium pH in LBG compared to those of LB (P < 0.001, Student's t test). These data show that UbiB is necessary for the full GIP of rig-A21 and rig-A258 mutants.
As with rig-13 described above, we used the crp-expressing plasmid pMQ185 to test whether a mutation of ubiB independent of the crp mutation maintained the ability to produce prodigiosin in glucose-rich medium. The rig-A21 strain (crp ubiB) with pMQ185 was insensitive to glucose with respect to prodigiosin production and slightly less sensitive to glucose with respect to pH (prodigiosin/pH in LBG, 0.6/4.2 for rig-A21 and pMQ185 and 0.01/3.55 for crp and pMQ185), supporting the conclusion that a ubiB mutation alone is sufficient for the phenotype.
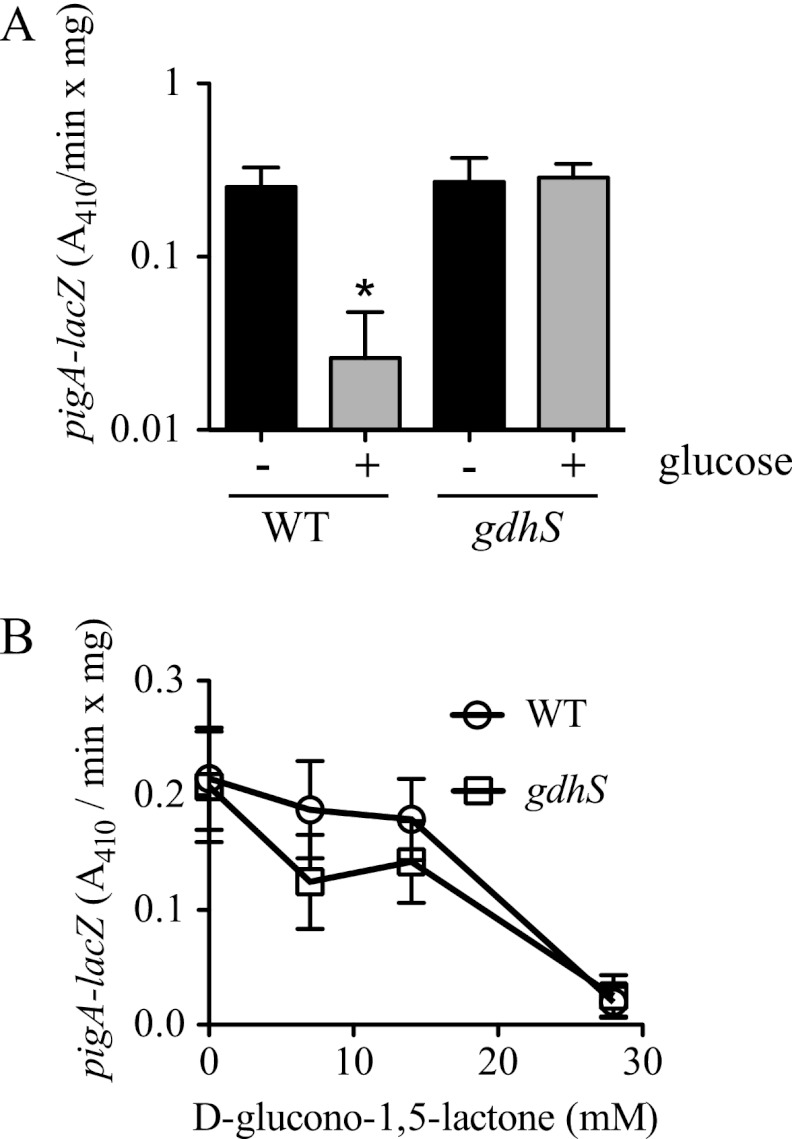
GIP acts at the transcriptional level and requires GdhS.
The pigA-N operon is responsible for biogenesis of prodigiosin (10, 23). We used a formerly described chromosomal pigA-lacZ reporter (27) to assess transcription from the prodigiosin biosynthetic operon in response to glucose. A >10-fold decrease was detected in the amount of pigA-lacZ-associated β-galactosidase activity between the WT culture grown in LBG to stationary phase and the same strain grown in LB medium (Fig. 7A).
Fig 7.
Altered pigA transcription in glucose-rich medium in the WT but not a gdhS mutant. (A and B) β-Galactosidase activity was measured from a chromosomal pigA promoter-lacZ fusion that measured expression of the prodigiosin biosynthetic operon after growth for 20 h. (A) β-Galactosidase activity was measured from WT (CMS376) and gdhS mutant (CMS1083) cultures grown in LBG (+ glucose) and LB (− glucose). Data from eight biological replicates per condition from two separate experiments are shown. (B) Expression of the PpigA-lacZ fusion was inhibited by growth in d-glucono-1,5-lactone at >20 mM in both the WT and the gdhS (rig-1 with restored crp) mutant strains. A representative experiment using 4 biological replicates per condition is shown and was consistent with a repetition of the experiment on a different day. Error bars indicate one standard deviation, and the asterisk indicates that the WT + glucose condition produced a different amount of β-galactosidase activity than all other conditions, as determined by an ANOVA with a Tukey posttest (P < 0.05).
As gdhS mutants generated prodigiosin even in LBG, transcription from the pigA-lacZ construct in LBG in these mutant strains was expected to be measurable. As predicted, equivalent levels of pigA promoter-driven β-galactosidase activity were measured in LB and LBG for the gdhS mutant (CMS1083) (Fig. 7A).
The effect of a d-glucono-1,5-lactone gradient on pigA-lacZ expression was performed to determine whether the effect was mediated at least partially through transcriptional inhibition of the prodigiosin biosynthetic operon. We observed a dose-dependent inhibition of d-glucono-1,5-lactone on pigA-lacZ expression in both the WT and a gdhS mutant (Fig. 7B).
DISCUSSION
The purpose of this study was to investigate the genetic basis of the glucose inhibition of prodigiosin production phenotype in S. marcescens. Previous studies have made observations regarding the inhibitory effect of glucose on growing (4, 8, 15, 27) and nonproliferating (50, 51, 52) cultures. However, this is the first study to show that GIP extends to clinical, environmental, and laboratory S. marcescens strains representative of multiple biotypes and to use genetic analysis to probe the mechanism by which glucose inhibits prodigiosin production.
As d-glucose, but not l-glucose or methyl-alpha-d-glucopyrannoside, confers the glucose inhibition of prodigiosin phenotype (GIP), it was concluded that the phenotype does not occur in response to a change in osmolarity or the direct inhibition of an enzyme involved in the prodigiosin biosynthetic process by d-glucose. No GIP effect was observed with glucose analogs, aldose, and other sugars, suggesting that enzymatic processing of d-glucose was key to establishment of the phenotype.
Here we established with growing cultures that glucose causes medium acidification and that buffering of the medium prevented the GIP phenotype, confirming work by Lorén and colleagues using a different culture system that GIP is mediated by reduced pH (50, 52). Consistently, another study by Lorén's group showed that prodigiosin production is strongly inhibited at a pH below 5.5 (51), and they concluded that pH reduction was the basis of GIP in nonproliferating cultures (50, 52), but the genetic basis of this phenotype was not determined.
The results of our genetic screen implicated quinoprotein glucose dehydrogenase as the major factor in GIP. This is based upon the observation that independent mutations in the gdhS gene prevent GIP and largely, but not completely, eliminate medium acidification, suggesting that GdhS is the major but not the sole factor in the glucose-mediated culture acidification. The gdhS mutant phenotypes were complemented by a known quinoprotein glucose dehydrogenase gene from E. coli, gcd, and by the S. marcescens gdhS gene, supporting that it is glucose dehydrogenase activity and not some other unknown function of the GdhS protein or another mutation that is responsible for GIP.
d-Glucono-1,5-lactone and d-gluconic acid, but not d-gluconate, inhibited prodigiosin production in the WT and the gdhS mutant, suggesting that the product of GdhS activity, d-glucono-1,5-lactone, is involved in GIP. d-Glucono-1,5-lactone, the cyclic ester of d-gluconic acid, spontaneously converts to d-gluconic acid in aqueous medium, and its hydrolysis is accompanied by an acidification of its environment (25, 55). Interestingly, Fineran et al. demonstrated that d-gluconate inhibits prodigiosin production by Serratia species ATCC 39006 (12), suggesting that the two species can use different environmental inputs to regulate prodigiosin production.
Data from this study support that genes involved in biosynthesis of PQQ and ubiquinone are important in GIP. The simplest reason for the ability of pqqE and ubiB mutants to produce pigment in glucose-rich medium is that they are defective in GdhS activity. The intermediate level of gluconic acid production in ubiB mutants (Fig. 4B) may account for the pink rather than red culture phenotype in LBG. The reason why ubiB mutants have an intermediate phenotype may be due to the ability of quinoprotein GDHs to utilize menaquinone in addition to ubiquinone as a cofactor (35). Alternatively, ubiB mutants in E. coli are able to produce ubiquinone in stationary phase but not in logarithmic-growth phase (40). Assuming a similar situation exists in Serratia, some ubiquinone production may be responsible for the observed intermediate phenotypes in ubiB mutants, as cultures in most assays in this study proceeded to stationary phase.
At the onset of this study, it was not clear that there was a role for transcriptional control in establishment of GIP, as the enzymes necessary for prodigiosin production may be inhibited by low pH. In support of a transcriptional control model, we observed that a transcriptional reporter to the pigment biosynthetic operon (pigA-N) is inhibited in LBG in the WT but not the gdhS mutant. This suggests that transcriptional control of pigA-N is a contributing factor for establishing GIP. As noted in Table 3, we found that the mutation of the transcription factor hexS is required for GIP. HexS from S. marcescens strain 274 was previously reported to directly bind to and inhibit expression from the pigA promoter (54), and this interaction has been verified for CMS376 (R. Lahr and R. Shanks, unpublished data). The role of HexS in establishment of GIP will be reported elsewhere.
Unlike the WT, gdhS mutants made prodigiosin when grown in LBG. More strikingly, the gdhS mutant produced greater amounts of prodigiosin in LBG than in LB (Fig. 1B). This may be expected, as glucose inhibits cAMP production in LBG (27) and cAMP inhibits prodigiosin production (28). Therefore, it is predicted that glucose should, in fact, increase prodigiosin production were it not for GdhS-mediated medium acidification. Consistent with the cAMP model, the increased prodigiosin evident in the gdhS mutant in LBG compared to that in LB (Fig. 1B) was absent in the crp gdhS double mutant with and without glucose (Fig. 3A), likely because CRP is required to respond to glucose-initiated changes in cAMP levels.
It is of interest that S. marcescens has complex systems by which glucose inhibits prodigiosin levels. As noted above, it is expected that glucose should increase prodigiosin levels through control of cAMP; however, this is only revealed when GDH activity is eliminated. These observations suggest that there are independent and antagonistic regulatory pathways by which glucose can mediate prodigiosin production. It is clear that the GDH-mediated pathway is dominant over CRP in regulation of prodigiosin under glucose-rich conditions. It may be energetically beneficial for the bacteria to inhibit prodigiosin production in glucose-rich environments. This is based upon the observations that quinoprotein GDH activity leads to the establishment of a proton gradient that can be used for the uptake of amino acids and other molecules (1, 32, 33) whereas prodigiosin promotes H+/Cl− symport across membranes (21, 41, 44), possibly uncoupling the proton gradient established through glucose oxidation. Consistent with this notion, Haddix and colleagues recently presented data suggesting that S. marcescens utilizes prodigiosin to reduce ATP production as a way to limit generation of damaging reactive oxygen species during stationary phase (21). Applied and environmental implications of this study include that gdhS or pqqE mutants may be useful in the production of secondary metabolites that are often inhibited by glucose, which is otherwise a useful carbon source for rapid bacterial growth. Supporting this scenario, not only was growth of gdhS or pqqE not inhibited by glucose, but these mutants were able to produce elevated levels of the secondary metabolite prodigiosin in glucose-rich medium.
Supplementary Material
ACKNOWLEDGMENTS
We give special thanks to Marissa Aston, Tara Veverka, and Aroba Sadaf for technical assistance and to Pryce Haddix and Kristin Arena for critical reading of the manuscript.
This study was funded by grant AI085570 from NIH/NIAID and a Research to Prevent Blindness Career Development Award to R.S. Additional funding was from the Eye and Ear Foundation of Pittsburgh, an unrestricted grant from Research to Prevent Blindness, and NIH grant EY08098.
Footnotes
Published ahead of print 29 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Anthony C. 2004. The quinoprotein dehydrogenases for methanol and glucose. Arch. Biochem. Biophys. 428: 2–9 [DOI] [PubMed] [Google Scholar]
- 2. Ben Farhat M, et al. 2009. Characterization of the mineral phosphate solubilizing activity of Serratia marcescens CTM 50650 isolated from the phosphate mine of Gafsa. Arch. Microbiol. 191: 815–824 [DOI] [PubMed] [Google Scholar]
- 3. Bouvet OM, Grimont PA. 1988. Extracellular oxidation of d-glucose by some members of the Enterobacteriaceae. Ann. Inst. Pasteur Microbiol. 139: 59–77 [DOI] [PubMed] [Google Scholar]
- 4. Bunting MI, Robinow CF, Bunting H. 1949. Factors affecting the elaboration of pigment and polysaccharide by Serratia marcescens. J. Bacteriol. 58: 114. [DOI] [PubMed] [Google Scholar]
- 5. Burger SR, Bennett JW. 1985. Droplet enrichment factors of pigmented and nonpigmented Serratia marcescens: possible selective function for prodigiosin. Appl. Environ. Microbiol. 50: 487–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang SC, Wei YH, Wei DL, Chen YY, Jong SC. 1991. Factors affecting the production of eremofortin C and PR toxin in Penicillium roqueforti. Appl. Environ. Microbiol. 57: 2581–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiang SL, Rubin EJ. 2002. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296: 179–185 [DOI] [PubMed] [Google Scholar]
- 8. Clements-Jewery S. 1976. The reversal of glucose repressed prodigiosin production in Serratia marcescens by the cyclic 3′5′-adenosine monophosphate inhibitor theophylline. Experientia 32: 421–422 [DOI] [PubMed] [Google Scholar]
- 9. Cleton-Jansen AM, Goosen N, Fayet O, van de Putte P. 1990. Cloning, mapping, and sequencing of the gene encoding Escherichia coli quinoprotein glucose dehydrogenase. J. Bacteriol. 172: 6308–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dauenhauer SA, Hull RA, Williams RP. 1984. Cloning and expression in Escherichia coli of Serratia marcescens genes encoding prodigiosin biosynthesis. J. Bacteriol. 158: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Espeso EA, Tilburn J, Arst HN, Jr, Penalva MA. 1993. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. EMBO J. 12: 3947–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fineran PC, Everson L, Slater H, Salmond GP. 2005. A GntR family transcriptional regulator (PigT) controls gluconate-mediated repression and defines a new, independent pathway for regulation of the tripyrrole antibiotic, prodigiosin, in Serratia. Microbiology 151: 3833–3845 [DOI] [PubMed] [Google Scholar]
- 13. Fineran PC, Williamson NR, Lilley KS, Salmond GP. 2007. Virulence and prodigiosin antibiotic biosynthesis in Serratia are regulated pleiotropically by the GGDEF/EAL domain protein, PigX. J. Bacteriol. 189: 7653–7662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gerber NN. 1975. Prodigiosin-like pigments. Crit. Rev. Microbiol. 3: 469–485 [DOI] [PubMed] [Google Scholar]
- 15. Giri AV, Anandkumar N, Muthukumaran G, Pennathur G. 2004. A novel medium for the enhanced cell growth and production of prodigiosin from Serratia marcescens isolated from soil. BMC Microbiol. 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grimont PA, Grimont F. 1978. Biotyping of Serratia marcescens and its use in epidemiological studies. J. Clin. Microbiol. 8: 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gristwood T, Fineran PC, Everson L, Salmond GP. 2008. PigZ, a TetR/AcrR family repressor, modulates secondary metabolism via the expression of a putative four-component resistance-nodulation-cell-division efflux pump, ZrpADBC, in Serratia sp. ATCC 39006. Mol. Microbiol. 69: 418–435 [DOI] [PubMed] [Google Scholar]
- 18. Gristwood T, Fineran PC, Everson L, Williamson NR, Salmond GP. 2009. The PhoBR two-component system regulates antibiotic biosynthesis in Serratia in response to phosphate. BMC Microbiol. 9: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gristwood T, McNeil MB, Clulow JS, Salmond GP, Fineran PC. 2011. PigS and PigP regulate prodigiosin biosynthesis in Serratia via differential control of divergent operons, which include predicted transporters of sulfur-containing molecules. J. Bacteriol. 193: 1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haavik HI. 1974. Studies on the formation of bacitracin by Bacillus licheniformis: role of catabolite repression and organic acids. J. Gen. Microbiol. 84: 321–326 [DOI] [PubMed] [Google Scholar]
- 21. Haddix PL, et al. 2008. Kinetic analysis of growth rate, ATP, and pigmentation suggests an energy-spilling function for the pigment prodigiosin of Serratia marcescens. J. Bacteriol. 190: 7453–7463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hameeda B, Reddy YH, Rupela OP, Kumar GN, Reddy G. 2006. Effect of carbon substrates on rock phosphate solubilization by bacteria from composts and macrofauna. Curr. Microbiol. 53: 298–302 [DOI] [PubMed] [Google Scholar]
- 23. Harris AK, et al. 2004. The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology 150: 3547–3560 [DOI] [PubMed] [Google Scholar]
- 24. Heinemann B, Howard AJ, Palocz HJ. 1970. Influence of dissolved oxygen levels on production of l-asparaginase and prodigiosin by Serratia marcescens. Appl. Microbiol. 19: 800–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hucho F, Wallenfels K. 1972. Glucono-lactonase from Escherichia coli. Biochim. Biophys. Acta 276: 176–179 [DOI] [PubMed] [Google Scholar]
- 26. James PD, Edwards C, Dawson M. 1991. The effects of temperature, pH and growth rate on secondary metabolism in Streptomyces thermoviolaceus grown in a chemostat. J. Gen. Microbiol. 137: 1715–1720 [DOI] [PubMed] [Google Scholar]
- 27. Kalivoda EJ, et al. 2010. Cyclic AMP negatively regulates prodigiosin production by Serratia marcescens. Res. Microbiol. 161: 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kalivoda EJ, Stella NA, O'Dee DM, Nau GJ, Shanks RM. 2008. The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl. Environ. Microbiol. 74: 3461–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krajewski V, et al. 2010. Metabolic engineering of Gluconobacter oxydans for improved growth rate and growth yield on glucose by elimination of gluconate formation. Appl. Environ. Microbiol. 76: 4369–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krishnaraj PU, Goldstein AH. 2001. Cloning of a Serratia marcescens DNA fragment that induces quinoprotein glucose dehydrogenase-mediated gluconic acid production in Escherichia coli in the presence of stationary phase Serratia marcescens. FEMS Microbiol. Lett. 205: 215–220 [DOI] [PubMed] [Google Scholar]
- 31. Kulasekara HD, et al. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55: 368–380 [DOI] [PubMed] [Google Scholar]
- 32. Matsushita K, Nonobe M, Shinagawa E, Adachi O, Ameyama M. 1987. Reconstitution of pyrroloquinoline quinone-dependent d-glucose oxidase respiratory chain of Escherichia coli with cytochrome o oxidase. J. Bacteriol. 169: 205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsushita K, Shinagawa E, Adachi O, Ameyama M. 1989. Quinoprotein d-glucose dehydrogenase of the Acinetobacter calcoaceticus respiratory chain: membrane-bound and soluble forms are different molecular species. Biochemistry 28: 6276–6280 [DOI] [PubMed] [Google Scholar]
- 34. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170: 2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mustafa G, et al. 2008. Menaquinone as well as ubiquinone as a bound quinone crucial for catalytic activity and intramolecular electron transfer in Escherichia coli membrane-bound glucose dehydrogenase. J. Biol. Chem. 283: 28169–28175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ordentlich A, Elad Y, Chet I. 1987. Rhizosphere colonization by Serratia marcescens for the control of Sclerotium rolfsii. Soil Biol. Biochem. 19: 747–751 [Google Scholar]
- 37. O'Toole GA, et al. 1999. Genetic approaches to the study of biofilms. Methods Enzymol. 310: 91–109 [DOI] [PubMed] [Google Scholar]
- 38. Perez-Tomas R, Vinas M. 2010. New insights on the antitumoral properties of prodiginines. Curr. Med. Chem. 17: 2222–2231 [DOI] [PubMed] [Google Scholar]
- 39. Pocker Y, Green E. 1973. Hydrolysis of d-glucono-delta-lactone. I. General acid-base catalysis, solvent deuterium isotope effects, and transition state characterization. J. Am. Chem. Soc. 95: 113–119 [DOI] [PubMed] [Google Scholar]
- 40. Poon WW, et al. 2000. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J. Bacteriol. 182: 5139–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rius N, Sole M, Fancia A, Loren JG. 1994. Buffering capacity of pigmented and nonpigmented strains of Serratia marcescens. Appl. Environ. Microbiol. 60: 2152–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rosenberg M. 1984. Isolation of pigmented and nonpigmented mutants of Serratia marcescens with reduced cell surface hydrophobicity. J. Bacteriol. 160: 480–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosenberg M, et al. 1986. Cell surface hydrophobicity of pigmented and nonpigmented clinical Serratia marcescens strains. Infect. Immun. 51: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sato T, et al. 1998. Prodigiosins as a new group of H+/Cl− symporters that uncouple proton translocators. J. Biol. Chem. 273: 21455–21462 [DOI] [PubMed] [Google Scholar]
- 45. Shah AJ, Tilburn J, Adlard MW, Arst HN., Jr 1991. pH regulation of penicillin production in Aspergillus nidulans. FEMS Microbiol. Lett. 61: 209–212 [DOI] [PubMed] [Google Scholar]
- 46. Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72: 5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shanks RM, Kadouri DE, MacEachran DP, O'Toole AG. 2009. New yeast recombineering tools for bacteria. Plasmid 62: 88–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shanks RM, et al. 2007. A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J. Bacteriol. 189: 7262–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Silverman MP, Munoz EF. 1973. Effect of iron and salt on prodigiosin synthesis in Serratia marcescens. J. Bacteriol. 114: 999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sole M, Francia A, Rius N, Loren JG. 1997. The role of pH in the “glucose effect” on prodigiosin production by non-proliferating cells of Serratia marcescens. Lett. Appl. Microbiol. 25: 81–84 [DOI] [PubMed] [Google Scholar]
- 51. Sole M, Rius N, Francia A, Loren JG. 1994. The effect of pH on prodigiosin production by non-proliferating cells of Serratia marcescens. Lett. Appl. Microbiol. 19: 341–344 [DOI] [PubMed] [Google Scholar]
- 52. Sole M, Rius N, Loren JG. 2000. Rapid extracellular acidification induced by glucose metabolism in non-proliferating cells of Serratia marcescens. Int. Microbiol. 3: 39–43 [PubMed] [Google Scholar]
- 53. Stella NA, Kalivoda EJ, O'Dee DM, Nau GJ, Shanks RM. 2008. Catabolite repression control of flagellum production by Serratia marcescens. Res. Microbiol. 159: 562–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tanikawa T, Nakagawa Y, Matsuyama T. 2006. Transcriptional downregulator HexS controlling prodigiosin and serrawettin W1 biosynthesis in Serratia marcescens. Microbiol. Immunol. 50: 587–596 [DOI] [PubMed] [Google Scholar]
- 55. Tarighi S, et al. 2008. The PA4204 gene encodes a periplasmic gluconolactonase (PpgL) which is important for fitness of Pseudomonas aeruginosa. Microbiology 154: 2979–2990 [DOI] [PubMed] [Google Scholar]
- 56. Williams RP, Goldschmidt ME, Gott CL. 1965. Inhibition by temperature of the terminal step in biosynthesis of prodigiosin. Biochem. Biophys. Res. Commun. 19: 177–181 [DOI] [PubMed] [Google Scholar]
- 57. Williams RP, Gott CL, Qadri SM, Scott RH. 1971. Influence of temperature of incubation and type of growth medium on pigmentation in Serratia marcescens. J. Bacteriol. 106: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Williamson NR, Fineran PC, Leeper FJ, Salmond GP. 2006. The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4: 887–899 [DOI] [PubMed] [Google Scholar]
- 59. Williamson NR, Fineran PC, Ogawa W, Woodley LR, Salmond GP. 2008. Integrated regulation involving quorum sensing, a two-component system, a GGDEF/EAL domain protein and a posttranscriptional regulator controls swarming and RhlA-dependent surfactant biosynthesis in Serratia. Environ. Microbiol. 10: 1202–1217 [DOI] [PubMed] [Google Scholar]
- 60. Witney FR, Failla ML, Weinberg ED. 1977. Phosphate inhibition of secondary metabolism in Serratia marcescens. Appl. Environ. Microbiol. 33: 1042–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.