Abstract
Bacterial Lon proteases play important roles in a variety of biological processes in addition to housekeeping functions. In this study, we focused on the Lon protease of Azorhizobium caulinodans, which can fix nitrogen both during free-living growth and in stem nodules of the legume Sesbania rostrata. The nitrogen fixation activity of an A. caulinodans lon mutant in the free-living state was not significantly different from that of the wild-type strain. However, the stem nodules formed by the lon mutant showed little or no nitrogen fixation activity. By microscopic analyses, two kinds of host cells were observed in the stem nodules formed by the lon mutant. One type has shrunken host cells containing a high density of bacteria, and the other type has oval or elongated host cells containing a low density or no bacteria. This phenotype is similar to a praR mutant highly expressing the reb genes. Quantitative reverse transcription-PCR analyses revealed that reb genes were also highly expressed in the lon mutant. Furthermore, a lon reb double mutant formed stem nodules showing higher nitrogen fixation activity than the lon mutant, and shrunken host cells were not observed in these stem nodules. These results suggest that Lon protease is required to suppress the expression of the reb genes and that high expression of reb genes in part causes aberrance in the A. caulinodans-S. rostrata symbiosis. In addition to the suppression of reb genes, it was found that Lon protease was involved in the regulation of exopolysaccharide production and autoagglutination of bacterial cells.
INTRODUCTION
Azorhizobium caulinodans ORS571 is a microsymbiont of a tropical legume, Sesbania rostrata, and is able to fix nitrogen in both the free-living and symbiotic states (15–17). Nitrogen-fixing nodules are formed by A. caulinodans on the stems, as well as on the roots, of S. rostrata. Stem nodules occur at the sites of adventitious root primordia located on the stems after crack entry invasion by A. caulinodans (61). During crack entry invasion, bacteria proliferate in the epidermal fissures at the adventitious root primordia on the stems (28). Cortical infection pockets are formed by local cell death, which is induced by the Nod factor that is synthesized by proteins encoded by nodulation genes (nod, nol, noe, and some other genes), and subsequent colonization by bacteria (28). From the infection pockets, infection threads guide bacteria toward the cells in the nodule primordia for symbiotic uptake (11).
Previously, the whole genome sequence of A. caulinodans was determined (38). Alongside genome sequence analysis, we performed a concurrent large-scale screening of rhizobial genetic factors involved in nodule development using A. caulinodans mutants created by random Tn5 mutagenesis (57). Among the Tn5 mutants screened, a mutant (Ao28-F11) with a transposon insertion in a gene (locus tag in the genome, AZC_1610) encoding a putative Lon protease was found to form stem nodules lacking nitrogen fixation activity.
Lon protease was first identified in Escherichia coli (58), and Lon-homologous proteins have been further identified in a wide range of living organisms, including not only eubacteria but also archaea (42), Saccharomyces cerevisiae (65–67), plants (3, 34, 51), and animals (7, 68). Lon protease is an ATP- and Mg2+-dependent protease belonging to the superfamily of AAA+ proteins (ATPases associated with different cellular activities) (43, 45). One of the functions of Lon protease is a housekeeping function that degrades misfolded proteins (29). In addition, Lon protease degrades some specific regulatory proteins and thus is also involved in the regulation of a variety of biological processes that include cell differentiation, sporulation, pathogenicity, survival under starvation conditions, quorum sensing, or acid resistance in different bacterial species (62).
Lon protease of bacteria also plays important roles in the interaction with plants. Many Gram-negative pathogens possess the type three secretion system (TTSS) that delivers effector proteins directly into the host cell cytosol. Lon proteases in some pathogens such as Salmonella enterica and Pseudomonas syringae are known to control the TTSS via degradation of TTSS regulatory proteins or effector proteins, and in same cases, pathogenicity or virulence is reduced by the mutation of lon (9, 36, 40, 59). In the case of Agrobacterium tumefaciens, Lon protease is thought to be required for the proper expression, assembly, or function of the VirB/D4-mediated T-DNA transfer system, and attenuated virulence has been reported for one lon mutant (55). Furthermore, in Sinorhizobium meliloti, a microsymbiont of alfalfa, Lon protease is thought to be involved in the regulation of exopolysaccharide (EPS) synthesis and is required for normal nodulation (56).
The EPS production of S. meliloti has been extensively analyzed. S. meliloti produces two kinds of EPSs, EPS I (succinoglycan) and EPS II (galactoglucan). S. meliloti contains two gene clusters (exo-exs and exp gene clusters) that direct the biosynthesis of EPS I and EPS II, respectively (5). A. caulinodans produces a linear homopolysaccharide of α-1,3-linked 4,6-O-(1-carboxyethylidene)-d-galactosyl residues (12), but there is much less detailed information about the biosynthetic pathway and regulatory mechanism of EPS in A. caulinodans than in S. meliloti. A. caulinodans does not possess a gene cluster corresponding to the S. meliloti exo-exs cluster. However, two gene clusters (exp clusters I and II) putatively corresponding to the S. meliloti exp gene cluster, which is composed of the operons expE, expA, and expD, as well as the expG-expC operon (4, 24), have been identified in the genome of A. caulinodans. exp cluster I (AZC_1831-1834), also known as the oac gene cluster, is composed of oac0, oac1, oac2, and oac3 (homologs of S. meliloti expA8, expA9, expA10, and expA7, respectively), and enzymes encoded by these genes are involved in biosynthesis of dTDP-l-rhamnose from glucose-1-phosphate (26). exp cluster II (AZC_3319-3332) is composed of genes homologous to expA5, expE1 to expE7, expD1, expD2, expG, expC, and expA4 of S. meliloti. AZC_3332 encodes a hypothetical protein. The expression levels of exp cluster I genes were not significantly changed among free-living bacteria in rich and minimal media and bacteroids in stem nodules. In contrast, the expression levels of the exp cluster II genes were higher in free-living cells grown in minimal medium than in those grown in rich medium, and the expression of these genes is more strongly induced in bacteroids than in free-living bacteria (63).
In this study, we made a lon deletion mutant of A. caulinodans and carried out microscopic and expression analyses. On the basis of the data obtained from these analyses, we propose that Lon protease of A. caulinodans possesses new functions in the relationship between bacteria and plants.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in this study are shown in Table 1. A. caulinodans ORS571 (16) and its derivatives were grown at 37°C in TY medium (6) or in synthetic L3 medium (1) containing 10 mM or no NH4Cl, respectively (designated L3+N and L3−N media, respectively). L3 medium is a modified LO medium (15), and except for NH4Cl, it consists of 10 mM potassium phosphate, 10 g liter−1 dl-sodium lactate, 100 mg liter−1 MgSO4 · 7H2O, 50 mg liter−1 NaCl, 40 mg liter−1 CaCl2 · 2H2O, 5.4 mg liter−1 FeCl3 · 6H2O, 5 mg liter−1 Na2MoO4 · 2H2O, 2 mg liter−1 biotin, 4 mg liter−1 nicotinic acid, and 4 mg liter−1 pantothenic acid. Unless otherwise noted, A. caulinodans strains were grown under aerobic (air; 21% O2) conditions with vigorous reciprocal shaking (250 rpm). When A. caulinodans strains were grown under microaerobic conditions, each test tube containing medium was sealed with a butyl rubber septum and the air in the tubes was replaced with N2 containing 3% O2. E. coli strains were grown in Luria-Bertani (LB) medium.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| A. caulinodans | ||
| ORS571 | Type strain | 16 |
| Anx7 | ORS571 derivative; ΔpraR | 1 |
| Anx171 | ORS571 derivative; Δlon | This study |
| Anx173 | Anx171 derivative; complemented with lon; Kmr | This study |
| Anx178 | Anx171 derivative; Δlon Δreb | This study |
| Anx185 | ORS571 derivative; rgs·his6-praR | This study |
| Anx186 | Anx171 derivative; Δlon rgs·his6-praR | This study |
| E. coli | ||
| S17-1 λpir | F− thi pro hsdR [RP4-2 Tc∷Mu Km∷Tn7 (Tpr Smr)] (λpir) | 53 |
| BLR-DE3 | F− ompT hsdSB(rB− mB−) gal dcm(DE3) Δ(srl-recA)306∷Tn10 (Tetr) | Novagen |
| Plasmids | ||
| pK18mobsacB | Suicide vector for gene disruption; lacZ mob sacB; Kmr | 52 |
| pTA-MTL | Suicide vector carrying lacZ reporter gene; Kmr | 33 |
| pCold I | Cold shock expression vector | TaKaRa |
| pTAC68 | pK18mobsacB carrying reb deletion fragment | 1 |
| pTAC89 | pK18mobsacB carrying lon deletion fragment | This study |
| pTAC93 | pTA-MTL carrying entire ORF of lon | This study |
| pTAC102 | pK18mobsacB carrying rgs·his6-praR | This study |
| pTAC103 | pCold I expressing His6-Lon | This study |
| pTAC112 | pCold I expressing His6-LonS684A | This study |
Kmr indicates resistance to kanamycin.
Plant growth and bacterial inoculation.
S. rostrata seeds were treated with concentrated sulfuric acid for 0.5 h, rinsed with sterile water, and soaked in sterile water on trays. The trays were placed at 37°C in an incubator in the dark for 3 days. After germination, S. rostrata plants were transferred into a commercial horticultural soil (Kureha Chemical) and grown at 35°C under a 24-h light regimen at an intensity of 50,000 lx as described previously (57). Although this soil contained 0.4 g N/kg, stem nodule formation was not inhibited by the use of this soil because S. rostrata can form stem nodules even under 6.5 mM NO3− conditions (2). A. caulinodans cultures grown overnight were inoculated onto the stems at 2 weeks after transplantation. The nodules formed on the second stem internode of each plant were used for analyses.
Sequence analysis.
The nucleotide sequence of the entire genome of A. caulinodans ORS571 is available in the DDBJ/EMBL/GenBank databases under accession number AP009384. Homology searches based on amino acid sequences were performed using the BLASTP program on the National Center for Biotechnology Information (NCBI) server (www.ncbi.nlm.nih.gov/BLAST/). Searches for protein signatures were performed with the InterProScan program on the European Bioinformatics Institute server (www.ebi.ac.uk/InterProScan/). Multiple-sequence alignments and phylogenetic analyses were carried out with the ClustalW programs (60).
DNA manipulation.
Genomic DNA isolation, digestion with restriction endonucleases, DNA ligation, E. coli transformation, and plasmid DNA isolation were performed according to standard protocols (50). PCR was performed using PrimeSTAR (TaKaRa). The plasmids and primers used in this study are shown in Tables 1 and 2, respectively.
Table 2.
PCR primer used in this study
| Primer | Sequence (5′–3′) | Reference |
|---|---|---|
| lon related | ||
| L1a | CGAATTCCCAGAACCTGCCTGTTTCAT | This study |
| L2 | ACCTCGGGAGACACCTTCTT | This study |
| L3b | AAGAAGGTGTCTCCCGAGGTGAAGGTCACCGCCAAGAAT | This study |
| L4c | CGGGATCCATGGGCGACGACTCAGTG | This study |
| L5 | AAGGAAGTGGTCGAGCAGAA | This study |
| L6 | AGATCTCGCTCTCCATCAGC | This study |
| L7 | GTGACCGTTCCTTCCATTGT | This study |
| L8a | CGAATCCATGACGAGCCCCAAGCAG | This study |
| L9d | GCTCTAGATGGGCGACGACTCAGTG | This study |
| L10 | GACGGACCAGCTGCAGGCGTCGCCATGGCCACC | This study |
| L11 | TGCAGCTGGTCCGTCCTTCGGGGTCGCGCCTTC | This study |
| praR related | ||
| P1a | CGAATTCTGCTATGAGGCGATCTTTCC | 1 |
| P5d | GCTCTAGAAAGCGTTCGTCAAATCGAA | 1 |
| P6 | TTTGGAAAGTATCGGCCTTG | 1 |
| P7 | CTTCTGGATCTGCTGGAAGG | 1 |
| P8 | GCACCTTGAGGATCTTGTGG | 1 |
| P9 | AACCCCATCGATAAGCATGT | 1 |
| P10c,e | GTGATGGTGATGGTGATGGGATCCTCTCATGATCTCTCCGGCTATGTC | This study |
| P11c,e | ATGAGAGGATCCCATCACCATCACCATCACGTCAAGAAGGCGCCGAAC | This study |
| P12 | CGACGCGTTCTGATTGTATG | This study |
| P13c | CGGGATCCGTCAAGAAGGCGCCGAAC | This study |
| reb related | ||
| R5 | GATCTGCACCCGCTCTCTAC | 1 |
| R6 | GACACTGGTGCTGTTGTTGG | 1 |
| R7 | AAGCCCGGATAGAGGTTGAT | 1 |
| R8 | AACCATTGCAACTCCAGAGG | 1 |
| R9 | ATTTCGTGGTGAGTGCCTTC | 1 |
| R10 | GAAGAAACCCCAGAATGCAA | 1 |
| R11 | GTTGTTCTGCGCGGTCAC | 1 |
| R12 | ACGATGCGCAACTCAGG | This study |
| R13 | CATTGAGGTTGTGCTGGTTC | This study |
| R14 | GCAAGGACCTAACCGAAGAA | This study |
| R15 | ACCGCGTTCTCGAACATC | This study |
| exp related | ||
| E1 | TGGAGTTTGCCGAGACCTAC | This study |
| E2 | CGAGGAAGGTCTTCTGTTCG | This study |
| E3 | CTGCATTTCTCCCAATCGTT | This study |
| E4 | GACAGCAGAACGAGGGTCTC | This study |
| E5 | GGTTCCACGACGTGATCTTC | This study |
| E6 | CGTCTCCTGAGTTTCCGAAC | This study |
| E7 | CGGTCATCAAGAACATCAGC | This study |
| E8 | TGGCCGTCCAGATAGATACC | This study |
| E9 | GTCATGCTGCTGTTCACCAT | This study |
| E10 | ACGATGTAATTGGCCTCCAG | This study |
| E11 | CTCCATCGACATCAAGCAGA | This study |
| E12 | TCAAGGAAAGCCTTCAGCTC | This study |
| 16S rRNA related | ||
| S1 | ACGGATTTCTTCCAGCAATG | 1 |
| S2 | ACCGGCAGTCCCTTTAGAGT | 1 |
| pTA-MTL related | ||
| T1 | ACAGTATCGGCCTCAGGAAG | 1 |
| T2 | TGACCTGAGACAGAGCATTAGC | 1 |
The EcoRI site is underlined.
The sequence complementary to L2 is underlined.
The BamHI site is underlined.
The XbaI site is underlined.
The rgs·his6 sequence is in bold.
Construction of lon and/or reb mutant strains.
Plasmids for gene deletion were constructed by using the splicing by overlap extension (SOEing) PCR method (32). The genomic DNA isolated from A. caulinodans ORS571 was used as the template in each first round of SOEing PCR.
To construct the plasmid used for the lon gene deletion, a fragment containing the 5′ end of lon and a fragment containing the 3′ end of lon were amplified by the first-round PCR using two primer pairs, L1-L2 and L3-L4, respectively. L3 contains the complementary sequence of L2, and the two amplified fragments were integrated by the second-round PCR using L1 and L4. The integrated fragment was digested with EcoRI and BamHI and cloned into a suicide vector, pK18mobsacB, which allows sucrose selection for vector loss (52). The resulting plasmid, designated pTAC89, was conjugated into ORS571 via E. coli S17-1 (λpir) (53) to introduce deletions by allelic exchange. The lon deletion mutant was designated Anx171.
To make a double mutant with lon and reb deleted, pTAC68 (1) was conjugated into Anx171 and the resulting strain was designated Anx178.
The deletion of lon was confirmed by PCR using the genomic DNA of each strain with two primer pairs, L5-L6 and L5-L7. The deletion of the reb locus was confirmed by PCR using the genomic DNA of each strain with two primer pairs, R5-R6 and R5-R7.
For complementation analyses, a fragment containing the entire open reading frame (ORF) of lon was amplified by PCR using primers L1-L4 and the genomic DNA of ORS571 as the template. The amplified fragment was digested with EcoRI and BamHI and cloned into pTA-MTL (33). The resulting plasmid, pTAC93, was conjugated into Anx171 via E. coli S17-1 (λpir), selecting for kanamycin resistance. Correct chromosomal integration of the plasmid was confirmed by PCR using vector-specific primers (T1 and T2) and chromosome-specific primers (L5 and L7). The complemented strain was designated Anx173.
Construction of strains expressing RGS·His6-tagged PraR.
For analyses of PraR protein expression, a plasmid containing the praR (AZC_0013) ORF with an insertion of 5′-AGAGGATCCCATCACCATCACCATCA-3′ (designated the rgs·his6 sequence) downstream of the start codon was constructed by SOEing PCR as follows. A fragment containing the upstream region and the 5′ end of praR and a fragment containing the entire ORF of praR were amplified by the first-round of PCR from genomic DNA of ORS571 with two primer pairs, P1-P10 and P11-P5, respectively. P11 and P10 contain the rgs·his6 sequence and its complement sequence at the 5′ end, respectively. The two amplified fragments were integrated by the second round of PCR using P1 and P5. The integrated PCR fragment was digested with EcoRI and XbaI and cloned into pK18mobsacB. The resulting plasmid, pTAC102, was conjugated into both ORS571 and Anx171. The mutants possessing rgs·his6-praR derived from ORS571 and Anx171 after allelic exchange were designated Anx185 and Anx186, respectively.
To confirm the correct integration of rgs·his6-tagged praR into the chromosome, the fragments amplified by PCR from genomic DNA of each strain with the chromosome-specific primer pair P6-P8 were digested with BamHI sites that are located in the rgs·his6 sequence and the 3′-end region upstream of the AZC_0012 ORF. Furthermore, the fragments amplified by PCR from the genomic DNA of each strain with primer pair P12-P7 flanking the rgs·his6 sequence were sequenced.
Purification of His6-tagged Lon.
To construct a plasmid expressing N-terminally His6-tagged Lon (His6-Lon), the lon ORF was amplified by PCR using primer pair L8-L9 and genomic DNA of ORS571, and the amplified fragment digested with EcoRI and XbaI was cloned into the pCold I vector (TaKaRa). The resulting plasmid was designated pTAC103. To convert the catalytically active serine residue to alanine (S684A), site-directed mutagenesis was performed by PrimeStar Max methods (TaKaRa) using primer pair L10-L11 and pTAC103. The resulting plasmid, expressing N-terminally His6-tagged mutant Lon (His6-LonS684A), was designated pTAC112. E. coli BLR-DE3 (Novagen) carrying pTAC103 or pTAC112 was grown in LB medium containing 50 μg/ml ampicillin at 37°C. When the optical density of the culture at 600 nm (OD600) reached 0.4, 1.0 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG) was added, the culture was further incubated at 15°C for 24 h, and then the cells were collected by centrifugation. The collected cells were suspended in binding/washing buffer (20 mM sodium phosphate buffer [pH 7.4], 500 mM NaCl, 60 mM imidazole) with 5 mM dithiothreitol, and sonicated three times for 15 s at 1-min intervals. The lysed cells were centrifuged at 20,000 × g and for 5 min at 4°C, and His6-tagged proteins were purified from the supernatant by using Ni+-charged magnetic beads (His Mag Sepharose Ni; GE Healthcare) according to the manufacturer's instructions. The beads capturing His6-tagged proteins were washed three times with binding/washing buffer, and His6-tagged proteins were eluted using elution buffer (20 mM sodium phosphate buffer [pH 7.4], 500 mM NaCl, 500 mM imidazole). The protein concentrations in the eluted samples were measured by the Bradford assay using bovine serum albumin as the standard.
To evaluate the purity of the eluted His6-Lon and His6-LonS684A, the eluted samples and total lysed cells were separated by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) using 10% polyacrylamide gels and the gel was stained with Coomassie brilliant blue (CBB) G-250. Furthermore, the eluted samples were fractionated by SDS-PAGE, electroblotted onto polyvinylidene difluoride (PVDF) membrane in EzFastBlot buffer (Atto), and incubated with mouse anti-His5 antibody (Qiagen). Horseradish peroxidase (HRP)-conjugated sheep anti-mouse antibodies (GE Healthcare) were used in combination with the ECL Advance detection kit (GE Healthcare) for chemiluminescence and visualization in an LAS3000 Luminescent Image Analyzer (Fujifilm).
Protease activity assay.
Protease activity was measured on the basis of the method of Twining (64) using the fluorescein isothiocyanate-labeled casein (FITC-casein) included in the Protease Fluorescent Detection kit (Sigma-Aldrich). Reaction buffer was made by mixing 20 μl of incubation buffer (20 mM phosphate buffer [pH 7.6], 150 mM NaCl [supplied with the kit]) with 2 μl of 100 mM ATP (final concentration, 4 mM), 5 μl of 100 mM MgCl2 (final concentration, 10 mM), and 20 μl of 0.5% FITC-casein. When ATP and/or MgCl2 was omitted, pure water was added instead. Five microliters of purified His6-Lon (320 ng/μl), purified His6-LonS684A (360 ng/μl), or elution buffer, which was used for purification of His6-tagged proteins, was added to the reaction buffer, and it was incubated at 37°C in the dark. After 8 h of incubation, the reaction was stopped by adding 50 μl of 0.6 N trichloroacetic acid and mixed well. After incubation for 30 min at 37°C in the dark, the reaction contents were centrifuged (10,000 × g for 10 min at room temperature) and 10 μl of the supernatant was mixed with 1 ml of 500 mM Tris-HCl (pH 8.5). For each sample, 200 μl of the mixture was transferred to a black 96-well plate and fluorescence intensity was measured in an Infinite F200 PRO multimode plate reader (TECAN) at wavelengths of 485 nm (excitation) and 535 nm (emission).
Acetylene reduction activities (ARAs) of bacterial cultures.
A. caulinodans strains grown in TY medium to an OD600 of 0.8 were collected by centrifugation and washed twice with L3−N medium. The washed bacteria were suspended in L3−N medium and cultured for 5 h. Bacteria were then collected and washed twice with L3−N medium. Bacteria were suspended in 2 ml of L3−N medium to an OD600 of 0.1 in a 32-ml test tube and grown under microaerobic conditions. After 16 h, acetylene was added to each tube up to 10% (vol/vol) and the bacteria were incubated for 2 h. After incubation, 400 μl of gas was sampled from the tube and the concentrations of acetylene and ethylene were measured with a gas chromatograph (model GC-17A; Shimadzu) equipped with a fused-silica column (Rt-U PLOT; Restek). The OD600 of each sample was measured after incubation with acetylene.
ARAs of stem nodules.
Ten stem nodules per plant were harvested and placed into a 20-ml vial sealed with a butyl rubber septum. Acetylene was added to each vial up to 10% (vol/vol), and the harvested stem nodules were incubated at 37°C for 2 h. After incubation, 100 μl of gas was sampled from the vial and the concentrations of acetylene and ethylene were measured.
Optical microscopic analysis.
Stem nodules were cut into two or three pieces with a razor blade, fixed in 0.25% glutaraldehyde and 2% paraformaldehyde in 50 mM HEPES buffer (pH 7.0) for 3 h at room temperature, and then kept at 4°C overnight. The fixed stem nodules were washed with 50 mM HEPES buffer (pH 7.0), dehydrated through a graded ethanol series (70% for 2 h, 96% for 2 h, and 100% for 1 h), and then embedded in Technovit 7100 (Heraeus Kulzer) according to the manufacturer's instructions. The embedded stem nodules were sliced into 5-μm sections with a microtome (RM-2125RT; Leica). The sections were stained with 0.05% toluidine blue O and observed using a bright-field microscope (DMLB; Leica).
Autoagglutination of bacterial cells.
A. caulinodans strains grown in TY medium to an OD600 of 0.8 were collected and washed twice with L3+N medium. The washed bacteria were suspended in 10 ml of L3+N medium to an OD600 of 0.01 in 50-ml tubes. The tubes containing bacterial cultures were positioned vertically in a rotary shaker and incubated with shaking (200 rpm). After 16 h, aliquots of bacterial cultures were observed with a bright-field microscope.
Quantification of EPSs produced by bacteria.
EPSs were quantified on the basis of the method of Gao et al. (23), with some modifications. A. caulinodans strains grown in TY medium to an OD600 of 0.8 were washed twice with L3+N medium and suspended in L3+N medium to an OD600 of 1.0. Fifteen-microliter aliquots of the bacterial suspensions were spotted onto TY or L3+N solid plates containing 0.8% agar. After incubation for 3 days, as many bacteria as possible were collected with a platinum loop and resuspended in 300 μl of λ buffer (10 mM Tris-HCl [pH 7.0], 10 mM MgSO4). The bacterial suspension was centrifuged (10,000 × g for 10 min at 4°C), and 250 μl of the supernatant was put aside while the pellet was washed with an additional 300 μl of λ buffer and centrifuged again and 250 μl of the supernatant was added to the first supernatant. A 400-μl volume of λ buffer was added to the pellet, and the OD600 of the suspended bacteria was measured. A 100-μl volume of the combined supernatant was mixed with 1 ml of 0.2% anthrone in concentrated sulfuric acid, incubated for 7 min at 100°C, and then quickly chilled on ice. The OD620 of the chilled mixture was measured. d-Glucose was used as a standard to calculate the glucose equivalents in the EPS samples. EPS amounts from each colony were evaluated by normalizing to the OD600 of the collected cell suspension.
RNA isolation and purification.
For the free-living bacterial culture, 1/10 volume of ice-cold stop solution (5% water-saturated phenol in ethanol) was added to it and mixed well immediately. Bacteria were harvested by centrifuging, and bacterial pellets were quickly frozen with liquid nitrogen and stored at −80°C until RNA isolation. In the case of stem nodules, the harvested nodules were quickly frozen with liquid nitrogen and stored at −80°C. Before RNA isolation, the frozen stem nodules were ground with a mortar and pestle. Crude RNA was isolated from each sample using TRIsure (Bioline), and degradation of contaminating DNA and purification of total RNA were performed using NucleoSpin RNA II (Macherey-Nagel) according to the manufacturer's instructions.
Quantitative reverse transcription (RT)-PCR.
Total RNA was isolated from free-living bacteria grown for 16 h in the appropriate medium and from stem nodules at 12 days postinoculation (dpi). The cDNA was synthesized from 400 ng of total RNA by using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa) according to the manufacturer's instructions. The synthesized cDNA was diluted exactly 20-fold and 400-fold with 10 mM Tris-HCl (pH 8.0) and used in the following quantitative PCR. The 400-fold-diluted cDNA was used for the analysis of 16S rRNA expression, while the 20-fold-diluted cDNA was used to determine the expression of other genes. Quantitative PCR was carried out with a LightCycler system (Roche) using a QuantiTect SYBR green PCR kit (Qiagen) with gene-specific primer pairs (R8-R9, R10-R11, R12-R13, and R14-R15 for reb genes AZC_3781, AZC_3782, AZC_3783, and AZC_3786, respectively; E1-E2, E3-E4, E5-E6, E7-E8, E9-E10, and E11-E12 for exp genes AZC_3319, AZC_3325, AZC_3326, AZC_3328, AZC_3329, and AZC_3331, respectively; P7-P9 for praR; S1-S2 for 16S rRNA) and 1-μl aliquots of the diluted cDNA. Genomic DNA isolated from the ORS571 strain was used as the template in constructing the standard curves to determine the copy number of the transcripts of each gene. To evaluate the expression level of each gene, the copy number of transcripts of each gene was normalized to that of the 16S rRNA.
Western blotting analyses of RGS·His6-PraR.
A. caulinodans strains grown in TY medium to an OD600 of 0.8 were washed twice with L3+N medium. The washed bacteria were suspended in L3+N medium to an OD600 of 0.01 and cultured for 16 h. The bacterial cells collected by centrifugation were sonicated (three times for 15 s at 1-min intervals) in phosphate-buffered saline (150 mM NaCl, 10 mM Na2HPO4, 20 mM NaH2PO4, pH 7.4) and fractionated by SDS-PAGE in 14% polyacrylamide gels. Fractionated proteins were electroblotted onto PVDF membrane and incubated with mouse anti-RGS·His4 antibody (Qiagen), followed by detection using HRP-conjugated sheep anti-mouse antibodies and an ECL Advance detection kit as described above. After detection, PVDF membrane was stained with CBB G-250 to confirm that equal amounts of total cell protein were loaded for SDS-PAGE.
RESULTS
Genetic organization of the lon gene in the genome and structure of Lon.
The lon gene (AZC_1610) is flanked by clpX (AZC_1609) and hupB (AZC_1611), which encode a putative ATP-dependent Clp protease and a putative histone-like DNA-binding protein, respectively. No lon-homologous genes other than AZC_1610 were found in the genome of A. caulinodans. The translation of the ORF of lon was previously predicted to start at a position 1,837,639 bp from the origin in the chromosome (38). However, in this study, we propose that the ORF of lon starts 150 bp downstream of the previous position (see Fig. S1 in the supplemental material). A BLASTP analysis revealed that the Lon protein of A. caulinodans was 78% and 64% identical to the Lon proteins of S. meliloti 1021 (NCBI reference no. NP_385363) and E. coli K-12 (NP_414973), respectively. An InterProScan analysis revealed that an “ATPase, AAA+ type, core” signature (accession no. IPR003959) and a “peptidase S16 active site” signature (accession no. IPR008268), conserved in all known Lon proteases, were also present in the Lon protein sequence of A. caulinodans (data not shown). The proteolytic domains of Lon proteases typically contain a serine-lysine dyad critical for catalysis, with the lysine appearing 43 residues downstream of the catalytic serine (8), and it was found that the Lon protein of A, caulinodans also contains a putative serine-lysine dyad (S684 and K727) in its sequence (data not shown).
Protease activity of recombinant Lon protease.
To examine whether the deduced Lon protein of A. caulinodans really has protease activity, we generated N-terminally His6-tagged wild-type Lon protein (His6-Lon) and N-terminally His6-tagged mutant Lon protein (His6-LonS684A) with conversion of the putative catalytic serine to alanine (S684A). His6-Lon and His6-LonS684A expressed in E. coli were purified and analyzed by SDS-PAGE and Western blotting with anti-His antibody. The molecular masses of purified His6-Lon and His6-LonS684A were in agreement with the calculated molecular masses (89 kDa; see Fig. S2A and B in the supplemental material). The protease activities of purified His6-Lon and His6-LonS684A were measured by degradation of FITC-casein in the presence of 4 mM ATP and/or 10 mM Mg2+ (see Fig. S2C in the supplemental material). The fluorescence intensity when His6-Lon was added to FITC-casein in the presence of both ATP and Mg2+ was higher than that measured under the other conditions lacking either ATP or Mg2+. This result suggests that the Lon protein of A. caulinodans is an ATP- and Mg2+-dependent protease, just like the other known Lon proteases. The fluorescence intensity was not increased when His6-LonS684A was added to FITC-casein, even in the presence of both ATP and Mg2+, suggesting that serine 684 is actually required for proteolytic activity.
Phenotypes of the lon deletion mutant in the free-living states.
In this study, we made a lon deletion mutant strain (Anx171) and an Anx171 derivative strain (Anx173) complemented with lon. For the genetic organization of Anx171 (Δlon) and Anx173 (Δlon+lon), see Fig. S1A in the supplemental material. The deletion of lon in Anx171 (Δlon) and the chromosomal integration of the plasmid carrying the entire ORF of lon in Anx173 (Δlon+lon) were confirmed by PCR (see Fig. S1C and D in the supplemental material).
The growth and nitrogen fixation activity of Anx171 (Δlon) in the free-living state were compared to those of ORS571. To avoid autoagglutination of bacterial cells (see below), these strains were cultured in test tubes containing liquid medium and incubated in an inclined position with vigorous reciprocal shaking. The growth of Anx171 (Δlon) was not significantly different from that of ORS571 during culture in TY medium under aerobic conditions and in L3−N medium under microaerobic conditions, but it was slower than that of ORS571 during culture in L3+N medium under aerobic conditions (see Fig. S3A in the supplemental material). The growth of Anx173 (Δlon+lon) was not significantly different from that of ORS571 under any of the conditions (data not shown). Significant differences between the ARAs of ORS571 and Anx171 (Δlon) were also not observed when the bacteria were grown in L3−N medium under microaerobic conditions (see Fig. S3B in the supplemental material), suggesting that Lon protease is not involved in nitrogen fixation in the free-living state.
When A. caulinodans ORS571 was cultured with rotary shaking in L3+N medium in test tubes positioned vertically, the bacterial cells showed autoagglutination, forming large clumps (Fig. 1). This autoagglutination was not observed when ORS571 was cultured in TY medium (data not shown). In contrast, Anx171 (Δlon) did not show autoagglutination even when cultured in L3+N medium under such mild shaking conditions (Fig. 1). Anx173 (Δlon+lon) showed autoagglutination similar to that of ORS571 (data not shown).
Fig 1.
Autoagglutination activity of the lon mutant and the lon reb double mutant. ORS571 (wild type [WT]), Anx171 (Δlon), Anx178 (Δlon Δreb), and Anx7 (ΔpraR) were grown in L3+N medium under aerobic conditions in vertical test tubes in a rotary shaker. After 16 h of incubation, cultures were observed.
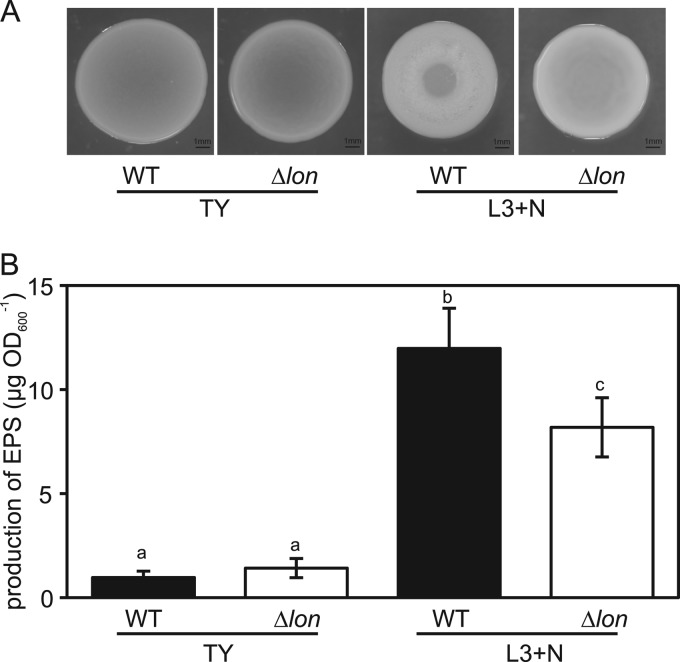
It is known that a lon mutant of S. meliloti produces larger amounts of EPS than the wild-type strain (56). Thus, the EPS production of Anx171 (Δlon) was compared to that of ORS571. To quantify the amounts of EPS released from bacterial cells, bacterial cultures grown in liquid medium were spotted onto TY and L3+N plates and incubated for 3 days (Fig. 2A). In both TY and L3+N plates, the diameters of spreading colonies of both strains were almost the same. However, the central region of each colony formed by ORS571 on L3+N plates appeared less dense than that of a colony formed by Anx171 (Δlon), a clear distinction between the phenotypes of Anx171 (Δlon) and ORS571. After incubation, most of the bacterial cells were scraped from the plates and the amounts of EPS were measured according to the method of Gao et al. (23). When Anx171 (Δlon) was grown on TY plates, its EPS production was not different from that of ORS571 (Fig. 2B). When grown on L3+N plates, these two strains produced more EPS than on TY plates; however, the amount of EPS produced by Anx171 (Δlon) was smaller than that produced by ORS571 (Fig. 2B).
Fig 2.
EPS production of the lon mutant. (A) Colonies of ORS571 (wild type [WT]) and Anx171 (Δlon) grown for 3 days on TY or L3+N medium plates containing 0.8% agar. A 15-μl aliquot of the bacterial suspension (OD600 of 1.0) was spotted onto each plate. (B) Measurement of EPS produced by ORS571 (wild type [WT]) and Anx171 (Δlon). The colonies on the plates were collected, and the EPSs released from bacterial cells were quantified. The EPS amount from each colony was evaluated by normalizing the collected cell suspension to the OD600. The values are means ± standard deviations of five replicate colonies. Different letters indicate significant differences (P < 0.05; Tukey-Kramer).
Phenotypes of stem nodules formed by the lon deletion mutant.
ORS571 and Anx171 (Δlon) were inoculated onto the stems of S. rostrata, and stem nodules formed by these strains were observed (Fig. 3A). The stem nodules formed by Anx171 (Δlon) were smaller than those formed by ORS571. The color of the inner region of the stem nodules formed by Anx171 (Δlon) was pale pink at 7 dpi and white at 12 dpi. This result suggests that Anx171 (Δlon) did not affect leghemoglobin induction in host plants at the early stage of nodule formation but the symbiotic process was aborted at the subsequent stage of nodule formation. To investigate the nitrogen fixation activities of the nodules formed by each strain, ARAs were measured chronologically after inoculation (Fig. 3B). The ARAs of the nodules formed by Anx171 (Δlon) were very low but detectable at 7 dpi and 10 dpi; however, they were not detected at 12 and 14 dpi. The size, inner-region color, and ARAs of Anx173 (Δlon+lon) and ORS571 nodules were almost identical to each other (data not shown).
Fig 3.

Stem nodules formed by the lon mutant and the lon reb double mutant. (A) Inner regions of stem nodules formed by ORS571 (wild type [WT]), Anx171 (Δlon), Anx178 (Δlon Δreb), and Anx7 (ΔpraR). Each strain was inoculated onto the stems of S. rostrata plants, and the stem nodules were observed at 7 and 12 dpi. (B) Nitrogen fixation activities of the stem nodules. The stem nodules formed by each strain were harvested at 7 to 14 dpi, and ARA was measured. The values are means ± standard deviations of five replicate plants. Different letters above the bars indicate significant differences (P < 0.05; Tukey-Kramer).
To investigate the phenotypes of stem nodules in detail, microscopic analyses were carried out (Fig. 4). The ORS571 nodules are shown filled with oval or elongated host cells that were infected with bacteria at both 7 and 12 dpi in Fig. 4A, E, and I. These oval or elongated cells were also observed in the Anx171 (Δlon) nodules at 7 dpi (Fig. 4B and F), but these host cells contained a lower number of bacteria than those of ORS571, and vacuoles were observed in such host cells (Fig. 4F). Furthermore, some oval or elongated cells were not stained, suggesting that vacuoles were fully expanded in such cells (Fig. 4F). In addition, shrunken host cells containing a larger number of bacterial cells than these oval or elongated cells were observed in Anx171 (Δlon) nodules at 7 dpi (Fig. 4F). These shrunken cells in Anx171 (Δlon) nodules were also observed at 12 dpi (Fig. 4J and M). Oval or elongated cells were also observed in Anx171 (Δlon) nodules at 12 dpi, but most of these cells were not stained (Fig. 4M). These results were very similar to those of an A. caulinodans mutant, Anx7, with a deletion of the praR gene encoding a putative transcription factor whose gene homologs are highly conserved in Alphaproteobacteria (1). To confirm the similarity, the Anx7 (ΔpraR) strain was inoculated onto stems and the phenotypes of the nodules formed by this mutant were compared to those of the nodules formed by Anx171 (Δlon). The nitrogen fixation activities of Anx7 (ΔpraR) nodules were lower than those of ORS571 nodules but higher than those of Anx171 (Δlon) nodules (Fig. 3B). The inner regions of nodules at 7 and 12 dpi were observed by microscopy (Fig. 4C, G, K, and N). Although most aspects of host cells in Anx7 (ΔpraR) and Anx171 (Δlon) nodules were similar, the number of shrunken cells containing bacteria seemed to be higher in Anx7 (ΔpraR) nodules than in Anx171 (Δlon) nodules (Fig. 4J and K).
Fig 4.
Optical microscope images of stem nodules of ORS571, Anx171 (Δlon), Anx7 (ΔpraR), and Anx78 (Δlon Δreb). Stem nodules at 7 dpi (A to H) and 12 dpi (I to O) were longitudinally sectioned and stained with toluidine blue O. Panels: A, E, and I, ORS571; B, F, J, and M, Anx171; C, G, K, and N, Anx7; D, H, L, and O, Anx178. Abbreviations: oc, oval or elongated host cell; sc, shrunken host cell; uc, unstained host cell; v, vacuole.
Quantitative RT-PCR analyses of reb and exp genes.
Previously, we found that shrunken host cells containing high-density bacteria were caused by high expression of the reb genes in Anx7 (ΔpraR) (1). The results of microscopic analyses suggest that reb genes are also highly expressed in Anx171 (Δlon). Thus, the expression levels of four reb genes (rebD1, AZC_3781; rebA1, AZC_3782; rebD2, AZC_3783; rebA2, AZC_3786) in Anx7 (ΔpraR) and ORS571 in the free-living and symbiotic states were analyzed by quantitative RT-PCR (Fig. 5A). In the case of free-living bacteria grown in L3+N or TY medium, the expression levels of these four genes were 3- to 4-fold higher in Anx171 (Δlon) than in ORS571. The expression levels of these four genes of ORS571 were lower in nodules than in free-living bacteria. However, the expression levels of these four genes in Anx171 (Δlon) nodules were much higher than in the free-living Anx171 (Δlon) strain, and these were 1,000- to 2,000-fold higher than in ORS571 nodules.
Fig 5.
Quantitative RT-PCR analyses of reb genes, exp genes, and praR in the free-living and symbiotic states. (A) Expression of reb genes (AZC_3781, AZC_3782, AZC_3783, and AZC_3786). (B) Expression of exp genes (AZC_3319, AZC_3325, AZC_3326, AZC_3328, AZC_3329, and AZC_3331). (C) Expression of praR (AZC_0013). Total RNAs were isolated from the free-living bacteria of strains ORS571 (wild type [WT]) and Anx171 (Δlon) grown in TY and L3+N media under aerobic conditions and from the stem nodules formed by ORS571 and Anx171 at 14 dpi. The amounts of transcripts of each gene and 16S rRNA in the total RNA from each sample were estimated by quantitative RT-PCR, and the expression levels of each gene were evaluated by normalization to the 16S rRNA level. The values are means ± standard deviations of three replicate cultures or plants and are presented relative to the results of free-living ORS571 bacteria grown in TY medium.
Because the amounts of EPS produced by Anx171 (Δlon) grown on L3+N medium plates were lower than those from the ORS571 strain, the expression levels of some exp genes putatively related to EPS production were also analyzed. In this study, we selected six genes (AZC_3319, expA5 like; AZC_3325, expE2 homolog; AZC_3326, expE1 homolog; AZC_3328, expD1 homolog; AZC_3329, expG homolog; AZC_3331, expA4 like) from among the exp cluster II genes (Fig. 5B). When bacteria were grown in TY medium, the expression levels of these Anx171 (Δlon) genes were almost the same as in ORS571, except for AZC_3325 (2.5-fold higher). The expression levels of the ORS571 genes were 3.7- to 35-fold higher in L3+N medium than in TY medium, but those of Anx171 (Δlon) were 1.2- to 3.7-fold higher. The expression of AZC_3325 and AZC_3326 in both strains was further induced in stem nodules, and AZC_3328 of Anx171 (Δlon) was rarely expressed in stem nodules.
Phenotypes of the lon reb double mutant.
In a previous study, we found that a double mutant with deletions of praR and reb formed wild-type nodules (1). To investigate whether the aberrant phenotypes of nodules in the Anx171 (Δlon) strain were caused by high expression of reb genes, we made a strain, Anx178, with deletions of both lon and reb (see Fig. S1B in the supplemental material). The deletions of lon and reb from Anx178 (Δlon Δreb) were confirmed by PCR (see Fig. S1C in the supplemental material).
There were no significant differences in growth, nitrogen-fixing ability, autoagglutination, or EPS production between free-living Anx178 (Δlon Δreb) and Anx171 (Δlon) when these mutants were grown under the same respective conditions employed in the comparison of the lon mutant and the wild-type strain (data not shown). Next, we investigated the phenotypes of Anx178 (Δlon Δreb) nodules. Anx178 (Δlon Δreb) stem nodules were larger than those formed by Anx171 (Δlon) but smaller than ORS571 stem nodules (Fig. 3A). The red color caused by leghemoglobin was detected in the inner region of the Anx178 (Δlon Δreb) stem nodules at 12 dpi, although it was lighter than in the ORS571 nodules (Fig. 3A). The ARAs of the Anx178 (Δlon Δreb) nodules were higher than those of the Anx171 (Δlon) nodules and were detectable even at 14 dpi, although these were lower than those of the ORS571 nodules (Fig. 3B).
By microscopic analyses, oval or elongated cells filled with bacteria were observed in the stem nodules formed by Anx178 (Δlon Δreb) at 7 dpi and some of these host cells contained expanded vacuoles (Fig. 4D and H). These oval or elongated cells were also observed in nodules at 12 dpi, but they contained fewer or no bacteria and more expanded vacuoles were observed (Fig. 4L and O). Shrunken host cells containing high-density bacteria, which were observed in Anx171 (Δlon) and Anx7 (ΔpraR) nodules, were not observed in Anx178 (Δlon Δreb) nodules.
Expression levels of the praR gene and PraR protein in the lon mutant.
To explore the causes of high expression of reb genes in Anx171 (Δlon), we analyzed the expression levels of the praR gene and the PraR protein. First, the expression levels of praR in Anx171 (Δlon) in the free-living and symbiotic states were compared to those in ORS571, but there were no significant differences in the expression levels of praR between these two strains under any condition (Fig. 5C). Next, to investigate the expression levels of the PraR protein, the native praR ORFs in the genomes of ORS571 and Anx171 (Δlon) were replaced with a modified praR ORF encoding RGS·His6-PraR. Two strains derived from ORS571 and Anx171 (Δlon) were designated Anx185 and Anx186, respectively (see Fig. S4 in the supplemental material). The presence of RGS·His6-PraR in the lysates of Anx185 (wild type, rgs·his6-praR), Anx186 (Δlon, rgs·his6-praR), ORS571, and Anx171 (Δlon) grown in L3 medium was estimated by Western blot analysis with anti-RGS·His4 antibody (Fig. S5 in the supplemental material). RGS·His6-PraR was detected in Anx185 (wild type, rgs·his6-praR) and Anx186 (Δlon, rgs·his6-praR) lysates, and it was found that the expression levels of RGS·His6-PraR were not different between these strains.
DISCUSSION
For establishment of a symbiosis between rhizobia and legumes, the power balance between the two partners should be adequately maintained in nodules. The expression of genes having harmful effects on each partner should be suppressed. The expression of the reb genes of A. caulinodans seems to be representative of this power play.
In this study, we found that the lon deletion mutant of A. caulinodans formed aberrant stem nodules and that this aberrance was in part caused by high expression of reb genes. The reb genes were originally identified from Caedibacter taeniospiralis (30). Caedibacter species are obligate endobiotic bacteria inhabiting paramecium hosts and are characterized by their ability to produce R-bodies (refractile inclusion bodies), which are insoluble protein ribbons that are seen coiled into cylindrical structures within the cell (47). It has been reported that paramecia that contain Caedibacter cells producing R-bodies kill paramecia that do not contain endobionts or that contain endobionts that are not of the same Caedibacter species (47, 48). Furthermore, a Caedibacter mutant that is defective in R-body production has also been observed as not exhibiting “killer traits” (13). The rebA, rebB, rebC, and/or rebD genes of C. taeniospiralis are present on plasmids and are involved in R-body synthesis and assembly (30, 35, 49). For a long time, the focus of research on reb genes and R-bodies had been only on the relationship between paramecia and Caedibacter bacteria. However, genomic sequencing analyses of different kinds of bacteria have revealed that some species that belong to the phylum Proteobacteria, as well as Kordia algicida OT-1 (54), which belongs to the phylum Bacteroidetes, possess reb-homologous genes, although reb-homologous genes have not been found in rhizobia other than A. caulinodans (1). Recently we carried out a functional analysis of the reb genes of A. caulinodans, which is the first example of such an analysis outside Caedibacter, and found that high expression of the reb genes caused aberrance in A. caulinodans-S. rostrata symbiosis (1). The expression of the reb genes is suppressed by the transcription factor PraR, and a praR reb double mutant formed normal stem nodules, although it is still unclear whether the expression of reb genes is regulated by PraR in a direct or an indirect manner.
Microscopic observations (Fig. 4) revealed that the nodule phenotypes of the lon mutant are similar to those of the praR mutant. It is noteworthy that shrunken host cells containing high-density bacteria were observed in the nodules formed by both mutants. These shrunken host cells were not observed in the stem nodules formed by the lon reb double mutant and the praR reb double mutant (Fig. 4). Taken together with the findings that the reb genes were highly expressed in the lon mutant (Fig. 5) and the praR mutant (1), these results strongly suggest that these shrunken host cells are the result of the high expression of the reb genes. As described above, the reb genes of Caedibacter could be responsible for the “killer trait” of the host paramecia. Thus, it is presumable that the shrunken host cells resulted from the attack of the bacteria highly expressing the reb genes. In addition to shrunken host cells, oval or elongated host cells were also observed in the nodules formed by the lon mutant and the praR mutant (Fig. 4). Bacteria were observed in such host cells at early stages of nodule formation (at 7dpi), but the bacterial density was lower than in shrunken host cells. In addition, expanded vacuoles were observed in these oval or elongated host cells. At a later stage of nodule formation (at 12 dpi), oval or elongated host cells rarely contained bacteria. It is presumed that the bacteria were lysed by the enlarged vacuoles in the oval or elongated cells; i.e., such host cells attack the bacteria, resulting in the elimination of the bacteria. From these observations, one of the most important functions of Lon protease in nodule formation is to maintain the power balance between A. caulinodans and S. rostrata by suppressing the expression of the reb genes.
Although the double mutation of praR and reb resulted in wild-type nodule formation (1), the double mutation of lon and reb did not (Fig. 3 and 4). This difference between the two sets of mutants suggests that the Lon protease controls some factors required for nodule formation in addition to suppression of the reb genes. It is possible that EPS production is one of these factors. The lon mutant in the free-living state produces a smaller amount of EPS than the wild type (Fig. 2), and one of the exp genes (AZC_3328, an expD1 homolog) was rarely expressed in the stem nodules of the lon mutant (Fig. 5). Rhizobial EPSs, especially of S. meliloti and Rhizobium leguminosarum, are known to play important roles in the establishment of symbiosis and are required for bacterial attachment to root hairs, curling of root hairs, elongation of infection threads, invasion of host cells, protection against plant defense responses, and so on (10, 14, 19, 20, 22, 25, 27, 31, 37, 39, 39, 44). In the case of A. caulinodans, two mutants, ORS571-X15 and ORS571-oac2 (23, 26), are known to be EPS-deficient mutants that have mutations in oac3 (AZC_1831) and oac2 (AZC_1832), respectively, These mutants are also known to have truncated lipopolysaccharides (LPSs) with an altered O-antigen structure (23). Based on a series of studies using these mutants, it is proposed that EPSs protect the bacteria against H2O2 produced by the plant in the infection pockets and infection threads in these early steps of nodulation (11, 12, 23, 41). Because there is little information about EPS-deficient mutants of A. caulinodans, especially in the case of the exp cluster II gene mutants whose LPS production is not thought to be altered, the real function of EPSs in A. caulinodans nodule formation is still unclear. We did not carry out further analyses of EPS production in this study because our major interest was in reb gene expression, but it is interesting to speculate how the Lon protease controls EPS production and the expression of exp genes in A. caulinodans. In contrast to A. caulinodans, an S. meliloti lon mutant overproduces EPSs and this overproduction has been speculated to be one of the causes of the defective symbiotic phenotype of this mutant (56). Thus, it seems likely that EPS production in each rhizobium is controlled differently by the Lon protease.
In this study, we found that the lon mutant did not show autoagglutination even when grown in L3+N medium, unlike the wild type (Fig. 1). The relationship between the Lon protease and autoagglutination has been well analyzed in Pseudomonas syringae pv. phaseolicola. A lon mutant of P. syringae develops larger aggregates than the wild type, which are caused by the increased expression and assembly of a functional TTSS apparatus (46). As A. caulinodans does not possess TTSS-related genes in its genome, autoagglutination of this bacterium is caused by other factors.
Both Lon and PraR are required for the suppression of reb genes. The interesting question is whether these two factors regulate the expression of reb genes in a cooperative or an independent manner. It is unlikely that PraR regulates the expression of reb genes via Lon (i.e., PraR regulates the expression or activities of Lon) because the praR reb double mutant forms wild-type stem nodules but the lon reb double mutant does not (Fig. 3 and 4). If PraR had an influence on Lon properties such as the expression and activity of lon and Lon, etc., and if the deletion of praR led to ineffectiveness of Lon, then the praR reb double mutant would form stem nodules similar to those of the lon reb double mutant. Furthermore, the praR mutant shows autoagglutination, just like the wild-type strain, but the lon mutant does not (Fig. 1). This fact also strongly suggests that Lon is not under the control of PraR. In addition, the previous microarray analysis showed that the deletion of praR had no influence on the expression levels of lon in the free-living state (1). So, is expression of the praR gene or the PraR protein under the control of Lon? We carried out expression analyses of both to answer this question, but the expression levels of the praR gene and the PraR protein are not influenced by the deletion of lon (Fig. 5; see Fig. S5 in the supplemental material). However, these results cannot negate the hypothesis that the PraR-mediated regulatory system is under the control of Lon. Recently it was reported that PraR is involved in quorum-sensing regulation in R. leguminosarum. PraR represses the expression of the rhiR and raiR genes, which encode transcription regulators involved in quorum-sensing regulation, and PraR is displaced from these promoters when PraR is bound to a small regulatory protein, CinS (21). Although cinS is present in only a few rhizobia (18) and not in A. caulinodans, it is possible that the activity of PraR in A. caulinodans might be controlled by binding to an unknown regulator other than CinS and that the properties of such a protein might be influenced by Lon protease activity. As another option, there still remains the possibility that Lon and PraR independently regulate the expression of reb genes. In any case, only Lon and PraR have been identified as regulation factors for the expression of reb genes, so further analyses are required to elucidate the reb expression system.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by the Japan Society for the Promotion of Science (grant 23780061).
Footnotes
Published ahead of print 29 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Akiba N, Aono T, Toyazaki H, Sato S, Oyaizu H. 2010. phrR-like gene praR of Azorhizobium caulinodans ORS571 is essential for symbiosis with Sesbania rostrata and is involved in expression of reb genes. Appl. Environ. Microbiol. 76: 3475–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aono T, Kanada N, Ijima A, Oyaizu H. 2001. The response of the phosphate uptake system and the organic acid exudation system to phosphate starvation in Sesbania rostrata. Plant Cell Physiol. 42: 1253–1264 [DOI] [PubMed] [Google Scholar]
- 3. Barakat S, Pearce DA, Sherman F, Rapp WD. 1998. Maize contains a Lon protease gene that can partially complement a yeast pim1-deletion mutant. Plant Mol. Biol. 37: 141–154 [DOI] [PubMed] [Google Scholar]
- 4. Becker A, et al. 1997. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J. Bacteriol. 179: 1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Becker A, et al. 2002. Regulation of succinoglycan and galactoglucan biosynthesis in Sinorhizobium meliloti. J. Mol. Microbiol. Biotechnol. 4: 187–190 [PubMed] [Google Scholar]
- 6. Beringer JE. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84: 188–198 [DOI] [PubMed] [Google Scholar]
- 7. Bota DA, Davies KJA. 2002. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 4: 674–680 [DOI] [PubMed] [Google Scholar]
- 8. Botos I, et al. 2004. The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site. J. Biol. Chem. 279: 8140–8148 [DOI] [PubMed] [Google Scholar]
- 9. Bretz J, Losada L, Lisboa K, Hutcheson SW. 2002. Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae. Mol. Microbiol. 45: 397–409 [DOI] [PubMed] [Google Scholar]
- 10. Cheng HP, Walker GC. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180: 5183–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. D'Haeze W, et al. 1998. Roles for azorhizobial Nod factors and surface polysaccharides in intercellular invasion and nodule penetration, respectively. Mol. Plant Microbe Interact. 11: 999–1008 [Google Scholar]
- 12. D'Haeze W, Glushka J, De Rycke R, Holsters M, Carlson RW. 2004. Structural characterization of extracellular polysaccharides of Azorhizobium caulinodans and importance for nodule initiation on Sesbania rostrata. Mol. Microbiol. 52: 485–500 [DOI] [PubMed] [Google Scholar]
- 13. Dilts JA, Quackenbush RL. 1986. A mutation in the R body-coding sequence destroys expression of the killer trait in P. tetraurelia. Science 232: 641–643 [DOI] [PubMed] [Google Scholar]
- 14. Doherty D, Leigh JA, Glazebrook J, Walker GC. 1988. Rhizobium meliloti mutants that overproduce the R. meliloti acidic calcofluor-binding exopolysaccharide. J. Bacteriol. 170: 4249–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dreyfus BL, Elmerich C, Dommergues Y. 1983. Free-living Rhizobium strain able to grow on N2 as the sole nitrogen source. Appl. Environ. Microbiol. 45: 711–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dreyfus B, Garcia JL, Gillis M. 1988. Characterization of Azorhizobium caulinodans gen. nov., sp. nov., a stem-nodulating nitrogen-fixing bacterium isolated from Sesbania rostrata. Int. J. Syst. Bacteriol. 38: 89–98 [Google Scholar]
- 17. Dreyfus B, Dommergues Y. 1981. Nitrogen-fixing nodules induced by Rhizobium on the stem of the tropical legume Sesbania rostrata. FEMS Microbiol. Lett. 10: 313–317 [Google Scholar]
- 18. Edwards A, et al. 2009. The cin and rai quorum-sensing regulatory systems in Rhizobium leguminosarum are coordinated by ExpR and CinS, a small regulatory protein coexpressed with CinI. J. Bacteriol. 191: 3059–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Finan TM, et al. 1985. Symbiotic mutants of Rhizobium meliloti that uncouple plant from bacterial differentiation. Cell 40: 869–877 [DOI] [PubMed] [Google Scholar]
- 20. Fraysse N, Couderc F, Poinsot V. 2003. Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur. J. Biochem. 270: 1365–1380 [DOI] [PubMed] [Google Scholar]
- 21. Frederix M, Edwards A, McAnulla C, Downie JA. 2011. Co-ordination of quorum-sensing regulation in Rhizobium leguminosarum by induction of an anti-repressor. Mol. Microbiol. 81: 994–1007 [DOI] [PubMed] [Google Scholar]
- 22. Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM. 2006. Investigations of Rhizobium biofilm formation. FEMS Microbiol. Ecol. 56: 195–206 [DOI] [PubMed] [Google Scholar]
- 23. Gao M, D'Haeze W, De Rycke R, Wolucka B, Holsters M. 2001. Knockout of an azorhizobial dTDP-l-rhamnose synthase affects lipopolysaccharide and extracellular polysaccharide production and disables symbiosis with Sesbania rostrata. Mol. Plant Microbe Interact. 14: 857–866 [DOI] [PubMed] [Google Scholar]
- 24. Glazebrook J, Walker GC. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56: 661–672 [DOI] [PubMed] [Google Scholar]
- 25. Glenn SA, Gurich N, Feeney MA, González JE. 2007. The ExpR/Sin quorum-sensing system controls succinoglycan production in Sinorhizobium meliloti. J. Bacteriol. 189: 7077–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goethals K, Leyman B, Van Den Eede G, Van Montagu M, Holsters M. 1994. An Azorhizobium caulinodans ORS571 locus involved in lipopolysaccharide production and nodule formation on Sesbania rostrata stems and roots. J. Bacteriol. 176: 92–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. González JEJ, York GMG, Walker GCG. 1996. Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function. Gene 179: 141–146 [DOI] [PubMed] [Google Scholar]
- 28. Goormachtig S, Capoen W, James EK, Holsters M. 2004. Switch from intracellular to intercellular invasion during water stress-tolerant legume nodulation. Proc. Natl. Acad. Sci. U. S. A. 101: 6303–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gottesman S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30: 465–506 [DOI] [PubMed] [Google Scholar]
- 30. Heruth DP, Pond FR, Dilts JA, Quackenbush RL. 1994. Characterization of genetic determinants for R body synthesis and assembly in Caedibacter taeniospiralis 47 and 116. J. Bacteriol. 176: 3559–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoang H, Becker A, Gonzalez J. 2004. The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J. Bacteriol. 186: 5460–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horton RM, Cai ZL, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8: 528–535 [PubMed] [Google Scholar]
- 33. Iki T, Aono T, Oyaizu H. 2007. Evidence for functional differentiation of duplicated nifH genes in Azorhizobium caulinodans. FEMS Microbiol. Lett. 274: 173–179 [DOI] [PubMed] [Google Scholar]
- 34. Janska H. 2005. ATP-dependent proteases in plant mitochondria: what do we know about them today? Physiol. Plant 123: 399–405 [Google Scholar]
- 35. Jeblick J, Kusch J. 2005. Sequence, transcription activity, and evolutionary origin of the R-body coding plasmid pKAP298 from the intracellular parasitic bacterium Caedibacter taeniospiralis. J. Mol. Evol. 60: 164–173 [DOI] [PubMed] [Google Scholar]
- 36. Lan L, Deng X, Xiao Y, Zhou J-M, Tang X. 2007. Mutation of Lon protease differentially affects the expression of Pseudomonas syringae type III secretion system genes in rich and minimal media and reduces pathogenicity. Mol. Plant Microbe Interact. 20: 682–696 [DOI] [PubMed] [Google Scholar]
- 37. Laus M, van Brussel A, Kijne J. 2005. Role of cellulose fibrils and exopolysaccharides of Rhizobium leguminosarum in attachment to and infection of Vicia sativa root hairs. Mol. Plant Microbe Interact. 18: 533–538 [DOI] [PubMed] [Google Scholar]
- 38. Lee K-B, et al. 2008. The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9: 271 doi:10.1186/1471-2164-9-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leigh JA, Signer ER, Walker GC. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. U. S. A. 82: 6231–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Losada LC, Hutcheson SW. 2005. Type III secretion chaperones of Pseudomonas syringae protect effectors from Lon-associated degradation. Mol. Microbiol. 55: 941–953 [DOI] [PubMed] [Google Scholar]
- 41. Mathis R, et al. 2005. Lipopolysaccharides as a communication signal for progression of legume endosymbiosis. Proc. Natl. Acad. Sci. U. S. A. 102: 2655–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maupin-Furlow JA, et al. 2005. Archaeal proteasomes and other regulatory proteases. Curr. Opin. Microbiol. 8: 720–728 [DOI] [PubMed] [Google Scholar]
- 43. Neuwald AF, Aravind L, Spouge JL, Koonin EV. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9: 27–43 [PubMed] [Google Scholar]
- 44. Niehaus K, Kapp D, Pühler A. 1993. Plant defence and delayed infection of alfalfa pseudonodules induced by an exopolysaccharide (EPS I)-deficient Rhizobium meliloti mutant. Planta 190: 415–425 [Google Scholar]
- 45. Ogura T, Wilkinson AJ. 2001. AAA+ superfamily ATPases: common structure—diverse function. Genes Cells 6: 575–597 [DOI] [PubMed] [Google Scholar]
- 46. Ortiz-Martín I, Thwaites R, Mansfield JW, Beuzón CR. 2010. Negative regulation of the Hrp type III secretion system in Pseudomonas syringae pv. phaseolicola. Mol. Plant Microbe Interact. 23: 682–701 [DOI] [PubMed] [Google Scholar]
- 47. Pond F, Gibson I, Lalucat J, Quackenbush R. 1989. R-body-producing bacteria. Microbiol. Rev. 53: 25–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Preer JR, Preer LB, Jurand A. 1974. Kappa and other endosymbionts in Paramecium aurelia. Bacteriol. Rev. 38: 113–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quackenbush RL, Burbach JA. 1983. Cloning and expression of DNA sequences associated with the killer trait of Paramecium tetraurelia stock 47. Proc. Natl. Acad. Sci. U. S. A. 80: 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 51. Sarria R, Lyznik A, Vallejos CE, Mackenzie SA. 1998. A cytoplasmic male sterility-associated mitochondrial peptide in common bean is post-translationally regulated. Plant Cell 10: 1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schäfer A, et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pk18 and pk19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145: 69–73 [DOI] [PubMed] [Google Scholar]
- 53. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1: 784–791 [Google Scholar]
- 54. Sohn JH, et al. 2004. Kordia algicida gen. nov., sp. nov., an algicidal bacterium isolated from red tide. Int. J. Syst. Evol. Microbiol. 54: 675–680 [DOI] [PubMed] [Google Scholar]
- 55. Su S, Stephens BB, Alexandre G, Farrand SK. 2006. Lon protease of the alpha-proteobacterium Agrobacterium tumefaciens is required for normal growth, cellular morphology and full virulence. Microbiology 152: 1197–1207 [DOI] [PubMed] [Google Scholar]
- 56. Summers ML, Botero LM, Busse SC, McDermott TR. 2000. The Sinorhizobium meliloti Lon protease is involved in regulating exopolysaccharide synthesis and is required for nodulation of alfalfa. J. Bacteriol. 182: 2551–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Suzuki S, et al. 2007. Rhizobial factors required for stem nodule maturation and maintenance in Sesbania rostrata-Azorhizobium caulinodans ORS571 symbiosis. Appl. Environ. Microbiol. 73: 6650–6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Swamy KH, Goldberg AL. 1981. E. coli contains eight soluble proteolytic activities, one being ATP dependent. Nature 292: 652–654 [DOI] [PubMed] [Google Scholar]
- 59. Takaya A, Kubota Y, Isogai E, Yamamoto T. 2005. Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to downregulation of Salmonella pathogenicity island 1 gene expression. Mol. Microbiol. 55: 839–852 [DOI] [PubMed] [Google Scholar]
- 60. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tsien HC, Dreyfus BL, Schmidt EL. 1983. Initial stages in the morphogenesis of nitrogen-fixing stem nodules of Sesbania rostrata. J. Bacteriol. 156: 888–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tsilibaris V, Maenhaut-Michel G, Van Melderen L. 2006. Biological roles of the Lon ATP-dependent protease. Res. Microbiol. 157: 701–713 [DOI] [PubMed] [Google Scholar]
- 63. Tsukada S, et al. 2009. Comparative genome-wide transcriptional profiling of Azorhizobium caulinodans ORS571 grown under free-living and symbiotic conditions. Appl. Environ. Microbiol. 75: 5037–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Twining SS. 1984. Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal. Biochem. 143: 30–34 [DOI] [PubMed] [Google Scholar]
- 65. van Dijl J, et al. 1998. The ATPase and protease domains of yeast mitochondrial Lon: roles in proteolysis and respiration-dependent growth. Proc. Natl. Acad. Sci. U. S. A. 95: 10584–10589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Van Dyck L, Langer T. 1999. ATP-dependent proteases controlling mitochondrial function in the yeast Saccharomyces cerevisiae. Cell. Mol. Life Sci. 56: 825–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Van Dyck L, Pearce D, Sherman F. 1994. PIM1 encodes a mitochondrial ATP-dependent protease that is required for mitochondrial function in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 269: 238–242 [PubMed] [Google Scholar]
- 68. Wang N, Gottesman S, Willingham MC, Gottesman MM, Maurizi MR. 1993. A human mitochondrial ATP-dependent protease that is highly homologous to bacterial Lon protease. Proc. Natl. Acad. Sci. U. S. A. 90: 11247–11251 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.