Abstract
Viruses of the Bacteria and Archaea play important roles in microbial evolution and ecology, and yet viral dynamics in natural systems remain poorly understood. Here, we created de novo assemblies from 6.4 Gbp of metagenomic sequence from eight community viral concentrate samples, collected from 12 h to 3 years apart from hypersaline Lake Tyrrell (LT), Victoria, Australia. Through extensive manual assembly curation, we reconstructed 7 complete and 28 partial novel genomes of viruses and virus-like entities (VLEs, which could be viruses or plasmids). We tracked these 35 populations across the eight samples and found that they are generally stable on the timescale of days and transient on the timescale of years, with some exceptions. Cross-detection of the 35 LT populations in three previously described haloviral metagenomes was limited to a few genes, and most previously sequenced haloviruses were not detected in our samples, though 3 were detected upon reducing our detection threshold from 90% to 75% nucleotide identity. Similar results were obtained when we applied our methods to haloviral metagenomic data previously reported from San Diego, CA: 10 contigs that we assembled from that system exhibited a variety of detection patterns on a timescale of weeks to 1 month but were generally not detected in LT. Our results suggest that most haloviral populations have a limited or, possibly, a temporally variable global distribution. This study provides high-resolution insight into viral biogeography and dynamics and it places “snapshot” viral metagenomes, collected at a single time and location, in context.
INTRODUCTION
As the most abundant and least well characterized biological entities on Earth, viruses have been described as the most significant untapped reservoir of biodiversity (66). Viruses contribute directly to biogeochemical cycles through cell lysis, and they have the potential to bring about catastrophic shifts in community structure over short timescales. In addition to their role as predators, some viruses can provide an auxiliary gene pool that may increase the fitness of their hosts (29, 64, 77). Despite a growing appreciation for the ecological role of viruses and an increase in viral genomic information in public databases, little is known about viral population dynamics in natural systems.
While a number of previous studies have investigated viral stability and persistence (11, 54, 60, 63, 71, 72, 78, 80), few metagenomic studies have evaluated viral population stability. In part, this is because low sampling depth has prevented tracking of genes or genomes and, also, because many metagenomic studies are “snapshots” of the community at a single time and location (52). In a metagenomic study from a San Diego (SD) halovirome, the use of tBLASTx similarity to known viruses suggested that the most abundant haloviral taxa were conserved across space and time, while modeling suggested that haloviral genotypic variants changed on the timescale of days (49). However, because most viral sequences do not have representatives in public databases, most viral populations cannot be identified in metagenomic data, nor can they be amplified in survey studies. In addition, well-documented high rates of horizontal gene transfer in viral populations (21) mean that tracking individual viral genes (that could be present in multiple, unrelated populations) could be misleading. For these reasons, genome-level resolution is important for a comprehensive understanding of viral population dynamics in natural systems.
Consistent with Santos et al. (58), we use the term “halovirus” to describe viruses found in hypersaline systems, including bacterial, archaeal, and (potentially) eukaryotic viruses. Haloviral isolates and communities have been the subject of a number of previous studies, which have been extensively reviewed (15, 30, 45, 46, 58). Outnumbering cells 10- to 100-fold (46), virus counts in hypersaline waters report at least 107 viruses per ml (15) and up to 2 × 109 per ml in crystallizer ponds (20). Several haloviral morphologies have been observed through transmission electron microscopy (TEM) analysis, including spindle shaped (the most abundant form), spherical, icosahedral, filamentous, and head-tail (41, 61). Recently, metagenomic techniques have been applied to the study of viral communities (17, 27), and in hypersaline systems, such studies have suggested that viral communities are relatively diverse (14, 49, 57, 61) and show some global conservation (57, 61). However, given the poor representation of viruses in public databases (52) and the lack of a universal marker gene for viruses, it is difficult to track viral populations across samples.
As in many previously studied hypersaline systems (12, 34, 39, 57), our study site, Lake Tyrrell (LT), Victoria, Australia, is dominated by halophilic Archaea, including Haloquadratum walsbyi (40; S. Podell, J. A. Ugalde, P. Narasingarao, J. F. Banfield, K. B. Heidelberg, and E. E. Allen, unpublished). Ultrasmall archaea (Nanohaloarchaea) represent 10 to 25% of the archaeal community, and halophilic Bacteria, such as Salinibacter ruber, represent approximately 20% of the total community (40). A small number of microbial eukaryotes are present, with populations dominated by the predatory flagellate Colpodella edax and the green alga Dunaliella salina (J. Holm, N. Eisenkolb, J. B. Emerson, K. Andrade, W. C. Nelson, and K. B. Heidelberg, unpublished). Here, we sought to characterize viral populations in the Lake Tyrrell system and place them in the context of other hypersaline systems. We tested the postulate that deep metagenomic sequencing would allow for extensive genomic reconstruction of the dominant viral populations in this moderately complex ecosystem, and we sought to determine whether tracking complete and near-complete genomes could provide insight into haloviral biogeography and population dynamics.
MATERIALS AND METHODS
Sample site.
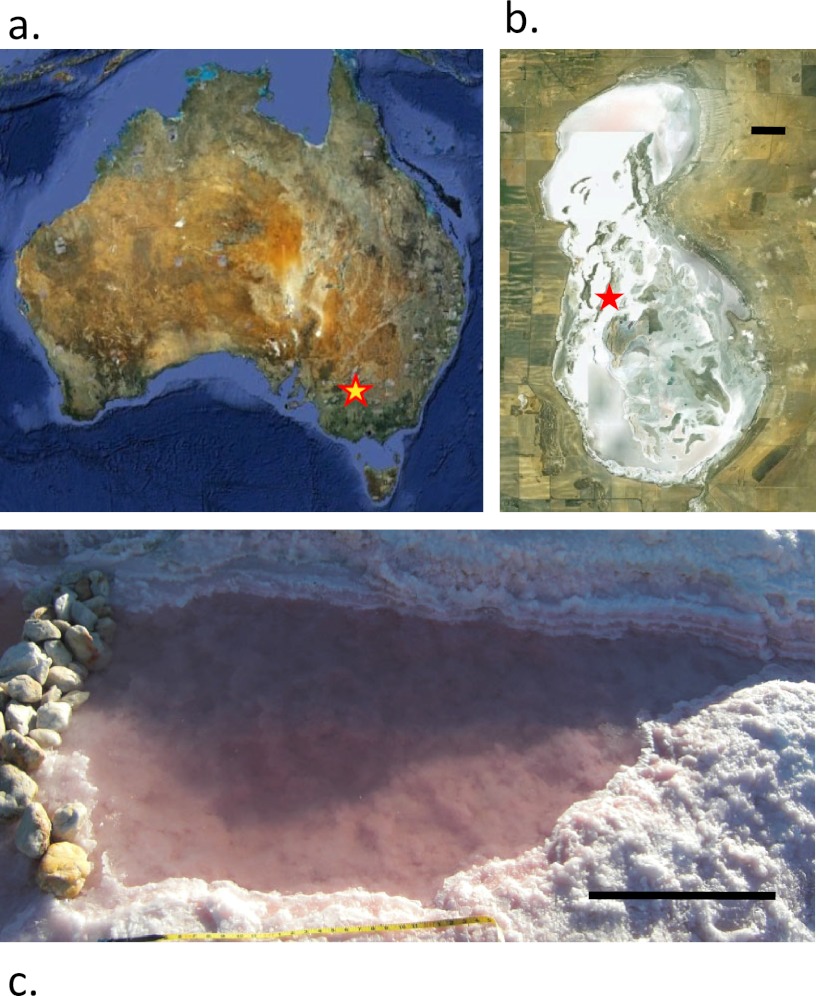
Samples were collected from Lake Tyrrell (LT), a hypersaline, thalassohaline lake in northwestern Victoria, Australia (Fig. 1), located 380 km east of Adelaide (35.309°S, 142.795°E). It has a surface area of ∼160 km2 and is the largest saline groundwater discharge lake in the Murray Basin. The system is hydrologically, geologically, and biogeochemically well characterized (31, 76). In winter, the lake contains ∼50 cm of water with an average salt content of >250 g · liter−1. In summer months (when sampling occurred for this study), the water evaporates, leaving an approximately 7-cm-thick halite crust and residual brines with salt concentrations generally >330 g · liter−1.
Fig 1.
Field site. (a) Star indicates location of Lake Tyrrell in Victoria, Australia. (Image reproduced from Google Earth [copyright 2010], copyright 2011 Cnes/Spot Image.) (b) Satellite image of Lake Tyrrell. Star indicates sampling location within the lake. Scale bar, 0.5 km. (Image reproduced from Google Earth [copyright 2010], copyright 2012 Cnes/Spot Image.) (c) Photograph of sampling site B in 2009. Scale bar, 0.5 m.
Sample collection.
Between 2007 and 2010, eight 10-liter surface water samples (0.3 m depth) were collected during the Austral summer from two pools, A and B, ∼300 m apart in Lake Tyrrell (Table 1). Pool A is ∼6 m in diameter and ∼2 m deep, but during the collection of sample 2007At1, pool A was connected to and surrounded by a shallow (∼10 cm) but expansive (∼100 m) body of water due to recent rainfall, which evaporated and/or was removed by wind prior to the collection of sample 2007At2. In 2009 and 2010, small pools at site B were isolated through artificial damming (Fig. 1c). In 2009, the site B sampling pool was ∼1.5 m in length and ∼20 cm deep; in 2010, the site B pool was ∼6 m in diameter and ∼0.5 m deep. Sites A and B are isolated in the summer, but the lake fills in the winter, resulting in annual mixing between the sites. Sample names include the year and site and, when part of a multiday time series, the time point (e.g., t1, t2, etc.).
Table 1.
Description of samples
| Sample | Date | Time | Site | TDS (wt%)a | Temp (°C) | pH | Sequencing method | Total sequencing (Mb) | Readsb | No. >10 kbc |
|---|---|---|---|---|---|---|---|---|---|---|
| 2007At1 | Jan. 23, 2007 | 15:00 | A | 31 | 22 | 7.23 | Illumina PE | 356 | 2,436,330 | 0 |
| 2007At2 | Jan. 25, 2007 | 15:00 | A | 31 | 28 | 7.09 | Illumina PE | 845 | 7,330,099 | 4 |
| 2009Bd | Jan. 5, 2009 | 7:21 | B | 24 | 18 | 6.86 | Illumina PE | 2,162 | 19,567,468 | 6 |
| 12:37 | 26 | 30 | 7.13 | |||||||
| 18:00 | 27 | 36 | 7.02 | |||||||
| 2010Bt1 | Jan. 7, 2010 | 7:45 | B | 32 | 20 | 7.23 | 454-Ti | 248 | 2,373,021 | 2 |
| 2010Bt2 | Jan. 7, 2010 | 20:00 | B | 36 | 32 | 7.25 | Illumina PE | 425 | 3,312,787 | 0 |
| 2010Bt3 | Jan. 8, 2010 | 8:00 | B | 34 | 21 | 7.2 | 454-Ti | 239 | 2,243,916 | 0 |
| 2010Bt4 | Jan. 10, 2010 | 0:36 | B | 32 | 33 | 7.16 | Illumina PE | 1,039 | 9,610,233 | 2 |
| 2010A | Jan. 10, 2010 | 12:50 | A | 35 | 37 | 7.05 | Illumina PE | 1,103 | 9,268,384 | 14 |
Total dissolved solids, weight percent.
Number of trimmed reads or 454 fragments included in the fragment recruitment analysis.
Number of genome fragments >10 kb long.
Sample 2009B is a pool of three samples, collected throughout a single day, pooled after DNA extraction.
Water samples were passed through a 20-μm Nytex prefilter and then sequentially filtered through 142-mm-diameter polyethersulfone membrane filters (Pall Corporation, NY) of decreasing porosities (3 μm, 0.8 μm, and 0.1 μm), using a peristaltic pump. We acknowledge that some viruses could be removed through 0.1-μm filtration, though to the best of our knowledge, all known haloviruses have capsids smaller than 0.1 μm (15). After passing through the 0.1-μm filter, filtrates were concentrated to 200 ml with a Pellicon II tangential flow filtration (TFF) device (Millipore, MA) fitted with a 30-kDa membrane. Molecular biology-grade glycerol was added to a final concentration of 10% by volume. Samples were stored on dry ice for a maximum of 6 days, followed by −80°C freezer storage for up to 3 years.
Epifluorescence microscopy.
Prior to DNA extraction, we confirmed successful removal of cells from viral concentrates through epifluorescence microscopy (65) on two samples (2007At1 and 2007At2), but unfortunately, the Anodisc 0.02-μm, 25-mm Al2O3 filters (Whatman, Inc., Piscataway, NJ) required for this analysis were not being produced when the remaining six samples were processed.
DNA extraction.
A detailed DNA extraction protocol is presented in the supplemental material. Briefly, after passing through the 0.1-μm filter, each TFF-concentrated sample was refiltered through a 0.2-μm filter and ultracentrifuged for 3 h at 12°C and 32,000 rpm. Pellets were resuspended in 250 μl of 1× TE (Tris-EDTA), followed by phenol-chloroform extraction and ethanol precipitation. The MoBio UltraClean 15 DNA purification kit (Carlsbad, CA) was used to purify the DNA according to the manufacturer's instructions. DNA amplification was not necessary. 16S rRNA genes were PCR amplified with universal primers for Bacteria (27F and 1492R) and Archaea (522F and 1354R) for all samples as a contamination check. Samples with high-quality DNA and no 16S rRNA gene amplification were sent to the J. Craig Venter Institute (JCVI, Rockville, MD) for library construction and sequencing. These methods should allow for sequencing of both single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) viruses, though without multiple-displacement amplification, ssDNA viruses are not likely to be enriched, as reported in some previous studies (e.g., see reference 25).
Library construction and sequencing.
For the two 454-Titanium (454-Ti), single-read pyrosequenced samples (2010Bt1 and 2010Bt3), library construction, emulsion PCR (emPCR), enrichment, and 454 sequencing were performed by the JCVI according to the standard operating procedures of 454 Life Sciences (Bradford, CT), with a few modifications. Specifically, quantitative PCR (qPCR) was used to determine the number of molecules needed for emPCR. In addition, automation (BioMek FX; Beckman Coulter, Brea, CA) was used to break the emulsions after emPCR, and butanol was added to enable easier sample handling during the breaking process. The REM (Robotic Enrichment Module) from Roche (Basel, Switzerland) was used to automate the bead enrichment process.
The six Illumina paired-end samples (2007At1, 2007At2, 2009B, 2010Bt2, 2010Bt4, and 2010A) underwent library construction and sequencing at the JCVI according to Illumina's (San Diego, CA) standard operating procedures, with a few modifications. Specifically, DNA was sheared using the Covaris (Woburn, MA) S2 or E210 systems, and all cleanup steps incorporated Agencourt AMPure XP beads (Beckman Coulter). The libraries were quantitated and quality controlled using the Agilent high-sensitivity DNA kit (Santa Clara, CA). Cluster generation and paired-end sequencing were completed according to Illumina's standard protocol. One hundred cycles were performed, resulting in 100-bp reads.
Assembly.
Each LT sample was independently assembled. First, reads were trimmed for quality, either with an in-house script that removes low-quality bases from Illumina reads or with sff_extract (454 Life Sciences) for 454 reads. Each of the two LT 454-Ti samples was assembled via Newbler with default parameters (33), and the same parameters were used to coassemble reads from four hypersaline virome samples from previous studies of a saltern near San Diego (SD), CA, reported by another group (14, 49). For the six LT Illumina samples, a variety of assembly algorithms and parameters were attempted and optimized, either to generate the best assembly for a given sample (i.e., the most large contigs with relatively uniform within-contig coverage; all Illumina samples, except for 2009B) or to generate the best assembly for a given genome (i.e., the greatest number of large contigs at the expected coverage depth for a given population; applied to sample 2009B in order to improve recovery of LTV2; coincidentally, this was also the best assembly for sample 2009B). The ABySS algorithm (62) with kmer size 45 and a minimum of 10 read pairs required to join contigs was used for samples 2007At1, 2007At2, 2010Bt2, 2010Bt4, and 2010A. The Velvet algorithm (81) with kmer size 95 was used for sample 2009B. Autoassembly correctness was evaluated manually, using Consed (19) and Tablet (35).
We applied previously described manual metagenomic assembly curation methods (e.g., see references 1, 2, 6, 38, 74) to our Illumina data. Details of manual assembly curation are in the supplemental material, but briefly, we took advantage of paired-read information and sequencing overlaps not utilized by the assembly algorithm, and we ensured that there was relatively uniform coverage throughout each genome or genome fragment.
Various degrees of manual curation were attempted for each sample, so the number of genomes or genome fragments that assembled from a given sample is not necessarily indicative of sample complexity. We used BLASTn to screen all 33 noncircularized contigs of >10 kb for similarity to each other and to the 7 complete genomes, resulting in 28 unique genome fragments (10,148 to 40,896 bp). The genome fragments share regions up to 595 bp in length (78% nucleic acid identity), and a few are technically scaffolds, connected by N′s generated by the assembly algorithm.
Annotation.
Genes were predicted from the 7 composite genome sequences by using Prodigal (22). Annotation of genes was performed using a series of sequence similarity searches to known sequence databases. We used BLASTp to compare predicted protein sequences to the KEGG and UniRef90 databases, giving priority to reciprocal best BLAST hits, and then we investigated protein motifs using InterProScan (48). The 28 genome fragments were not annotated, but we did conduct BLAST searches to verify that their sequences are consistent with viruses or virus-like entities (VLEs) (data not shown).
Fragment recruitment.
Accounting for different biases in the two sequencing technologies was beyond the scope of this study, but we did make the read lengths comparable. To approximate Illumina-sized reads, 454 reads were cut in silico with an in-house script, which generates consecutive 100-bp fragments from the start of each 454 read. The last sequence increment was retained if it was ≥72 bp. For simplicity, these read fragments are called reads throughout the text. Using gsMapper (Newbler [33]) with a minimum overlap length of 40 and a minimum overlap identity of 90%, unique reads from each of the eight samples were used as queries against the 7 genomes or the 28 genome fragments as references (duplicate reads were removed, though nearly identical mapping counts were obtained when duplicate reads were included). Similarly, reads from three previously described haloviral metagenomes (14, 49, 57, 61) were used as queries against the 35 LT genomes and genome fragments, and 10 previously described haloviral genome sequences (His1 and His2 [10]; HF1 [67]; HF2 [68]; HRPV-1 [44]; HHPV-1 [51]; SH1 [8]; BJ1 [42]; ϕCh1 [26]; and EHP-1 [55]) were used as references for mapping reads from each of our eight samples. The same fragment recruitment parameters were applied to data from the previously published San Diego halovirome (14, 49), using two sets of references (first, the 10 largest contigs assembled directly from the San Diego samples, and second, the 10 previously sequenced haloviruses described above). Ace files were analyzed in Consed (19) to determine the number of reads that mapped to each genome or genome fragment by sample.
Time series analysis was applied to the 35 complete and near-complete genomes from LT and to the 10 contigs from San Diego (SD). We counted the number of reads that mapped to each genome or genome fragment in each sample (see mapping counts in Tables S1 and S2 in the supplemental material) and then normalized by genome size or contig length and the total number of reads in the sample (normalization details are in the supplemental material). For the 7 complete LT genomes and 10 contigs from SD, at least 1× coverage across at least 30% of the genome was required for detection. Manual determination of percent coverage was not practical for the 28 LT genome fragments, so we set their detection limit as equal to or greater than the smallest normalized mapping count for any genome fragment in the sample from which it assembled (a proxy for the smallest number of reads that could generate a genome fragment). This could lead to underestimates of detection of low-abundance populations.
Bioinformatic assessments of library contents.
After assembly, Prodigal-predicted genes (22) from all contigs larger than 500 bp from all viral concentrate libraries (30,525 nonredundant genes, clustered at 95% nucleotide identity) were run through InterProScan (48) to identify domains and hidden Markov models (HMMs), through BLASTp against NCBI's nonredundant protein database to identify the best BLAST hits, and through BLASTn against the SILVA 16S rRNA gene database (47) to identify potential cellular contamination. We also used fragment recruitment with the same detection cutoffs as described above (at least 1× coverage across at least 30% of the genome) to search the LT unassembled reads for 19 previously sequenced halophilic plasmids available from the ACLAME database (28). We compared their presence in the eight LT viral concentrates to their presence in libraries from three LT 0.8-μm filters sequenced from the same samples (unpublished data collected prior to TFF concentration from samples 2007At1, 2009B, and 2010Bt3).
Nucleotide sequence accession number.
Sequencing reads from each of the eight samples and assembled nucleotide sequences for the 7 composite genomes and 28 genome fragments have been submitted to GenBank (BioProject accession number PRJNA81851).
RESULTS
Through extensive manual curation of the LT Illumina assemblies, we reconstructed complete composite genomes from six LT viruses (LTV1, LTV2, and LTV4 to -7) and one virus-like entity (LTVLE3). By virus-like entity (VLE), we mean virus or plasmid (see below). Because in silico circularization is an added assurance of assembly correctness, each genome was considered closed when it circularized. However, from our data set, only circular or circularly permuted viruses or plasmids should circularize. In order to observe trends for a larger number of populations and genome types, we considered all unique contigs >10 kb (the 7 circular genomes and 28 additional genome fragments) to be population representatives. Because these genomes and genome fragments were generated from metagenomic data, they are composite sequences that represent heterogeneous, nonclonal populations.
It should be noted that our DNA extraction protocol could result in the inclusion of extracellular plasmids and other free DNA in our metagenomic libraries because we did not include a DNase treatment prior to virion lysis (a DNase treatment was attempted but resulted in complete degradation of all DNA; see the supplemental material). Not surprisingly, only ∼15% of the 30,525 viral concentrate genes from all samples had functional predictions, and of those, nearly all would be consistent with but not necessarily exclusive to viruses. We searched the InterProScan annotations for genes that could be linked relatively (though not entirely) unambiguously to plasmids (i.e., conjugative transfer genes) or viruses (i.e., structural proteins, terminases, and cell surface recognition/degradation proteins). We found one conjugative transfer gene and 566 virus-specific genes. Of 49 ParB domain-containing proteins, which are known to be present in both viruses and plasmids, 25 had the best BLASTp hits to predicted proteins from a Spanish halovirome (57). One gene of the 30,525 had significant BLASTn similarity to a 16S rRNA gene from a halophilic archaeon. Of the 19 plasmids from the ACLAME database, 12 were detected in LT. All 12 were detected on the 0.8-μm filters. Two were also detected in the viral concentrates (one in all eight samples and a second only in sample 2010Bt4). The plasmid detected in all eight viral concentrate samples was 10 to 115 times (average 55 times) more abundant on the 0.8-μm filters, and several ∼5- to 10-kb regions of the plasmid were not detected in the viral concentrates, though nearly the entire sequence was present on the 0.8-μm filters. The plasmid detected in sample 2010Bt4 was nine times more abundant on the 0.8-μm filter from the same time series (sample 2010Bt3; no 0.8-μm filter DNA was sequenced from exactly the same sample). It is certainly possible that some of the regions detected within these plasmids could be contained on unknown viruses, but conservatively, we assume that they are indicative of a small amount of plasmid contamination.
We found viral structural genes in LTV1, -2, and -4 to -7, so we call them viruses. LTVLE3 had annotation that would be consistent with both viruses and plasmids, so we categorize it as a VLE. The 28 genome fragments often did not represent genome portions large enough to contain genes specific to viruses or plasmids, but given that the libraries appear to be mostly viral, we suspect that they are mostly or exclusively viral as well. Still, to ensure correctness, we refer to all LT populations together as viruses and VLEs. Because the general trends that we observed across the 35 LT populations were also observed for populations that we can confidently identify as viral (Fig. 2a, b, and d to g), and because similar trends were also observed in a viral metagenomic data set from San Diego, CA (see below), we are confident that our findings accurately represent haloviral population dynamics, regardless of any potential contamination in our libraries.
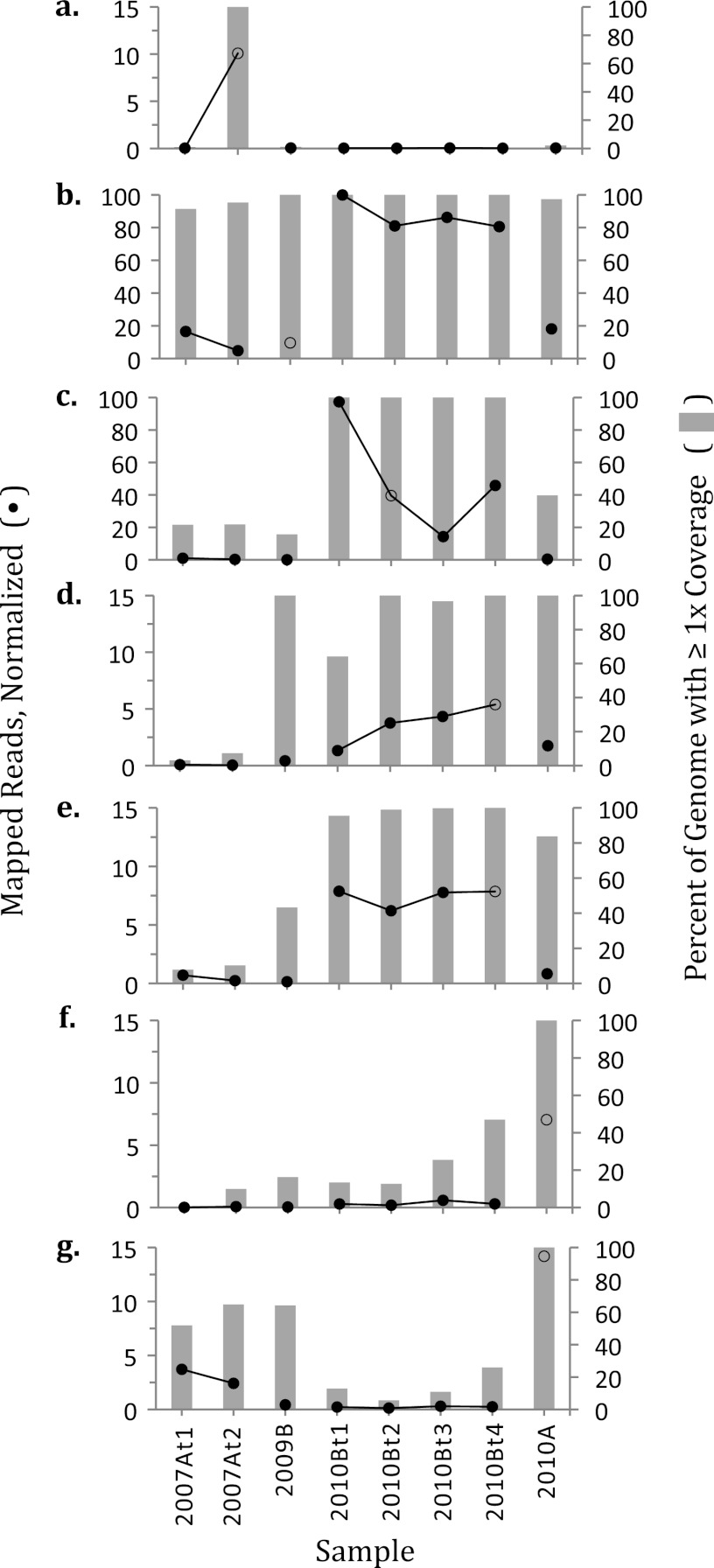
Fig 2.
Presence and abundance of seven LT viruses and VLEs through time. (a) LTV1, (b) LTV2, (c) LTVLE3, (d) LTV4, (e) LTV5, (f) LTV6, (g) LTV7. Samples are along the x axis. The y axis on the right is the percentage of the genome to which at least one read maps, and it corresponds to the gray bar graphs. The y axis on the left corresponds to the black data points and is the normalized number of mapped reads, calculated as follows (for more detail, see the supplemental material): the number of reads or 454 read fragments recruited to a given genome from a given sample was multiplied by normalization factors for (i) the number of reads in the sample, (ii) the genome size, and (iii) scale, with the largest data point equal to 100. Note that there are two left y axis maxima: 15 for LTV1 and LTV4 to -7 and 100 for LTV2 and LTVLE3. Black data points connected by a line are from sequential day scale time series samples. Open black circles correspond to the sample from which a given genome was assembled.
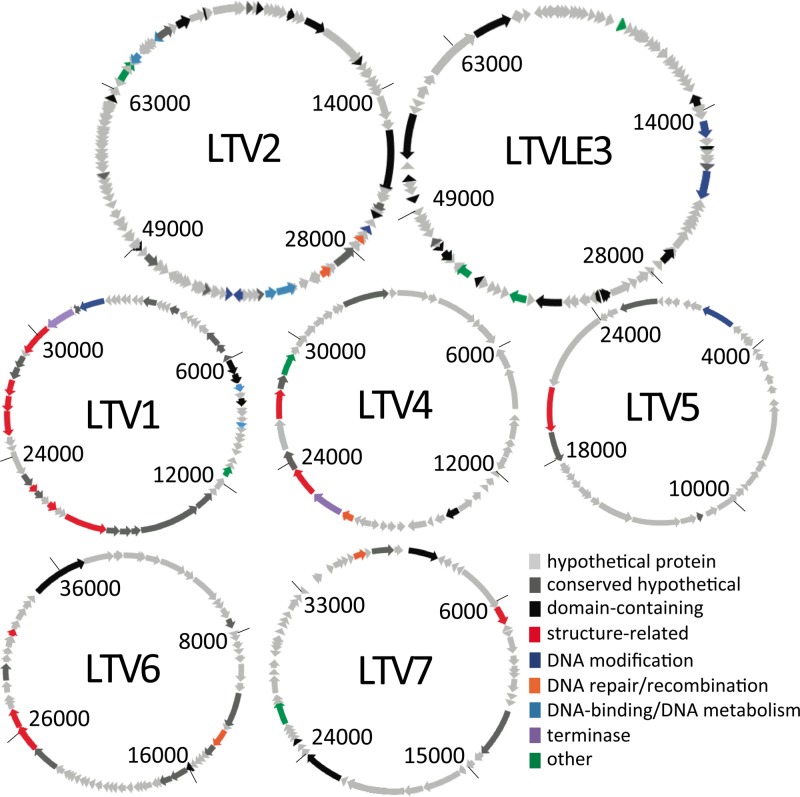
Features of each of the seven composite genomes are in Table 2, annotations are in Table 3, and genome representations are in Fig. 3. In the following sections, abundance descriptions are based on relative representation in our libraries. For example, low abundance indicates presence above the detection limit but representation by a relatively small number of recruited reads.
Table 2.
LT viral and VLE genome summaries
| Genome | Length (bp) | % GC | % coding | Predicted genes | CHPa | Assembly sampleb | Coveragec | Assembly reads (%)d |
|---|---|---|---|---|---|---|---|---|
| LTV1 | 34,489 | 42.4 | 97 | 66 | 11 | 2007At2 | 26.3 | 0.12 |
| LTV2 | 76,716 | 46.7 | 92 | 131 | 4 | 2009B | 66.8 | 0.26 |
| LTVLE3 | 71,341 | 40.3 | 87 | 107 | 2 | 2010Bt2 | 46.7 | 1 |
| LTV4 | 35,497 | 57.6 | 92 | 47 | 2 | 2010Bt4 | 18.4 | 0.068 |
| LTV5 | 26,538 | 45.2 | 94 | 44 | 2 | 2010Bt4 | 26.9 | 0.074 |
| LTV6 | 39,362 | 34.5 | 97 | 66 | 7 | 2010A | 23.2 | 0.099 |
| LTV7 | 38,139 | 43.4 | 91 | 60 | 1 | 2010A | 46.8 | 0.19 |
Conserved hypothetical proteins.
Sample from which the genome was assembled.
Average depth of coverage in the sample from which the genome was assembled.
Percentage of reads from the assembly sample used to assemble the genome.
Table 3.
Genes with predicted functions from complete LT genomes
| Genome | Annotationa | Organism (best BLAST hit) | E value | % coverageb | Startc | Stopc |
|---|---|---|---|---|---|---|
| LTV1 | ERF superfamily protein | 2E-14 | 5934 | 6635 | ||
| Single-stranded DNA-binding protein | Bartonella rochalimae | 3E-33 | 98.5 | 7117 | 7533 | |
| NinB recombinase superfamily protein | 8E-9 | 7832 | 8212 | |||
| DNA-binding protein | Nitratiruptor sp. | 2E-14 | 96 | 9009 | 9305 | |
| Lysozyme | Phaeospirillum molischianum | 5E-25 | 76.2 | 11138 | 11629 | |
| Minor tail protein | Vibrio mimicus | 8E-21 | 44 | 18099 | 20117 | |
| Phage major tail protein | Sphingomonas sp. | 2E-26 | 94 | 20664 | 21095 | |
| Putative phage head-tail adaptor | Aggregatibacter aphrophilus | 2E-10 | 74 | 21909 | 22277 | |
| HK97 family major capsid protein | Xanthobacter autotrophicus | 9E-68 | 74.2 | 24885 | 26081 | |
| Prohead protease | Gemmata obscuriglobus | 1E-35 | 73.4 | 26044 | 26766 | |
| Putative head morphogenesis protein | Gemmata obscuriglobus | 4E-10 | 54 | 26798 | 27547 | |
| HK97 family phage portal protein | Wolbachia sp. | 8E-56 | 70.9 | 28977 | 30566 | |
| Phage terminase, large subunit | Burkholderia sp. | 3E-97 | 98 | 30563 | 31762 | |
| DNA methylase | Hydrogenophaga sp. | 1E-88 | 95.5 | 32314 | 33522 | |
| LTV2 | RuvA domain 2-like superfamily protein | 1E-5 | 1226 | 1513 | ||
| Nucleotide-diphosphosugar-transferase family protein | 4E-8 | 3607 | 4296 | |||
| Concanavalin A-like lectins/glucanases family protein | 5E-21 | 5174 | 7078 | |||
| A2M_N family protein | 1E-7 | 10981 | 11301 | |||
| AlbA-like superfamily protein | 6E-8 | 24852 | 25115 | |||
| DNA methyltransferase | Environmental halophage eHP-11 | 5E-71 | 91 | 25875 | 26288 | |
| Repair endonuclease XPF | Aeropyrum pernix | 2E-17 | 83.3 | 26785 | 27396 | |
| DNA repair and recombination protein RadA | Acidilobus saccharovorans | 1E-49 | 95.5 | 30696 | 31688 | |
| Ribonucleotide reductase, alpha subunit | Candidatus “Nanosalina” | 0 | 98.8 | 34023 | 35708 | |
| Ribonucleotide reductase, beta chain | Candidatus “Nanosalina” | 2E-153 | 99.4 | 35708 | 36706 | |
| Site-specific DNA methyltransferase | Haloquadratum walsbyi | 2E-100 | 94.6 | 38607 | 39443 | |
| DNA modification protein | Paenibacillus larvae | 2E-11 | 75.9 | 39467 | 39952 | |
| YonJ-like protein (putative DNA polymerase) | Halovirus HF2 | 4E-58 | 92.2 | 63887 | 65083 | |
| Phosphodiesterase/nucleotide pyrophosphatase | Halothermothrix orenii | 1E-14 | 78.9 | 65595 | 66563 | |
| Pyrophosphatase | Mycobacterium phage Bongo | 2E-28 | 100 | 68303 | 68617 | |
| Phage portal domain-containing protein | 1E-7 | 70635 | 71861 | |||
| DNA ligase superfamily protein | 6E-8 | 73231 | 73719 | |||
| LTVLE3 | Rhodopsin | Candidatus “Nanosalina” | 3E-17 | 96 | 3806 | 4426 |
| Phage integrase domain-containing protein | 9E-11 | 12797 | 13804 | |||
| DNA methylase | Halovirus phiCh1 | 6E-75 | 89 | 15034 | 16338 | |
| Putative methyltransferase | Environmental halophage eHP-35 | 4E-68 | 57 | 18945 | 21176 | |
| Nucleoside triphosphate hydrolase superfamily protein | 4E-19 | 25637 | 26908 | |||
| Transglutaminase-like family protein | 1E-5 | 32396 | 32980 | |||
| ATP-binding protein | Haloferax volcanii | 2E-99 | 92.4 | 38028 | 39371 | |
| Archaeal cell division control protein 6 | Haloarcula marismortui | 3E-126 | 96.6 | 42727 | 43971 | |
| ParA domain-containing protein | Haloarcula marismortui | 2E-78 | 91.9 | 44539 | 45390 | |
| SpoVT/AbrB family protein | 2E-7 | 45500 | 45799 | |||
| Nucleic acid-binding superfamily protein | 6E-12 | 51642 | 52139 | |||
| DNA primase catalytic core domain-containing protein | 3E-23 | 53315 | 56917 | |||
| LTV4 | MurNAc-LAA superfamily (putative endolysin) | 3E-14 | 14762 | 15325 | ||
| Exonuclease family protein | Marinobacter adhaerens | 2E-18 | 87.6 | 19892 | 20494 | |
| Phage terminase, large subunit | Clostridium botulinum | 1E-23 | 88 | 20615 | 22066 | |
| HK97 family phage portal protein | Ahrensia sp. | 2E-23 | 74.9 | 22183 | 23607 | |
| HK97 family major capsid protein | Pseudomonas putida | 2E-47 | 80.3 | 26203 | 27615 | |
| Putative N-acetylglycosaminyltransferase | Rhodopirellula baltica | 3E-14 | 47.9 | 28333 | 29454 | |
| LTV5 | DNA methylase | Paenibacillus elgii | 1E-69 | 88.6 | 1560 | 2825 |
| Peptidase U35 phage prohead HK97 | Natrialba magadii | 3E-13 | 28 | 19156 | 20850 | |
| LTV6 | RecT family protein | Ruminococcus sp. | 3E-39 | 76.1 | 13164 | 14180 |
| DNA-binding domain protein, excisionase family | 1E-9 | 15951 | 16121 | |||
| Phage portal protein | Bacillus sp. | 3E-79 | 91 | 24995 | 26422 | |
| Phage-related GP7 minor head protein | Deep sea thermophilic phage D6E | 2E-25 | 71.6 | 26419 | 27474 | |
| Phage virion morphogenesis | Roseburia intestinalis | 1E-14 | 90 | 31494 | 31895 | |
| Phage tail tape measure domain-containing protein | 2E-36 | 34232 | 37249 | |||
| LTV7 | Phage portal domain-containing protein | 4E-16 | 732 | 2261 | ||
| Major capsid protein, gp5 superfamily | 7E-6 | 6216 | 7139 | |||
| YonJ-like protein (putative DNA polymerase) | Halovirus HF2 | 8E-39 | 78.6 | 25874 | 27067 | |
| Repair endonuclease XPF | Archaeglobus veneficus | 2E-13 | 90.8 | 36078 | 36731 |
Some domain-containing proteins (e.g., PKD and winged-helix DNA-binding domain proteins) are not included in this list.
Percentage of the predicted protein covered by the BLAST hit.
Start and stop locations (not indicative of gene direction) correspond to locations on the genome in Fig. 3.
Fig 3.
Genome figures. Circular representations of the seven composite genomes, to scale within but not among genomes. Numbers inside each circle indicate positions within the linear contig (origins of replication were not determined). Arrows indicate the positions of genes.
Characteristics and time series analysis of the seven LT genomes.
LTV1 was observed in a single sample, 2007At2 (Fig. 2a), which was collected after the sampling pool at site A had returned to its normal size following a rainfall event prior to the start of the two-sample time series. Despite the change in pool size, no obvious geochemical differences were observed between the two samples (Table 1; also see Table S3 in the supplemental material). LTV2 was present in all eight samples and especially abundant in the 2010 site B 4-day time series, in which it represented 2.2 to 2.7% of the reads (see Fig. S1 in the supplemental material), though it was assembled from sample 2009B, in which it accounts for only 0.3% of the reads. Nearly the entire LTV2 genome was detected in all samples (Fig. 2b). The remaining five complete genomes were detected in more than one sample but not in all samples in the 3-year study period (Fig. 2).
The functional annotation of LTVLE3 would be consistent with plasmids or viruses (Table 3). Of its predicted proteins, the one perhaps most often associated with plasmids is a ParA domain-containing protein, which, notably, has a significant BLASTp hit to a protein in halovirus ϕCh1 (E value 8e-43, 83% coverage). In addition, two of LTVLE3's best BLAST hits are to haloviruses. Interestingly, LTVLE3 contains a rhodopsin with high similarity to the nanohaloarchaeal “xenorhodopsins” (75).
LTV4 was assembled from the smallest fraction of a sample, representing only 0.068% of the reads from sample 2010Bt4. BLAST hits from LTV4 were to a variety of bacteriophages and bacteria, including two conserved hypothetical proteins with hits to Salinibacter ruber (37). LTV4 was present at relatively low abundance in all six samples from 2009 to 2010 but was not detected in the 2007 samples (Fig. 2d), a pattern shared with LTV5 (Fig. 2e). The vast majority of the LTV5 genome is novel, but three consecutive predicted genes had their best hits to Natrialba magadii, a haloalkaliphilic archaeon (24).
LTV6 was the only one of the 7 genomes to show dynamic presence in the 2010 4-day time series, in which it was not detected in 2010Bt1 to -3 but was present at low abundance in sample 2010Bt4. Though below the 30% coverage cutoff required for detection, it may be significant that we observed 12.7 to 25.5% coverage of LTV6 in 2010Bt1 to -3. LTV6 was also relatively abundant in sample 2010A, from which it was assembled (Fig. 2f). LTV7 was detected in at least some samples from each sampling trip (2007, 2009, and 2010) but not all samples in the study (Fig. 2g), a pattern not observed for any of the other LT populations.
Time series analysis of the 28 LT genome fragments.
The 28 LT genome fragments displayed 16 different presence/absence patterns across the eight time points (Table 4). While we describe the LT genomes and genome fragments as 35 populations for simplicity, it is important to consider that multiple genome fragments could represent different regions of the same genome. Nine of the 14 genome fragments assembled from sample 2010A (see Table S2 in the supplemental material) were from populations detected only in that sample, and detection only in 2010A is the most common pattern reported in Table 4. The second-most-common pattern is presence only in sample 2007At2, exhibited by 3 of the 4 genome fragments generated from that sample. Only 2 genome fragments were detected in all eight samples, while 16 were either present in a single sample or present exclusively in a day scale time series. Within each of the day scale time series, the 28 genome fragments were typically consistently detected (4 in the 2007 2-day time series and 9 in the 2010 4-day time series) or not detected (17 in 2007 and 17 in 2010), for a total of 21 and 26 contigs, respectively. However, of the 11 genome fragments detected in the 2007 series, 7 were detected in only one of the two samples.
Table 4.
Detection patterns for the 28 genome fragments
| Detection patterna | No. of fragmentsb |
|---|---|
| 2010A only | 9 |
| 2007At2 only | 3 |
| All samples | 2 |
| All 2010 4-day time series only | 2 |
| Both 2007 only | 1 |
| 2009B only | 1 |
| All 2009 to 2010 only | 1 |
| All 2010 only | 1 |
| 2007At1 and 2009B | 1 |
| 2007At1 and all 2010 4-day time series | 1 |
| 2007At1 and all 2010 | 1 |
| 2007At2 and 2010A | 1 |
| All except 2009B | 1 |
| 2009B and 2010Bt4 | 1 |
| 2009B and 2010A | 1 |
| 2010A and 2010Bt1-3 | 1 |
Sample(s) in which a given genome fragment was detected (some fragments share the same detection pattern).
Number of genome fragments that exhibited the detection pattern.
Detection of previously sequenced haloviruses in LT samples.
Based on fragment recruitment at 90% nucleotide identity, none of 10 previously sequenced haloviruses achieved the 30% coverage cutoff required for detection in any LT sample. However, up to 18% of the archaeal BJ1 virus genome (42) was present at low abundance (up to 0.03% of the reads) in all LT samples, and a small number of reads mapped to localized regions within EHP-1 (55) and ϕCh1 (26) (see Table S4 in the supplemental material). The vast majority of EHP-1 hits were to two ribonucleotide reductase genes that are highly similar at the nucleotide level to the LTV2 ribonucleotide reductases, but interestingly, when reads from sample 2009B were competitively mapped to LTV2 and EHP-1, many reads still mapped to the EHP-1 ribonucleotide reductase genes. We observed essentially no detection of the remaining seven previously sequenced haloviruses in our samples, though mapping of a very small number of reads was observed for some viruses in some samples (data not shown).
Detection of the 35 LT populations in other haloviral metagenomes.
None of the 35 LT virus and virus-like populations was detected through fragment recruitment at 90% nucleotide identity in any of three previously sequenced haloviromes, even when the virome sequences were cut to 72- to 100-bp fragments in silico. In a haloviral metagenomic data set from a saltern near San Diego, CA (14, 49), consisting of 509,673 ∼100-bp 454 reads from purified viral fractions from high-salinity ponds (28 to 30% salt), up to 51 reads mapped to isolated regions within a small number of LT genomes and genome fragments, but less than 5% of any LT genome or genome fragment was covered. For the remaining viromes, a haloviral metagenomic data set from hypersaline Lake Retba, Senegal (993 Sanger reads [61]), and 380 kb of haloviral metagenomic sequence from Santa Pola in southeast Spain (57), the most reads to map to any of the 35 LT populations was three.
Corroboration of our results in a previously reported halovirome.
We acknowledge that the sequencing throughput in previously reported haloviromes is significantly lower than the 6.4 Gb that we report from Lake Tyrrell viral concentrates, which could affect our comparisons with other data sets. However, of the four samples collected from the hypersaline virome near San Diego, CA, described above (48.5 Mb [14, 49]), two report an amount of sequencing within one order of magnitude of our samples, so we expect that trends for the most abundant populations should be observable. Using the same fragment recruitment detection limits as described above (at least 1× coverage at 90% nucleotide identity across at least 30% of the genome), we found that none of the 10 previously sequenced haloviruses was detected in any of the San Diego (SD) samples. The percent genome coverage was even lower than the very small amount of coverage detected in LT.
We also assembled the SD reads, using the default Newbler parameters described in Materials and Methods. To the best of our knowledge, assembly was not attempted in the published work, apart from the assemblies generated through the PHACCS and MaxiΦ programs for modeling alpha and beta diversity, respectively (49). While only one of the resulting contigs achieved our 10-kb cutoff required to represent a population, we characterized detection patterns for the 10 largest contigs (3 to 13 kb) across three of the four SD samples (a fourth SD sample had a very small number of reads; see below), and we also searched for the SD contigs in the LT data through fragment recruitment. The SD contigs were not detected in any of the LT samples, though as many as 1,319 LT reads did map to SD contigs in isolated regions, covering up to 20% of one contig. Across three of the SD samples, collected on 16 November 2005 (sample L), 7 December 2005 (sample N), and 20 December 2005 (sample O), the 10 SD contigs displayed a variety of detection patterns. Six contigs were detected in a single sample (2, 1, and 3 contigs in samples L, N, and O, respectively), two were detected in both L and N, and two were detected in both N and O. No contigs were detected in all three SD samples. In a fourth SD sample, collected on Nov. 28, 2005 (between samples L and N), no SD contigs were detected, but that is most likely due to sampling bias, as that sample contained only 4,645 reads.
Haloviral population dynamics, based on a less stringent detection threshold.
Because no viral or VLE populations were detected in multiple systems in this study, in contrast to results from another study (18), we sought to determine whether different fragment recruitment thresholds would affect our results. We repeated our fragment recruitment at 75% nucleotide identity, keeping all other parameters the same, using three sets of references (the 7 complete LT genomes, the 10 previously sequenced haloviruses, and the 10 largest contigs from SD) and reads from all LT and SD samples separately as queries. No significant differences in detection patterns were observed for any within-system analyses, although slight increases in percent genome coverage (from ∼20% to just above 30%) technically pushed a small number of populations above the detection threshold in a few cases. Still, none of the LT genomes or 10 previously sequenced haloviruses was detected in SD. However, interestingly, two SD contigs were detected in LT but only in one sample (2009B). Similarly, 3 of the 10 previously sequenced haloviruses were detected in LT but only in some samples, as follows: BJ1 (both 2007 samples, 2009B, 2010Bt4, and 2010A; nearly 30% coverage of BJ1 was also observed in the remaining samples), ϕCh1 (2007At2, 2009B, 2010Bt4, and 2010A), and SH1 (2007At2, 2009B, 2010Bt4, and 2010A).
DISCUSSION
Most prior viral metagenomic studies from short-read sequencing data have relied upon unassembled read or short-contig analysis because significant assembly was precluded by high viral diversity and relatively low sequencing throughput. In this study, we have leveraged deep metagenomic sequencing (6.4 Gb) in combination with careful manual assembly curation to reconstruct 28 near-complete and 7 complete composite genomes from viruses and virus-like entities (VLEs) in hypersaline Lake Tyrrell, Victoria, Australia. This has allowed us to uncover previously unrecognized patterns of viral population dynamics on relatively short timescales. We have nearly doubled the number of haloviruses and VLEs with sequenced representatives (15, 18, 55), and the assembly of genomes from as little as 0.068% of the reads in a sample suggests that comprehensive viral genomic reconstruction may be possible in more complex systems through deep metagenomic sequencing. Importantly, we were able to obtain enough DNA for metagenomic sequencing without amplification, avoiding potential biases associated with multiple-displacement amplification (MDA) and presumably achieving more accurate relative abundance estimates. The 35 LT virus and VLE populations showed a range of stabilities across 8 LT viral metagenomic samples collected from 2007 to 2010, from detection at one time in one location (14 populations) to presence at all times and locations studied (3 populations). The results indicate a diverse and dynamic viral consortium within Lake Tyrrell, and the time series results have been corroborated in a halovirome from San Diego, CA, suggesting that viral populations may be highly dynamic on relatively short timescales in hypersaline systems worldwide.
Many of our results are based on the assembly of short-read metagenomic sequencing data. While we have taken extensive precautions to ensure assembly correctness through manual assembly curation (see the supplemental material), particularly for the seven genomes, we acknowledge that some misassemblies are still possible. It is encouraging that the 7 genomes circularized, that they have a relatively uniform depth of coverage in the samples from which they assembled, and that many genes were predicted from these assemblies with database matches that are consistent with viruses, VLEs, and halophiles. The results of our temporal and biogeographical analyses demonstrate a shift in coverage depth from ∼20× to 50× for a given population in the sample from which it was assembled (Table 2) to essentially zero coverage in samples in which that population was not detected (Fig. 2). This means that the observed temporal and biogeographical trends are significant, even if the assemblies are not 100% correct.
Features of the 7 complete composite genomes.
Within the 6 complete viral genomes and a 7th VLE genome, database matches to phages, archaeal viruses, bacteria, and archaea suggest that both phage and archaeal viral genomes were reconstructed. Host range is difficult to determine without isolates, but some inferences can be made. For example, a significant hit to lysozyme, which is used to break down peptidoglycan in bacterial cell walls, along with BLAST hits to bacteria suggest that LTV1 is a bacteriophage (Table 3). As no bacteriophages have been isolated from hypersaline systems, it is not surprising that the bacterial BLAST hits are to a variety of organisms. A preponderance of hits to bacteriophages and bacteria suggests that LTV4 is also likely to be a bacteriophage, possibly infecting Salinibacter ruber, on account of two best BLAST hits to S. ruber. The abundance of archaea in the system (∼80% or more) suggests that some of the LT viruses target archaea. In particular, LTV2 is likely to be an archaeal virus and LTVLE3 is likely to be an archaeal virus or plasmid, based on a number of top hits to archaea.
In addition to host range, gene content from the 7 viral and VLE genomes may provide information about lifestyle and function. Annotations that could be associated with lysogeny include a putative phage integrase in LTVLE3 and a putative excisionase in LTV6. LTV2 encodes ribonucleotide reductases, which are more often associated with lytic than temperate viruses (55). Of the functions that could be identified, the most prevalent is methyltransferase or DNA methylase, found in LTV1, -2, and -5 and LTVLE3. The abundance of methyltransferases suggests that evasion of host restriction modification systems is (or has been) important in the Lake Tyrrell system, though methyltransferases have also been shown to contribute to DNA mismatch repair, gene expression regulation, and replication initiation (9).
Interestingly, LTVLE3 contains a rhodopsin with high similarity to the nanohaloarchaeal “xenorhodopsins,” which cluster with rhodopsins of bacteria from diverse environments and are phylogenetically distinct from bacteriorhodopsins of the class Halobacteria (75). The function of the xenorhodopsins is unknown, as they cluster separately from rhodopsins with known functions (i.e., proton pumps, chloride pumps, and sensory rhodopsins), though it has been suggested that they may be sensory rhodopsins, based on conserved amino acid motifs (23, 75). As with the other xenorhodopsins, the LTVLE3 rhodopsin contains all of the conserved amino acid motifs predicted to be required for binding the retinal chromophore for light absorption (J. Ugalde, personal communication). Bacterio-opsins have been reported on plasmids of halophilic archaea (e.g., see reference 7) but not on any viruses, though auxiliary metabolic genes, including photosynthesis genes (e.g., see reference 32), are commonly found in viral genomes (52). If LTVLE3 is a virus, this rhodopsin would extend the presence of host photosensory and/or energy-harvesting genes, well documented in cyanophages (29, 64, 70), to viruses (putatively) of the Archaea.
Diversity of viruses and VLEs in hypersaline systems.
While LTV2 and LTVLE3 represent up to 2.7 and 2.5% of the reads in any sample, respectively, it is noteworthy that most of the 7 LT genomes were assembled from approximately 0.1% of the reads from a sample (Table 2). We infer that LTV2 and LTVLE3 were among the dominant populations in Lake Tyrrell at the times and locations sampled by this study, whereas populations detected at the 0.1% level were subdominant (of course, potential contamination in unassembled reads could affect these abundance estimates). These findings highlight the high diversity of the LT viral community. Although it is possible that more abundant populations were present but missed, for example, due to genome fragmentation as a result of within-population variation, these results suggest that approximately 1,000 subdominant-to-dominant viral and VLE populations may be present in the LT system at any given time. This is orders of magnitude more than the number of haloviral groups observed in previous studies through other methods (e.g., 1 to 8 groups based on pulsed-field gel electrophoresis [PFGE] bands from a haloviral community [13] and 5 groups based on dinucleotide frequencies from a halovirome [57]). To be clear, the previously cited work did not suggest that these groupings represented individual viral populations, but our results would suggest that each of the groups could contain hundreds of populations. The number of populations predicted from LT is similar to the number of viral types estimated through modeling in other aqueous systems, including hot springs, from which ∼1,300 to 1,400 viral types were predicted (59). Modeling estimates have also predicted that the most abundant viral genotypes in marine systems comprise ∼2.3 to 13.3% of the viral community (4), so abundances of ∼2.5 to 2.7% in LT suggest similar levels of dominance in hypersaline and marine viral assemblages. Of course, methodological differences in the calculation of these estimates should be considered when interpreting these data.
Temporal dynamics of viral and VLE populations in hypersaline systems.
In general, on the timescale of days, LT viral and VLE populations were relatively stable. This result is consistent with a PFGE-based study in a Spanish solar saltern, in which no significant differences were observed in viral DNA band patterns from samples collected 6 days apart (53). In the 2010 LT 4-day time series, 6 of the 7 complete genomes and 26 of the 28 genome fragments exhibited the same detection pattern (i.e., uniform presence or absence) in all four samples in the series (Fig. 2 and Table 4).
However, LTV1 and 7 genome fragments were present in only one sample of the 2007 2-day time series. We suspect that the viral community changed as a result of hosts' responses to significant lake evaporation between samplings, perhaps including generational timescale changes in host immunity (73) and/or the induction of lysogens. The results of a laboratory experiment by Santos and colleagues (56), in which the active viral community from a hypersaline lake changed following dilution with distilled water, suggest that differences in the LT lake level before and during our time series could have affected the viral community. However, no change in salinity or significant change in geochemistry was observed between the two 2007 LT samples (Table 1; also see Table S3 in the supplemental material), so the significance of the dilution and/or subsequent evaporation is unclear. The pool area for sample 2007At1 was much larger than it was for any other LT sample, including 2007At2 (see Materials and Methods), so spatial heterogeneity at the sampling site may equally well explain the differences observed.
On timescales of less than 3 years, LT viral and VLE populations tended to be highly dynamic. Six of the 7 complete genomes and 26 of the 28 genome fragments were present in some but not all eight samples, indicating transience from 2007 to 2010. However, the detection of 2 genome fragments and 91 to 100% of the LTV2 genome in all eight samples (Fig. 2b) suggests that some viral populations may be stable and maintain infectivity in the LT system for 3 years.
In a previously reported study from a saltern near San Diego (SD), CA (14, 49), a multiweek time series was sampled. Of the 10 largest contigs from that system, based on assemblies that we generated in this study, 4 were present in samples collected 2 to 3 weeks apart, 6 were present in single samples, and none were present in all three samples collected over approximately 1 month. As such, we propose a slightly modified interpretation of the original study, which suggested that haloviral taxa were stable while genotypes varied on short timescales (49). Because the most common haloviral taxa in that system (i.e., the 10 SD contigs that we assembled) appear not to have been represented in public databases and because the viral taxa reported to be most abundant in the original study, based on tBLASTx comparisons to previously sequenced viral genomes, were actually not detected in that system through fragment recruitment in this study, we suggest that trends for the dominant haloviral taxa were actually not observed. A similar possible interpretation was presented as a caveat in the published work. We suggest that the variation observed at the genotype (“strain”) level through modeling was actually an observation of the population level shifts that we see in this study.
Ecological models of virus-host dynamics (e.g., Lotka-Volterra-like “kill-the-winner” [69] and constant-diversity dynamics [50]) have predicted shifts in the presence and abundance of viral groups, as demonstrated in this study, but the timescales on which these dynamics were predicted to occur have not been well constrained. In addition, viral dynamics have been presumed to occur at the genotype (subpopulation or strain) level. While viral genotypes could also exhibit interesting dynamics, this study demonstrates dramatic shifts in the presence and relative abundance of viral populations. We also provide potential timescale constraints for ecological models. Assuming that viral population dynamics occur on similar timescales within each of the two systems studied (LT and SD), our results would suggest that haloviral populations are generally stable on the timescale of days, dynamic or stable in relatively equal proportions on timescales of 2 to 3 weeks, and generally dynamic on timescales of 1 month to 3 years. Other temporal patterns, such as seasonal shifts (43, 78), could occur in hypersaline systems but may have been missed due to the frequency of sampling. Indeed, the most likely explanation for the presence of some populations in single samples is that they cycle within a given system on timescales not captured by these studies. While we assume that the populations that we characterized are among the more dominant populations because they assembled, we recognize that their dynamics may not reflect the behavior of less abundant and/or less genomically cohesive populations.
Spatial heterogeneity of haloviral and VLE populations.
Within LT, spatial heterogeneity was observed between sites A and B (isolated pools ∼300 m apart) during the same week in 2010, in which 15 genome fragments and LTV7 exhibited different detection patterns. For our cross-system comparisons, while we cannot discount the possibility that our results could be influenced by differences in methodology (for example, different sample collection and virus isolation techniques, potential biases associated with whole-genome amplification in other studies, and differences in sequencing technology and throughput), given the temporal and spatial heterogeneity of viral populations within individual systems, it is not surprising that we did not detect most viral populations across geographically diverse hypersaline systems. However, this result is surprising in the context of some previous studies that have supported conservation across hypersaline systems in both host and viral communities. For example, one of the most dominant LT microbial species, Haloquadratum walsbyi, is suggested to be abundant and highly conserved in hypersaline systems worldwide, based on comparative genomics of isolates (16). Also, a recent study shows that many haloviral isolates can infect hosts from diverse geographic locations (5), and read- and small contig-based comparisons show some sequence similarity among globally distributed haloviral communities (57, 61). Another recent study suggests some conservation of nearly complete haloviral genomes across systems (18). While the aforementioned methodological differences may have precluded the detection of some LT populations in other systems, and while most previously sequenced haloviruses are isolates that may not represent the dominant members of their communities, it is interesting that the 35 LT viral and VLE populations were not detected in other hypersaline systems sampled by metagenomics, nor were previously sequenced haloviruses detected in LT, based on the 90% nucleotide identity detection limit set in this study. Similarly, the largest contigs in the SD system were not detected in LT, nor were previously sequenced haloviruses detected in the SD metagenomic data.
Some genome-level conservation across systems was observed when we reduced the fragment recruitment detection threshold from 90% to 75% nucleotide identity. Namely, three previously sequenced haloviruses and two SD contigs were detected in some LT samples. This most likely suggests that similar populations with divergent nucleotide sequences were detected across systems, but it could mean that reducing the detection threshold allows for similar genes from different populations to yield false-positive identification. While some viral genomes reconstructed from Spain were also detected in San Diego in a study based on similar detection thresholds (75% nucleotide identity across at least 50% of each read), most populations were not detected in both systems (18). Together, these studies show that, while there is clearly some conservation of haloviruses at the nucleotide level in geographically diverse locations, most haloviral populations are not globally conserved, at least on the temporal and spatial scales sampled thus far. While these results would seem to imply a limited distribution of most haloviral populations, they would also be consistent with a temporally variable global distribution. The latter would be supported by the detection of haloviruses from other systems in only some LT samples, as observed at 75% nucleotide identity, and by the fact that haloviruses maintain infectivity against hosts from geographically diverse locations (4).
Implications for future viral metagenomic analyses.
Many previous viral metagenomic studies have relied upon representative sequences in public databases to predict viral function (e.g., see reference 14) and/or taxonomy (e.g., see reference 49) and/or have been limited to indirect analyses, like modeling (e.g., see reference 3), to characterize viral communities. One of the most exciting results of this study is that, with deep metagenomic sequencing and assembly, it is now possible to study natural viral populations directly, based on genomic information generated from a given system. While extensive manual assembly curation was required to close the 7 genomes in this study from 6.4 Gb of metagenomic data, this may not always be necessary. For example, a recent study reported autoassembly of 78 circularized genomes from a 48-Gb virome from the human gut (36), suggesting that assembly may be more facile in some systems and/or with increased sequencing throughput.
It is well appreciated that viruses are woefully underrepresented in public databases (52, 79), and in this study, we confirmed that the identification of viral taxa based on BLAST hits to previously sequenced viruses may not be reliable. Specifically, we showed that three viruses (BJ1, ϕCh1, and HF2) reported to be among the five most dominant viral taxa in each of the samples from the San Diego system, based on tBLASTx searches (E value 0.001) against complete viral genomes (49), were actually not present in the metagenomic libraries from that system. We suggest that BLAST searches are an appropriate means of identifying candidate viral populations that may be present in a given system but that if the detection of a given virus is relevant to the results of a study, its presence should be confirmed by mapping to the reference genome or through BLAST searches along the full length of the reference genome, as in reference 18.
This study represents the first genome-based characterization of natural viral population dynamics on a variety of timescales. The observation of highly dynamic viral populations in individual systems on timescales ranging from weeks to years exposes a need for cautious interpretation of “snapshot” viral metagenomes collected at a single time and location. This de novo assembly and genomic reconstruction-based tracking of viral (and virus-like) populations provides a foundation for future high-resolution metagenomic efforts to characterize viral biogeography and to constrain ecological models of virus-host population dynamics in a variety of environments. It will be interesting to see what impact viral population dynamics may have on ecosystem-scale processes, including viral production, infectivity rates, and host community structure, and we recommend that future efforts consider characterizing these processes across time and space.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this work was provided by National Science Foundation award 0626526 and grant DE-FG02-07ER64505 from the Department of Energy.
We thank Cheetham Salt Works (Victoria, Australia) for permission to collect samples, John Moreau, Jochen Brocks, and Mike Dyall-Smith for assistance in the field and generous access to reagents and laboratory equipment, Matt Lewis and the J. Craig Venter Institute (JCVI) for library construction and sequencing, and Shannon Williamson and Doug Fadrosh (JCVI) for training J.B.E. in virus-related laboratory techniques.
Footnotes
Published ahead of print 6 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Allen EE, et al. 2007. Genome dynamics in a natural archaeal population. Proc. Natl. Acad. Sci. U. S. A. 104: 1883–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersson AF, Banfield JF. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047–1050 [DOI] [PubMed] [Google Scholar]
- 3. Angly F, et al. 2005. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics 6: 41 doi:10.1186/1471-2105-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Angly FE, et al. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4: e368 doi:10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM. 2011. Global network of specific virus-host interactions in hypersaline environments. Environ. Microbiol. 2: 426–440 [DOI] [PubMed] [Google Scholar]
- 6. Baker BJ, et al. 2006. Lineages of acidophilic Archaea revealed by community genomic analysis. Science 314: 1933–1935 [DOI] [PubMed] [Google Scholar]
- 7. Baliga NS, et al. 2004. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 14: 2221–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bamford DH, et al. 2005. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J. Virol. 79: 9097–9107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baranyi U, Klein R, Lubitz W, Krüger DH, Witte A. 2000. The archaeal halophilic virus-encoded Dam-like methyltransferase M. ϕCh1-I methylates adenine residues and complements dam mutants in the low salt environment of Escherichia coli. Mol. Microbiol. 35: 1168–1179 [DOI] [PubMed] [Google Scholar]
- 10. Bath C, Cukalac T, Porter K, Dyall-Smith ML. 2006. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology 350: 228–239 [DOI] [PubMed] [Google Scholar]
- 11. Bratbak G, Heldal M, Norland S, Thingstad TF. 1990. Viruses as partners in spring bloom microbial trophodynamics. Appl. Environ. Microbiol. 56: 1400–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML. 2004. Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl. Environ. Microbiol. 70: 5258–5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diez B, Anton J, Guixa-Boixereu N, Pedros-Alio C, Rodriguez-Valera F. 2000. Pulse-field gel electrophoresis analysis of virus assemblages present in a hypersaline environment. Int. Microbiol. 3: 159–164 [PubMed] [Google Scholar]
- 14. Dinsdale EA, et al. 2008. Functional metagenomic profiling of nine biomes. Nature 452: 629–632 [DOI] [PubMed] [Google Scholar]
- 15. Dyall-Smith M, Tang S-L, Bath C. 2003. Haloarchaeal viruses: how diverse are they? Res. Microbiol. 154: 309–313 [DOI] [PubMed] [Google Scholar]
- 16. Dyall-Smith ML, et al. 2011. Haloquadratum walsbyi: limited diversity in a global pond. PLoS One 6: e20968 doi:10.1371/journal.pone.0020968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edwards RA, Rohwer F. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3: 504–510 [DOI] [PubMed] [Google Scholar]
- 18. Garcia-Heredia I, et al. 2012. Reconstructing viral genomes from the environment using fosmid clones: the case of haloviruses. PLoS One 7: e33802 doi:10.1371/journal.pone.0033802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gordon D, Abajian C, Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8: 195–202 [DOI] [PubMed] [Google Scholar]
- 20. Guixa-Boixareu N, Calderon-Paz JI, Heldal M, Bratbak G, Pedros-Alio C. 1996. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat. Microb. Ecol. 11: 215–227 [Google Scholar]
- 21. Hatfull GF. 2008. Bacteriophage genomics. Curr. Opin. Microbiol. 11: 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119 doi:10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jung K-H, Trivedi VD, Spudich JL. 2003. Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 47: 1513–1522 [DOI] [PubMed] [Google Scholar]
- 24. Kamekura M, Dyall-Smith ML, Upasani V, Ventosa A, Kates M. 1997. Diversity of alkaliphilic halobacteria: proposals for transfer of Natronobacterium vacuolatum, Natronobacterium magadii, and Natronbacterium pharaonis to Halorubrum, Natrialba, and Natronomonas gen. nov., respectively, as Halorubrum vacuolatum comb. nov., Natrialba magadii comb. nov., and Natronomonas pharaonis comb. nov., respectively. Int. J. Syst. Bacteriol. 47: 853–857 [DOI] [PubMed] [Google Scholar]
- 25. Kim K-H, et al. 2008. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl. Environ. Microbiol. 74: 5975–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klein R, et al. 2002. Natrialba magadii virus ϕCh1: first complete nucleotide sequence and functional organization of a virus infecting a haloalkaliphilic archaeon. Mol. Microbiol. 45: 851–863 [DOI] [PubMed] [Google Scholar]
- 27. Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. 2010. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 18: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leplae Rl, Lima-Mendez G, Toussaint A. 2010. ACLAME: a classification of mobile genetic elements, update 2010. Nucleic Acids Res. 38: D57–D61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. 2005. Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438: 86–89 [DOI] [PubMed] [Google Scholar]
- 30. Ma Y, Galinski EA, Grant WD, Oren A, Ventosa A. 2010. Halophiles 2010: life in saline environments. Appl. Environ. Microbiol. 76: 6971–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Macumber PG. 1992. Hydrological processes in the Tyrrell Basin, southeastern Australia. Chem. Geol. 96: 1–18 [Google Scholar]
- 32. Mann NH, Cook A, Millard A, Bailey S, Clokie M. 2003. Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424: 741. [DOI] [PubMed] [Google Scholar]
- 33. Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maturrano L, Santos F, Rossello-Mora R, Anton J. 2006. Microbial diversity in Maras Salterns, a hypersaline environment in the Peruvian Andes. Appl. Environ. Microbiol. 72: 3887–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Milne I, et al. 2010. Tablet: next generation sequence assembly visualization. Bioinformatics 26: 401–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Minot S, Grunberg S, Wu GD, Lewis JD, Bushman FD. 2012. Hypervariable loci in the human gut virome. Proc. Natl. Acad. Sci. U. S. A. 109: 3962–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mongodin EF, et al. 2005. The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. U. S. A. 102: 18147–18152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morowitz MJ, et al. 2011. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc. Natl. Acad. Sci. U. S. A. 108: 1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mutlu MB, et al. 2008. Prokaryotic diversity in Tuz Lake, a hypersaline environment in inland Turkey. FEMS Microbiol. Ecol. 65: 474–483 [DOI] [PubMed] [Google Scholar]
- 40. Narasingarao P, et al. 2012. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 6: 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oren A, Bratbak G, Heldal M. 1997. Occurrence of virus-like particles in the Dead Sea. Extremophiles 1: 143–149 [DOI] [PubMed] [Google Scholar]
- 42. Pagaling E, et al. 2007. Sequence analysis of an archaeal virus isolated from a hypersaline lake in Inner Mongolia, China. BMC Genomics 8: 410 doi:10.1186/1471-2164-8-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parsons RJ, Breitbart M, Lomas MW, Carlson CA. 2011. Ocean time-series reveals recurring seasonal patterns of virioplankton dynamics in the northwestern Sargasso Sea. ISME J. 2: 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pietilä MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. 2009. An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol. Microbiol. 72: 307–319 [DOI] [PubMed] [Google Scholar]
- 45. Pina M, Bize A, Forterre P, Prangishvili D. 2011. The archeoviruses. FEMS Microbiol. Rev. 6: 1035–1054 [DOI] [PubMed] [Google Scholar]
- 46. Porter K, Russ BE, Dyall-Smith ML. 2007. Virus-host interactions in salt lakes. Curr. Opin. Microbiol. 10: 418–424 [DOI] [PubMed] [Google Scholar]
- 47. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35: 7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Quevillon E, et al. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33: W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodriguez-Brito B, et al. 2010. Viral and microbial community dynamics in four aquatic environments. ISME J. 4: 739–751 [DOI] [PubMed] [Google Scholar]
- 50. Rodriguez-Valera F, et al. 2009. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7: 828–836 [DOI] [PubMed] [Google Scholar]
- 51. Roine E, et al. 2010. New, closely related haloarchaeal viral elements with different nucleic acid types. J. Virol. 84: 3682–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rosario K, Breitbart M. 2011. Exploring the viral world through metagenomics. Curr. Opin. Virol. 1: 289–297 [DOI] [PubMed] [Google Scholar]
- 53. Sandaa R-A, Foss Skjoldal E, Bratbak G. 2003. Virioplankton community structure along a salinity gradient in a solar saltern. Extremophiles 7: 347–351 [DOI] [PubMed] [Google Scholar]
- 54. Sandaa R-A, Larsen A. 2006. Seasonal variations in virus-host populations in Norwegian coastal waters: focusing on the cyanophage community infecting marine Synechococcus spp. Appl. Environ. Microbiol. 72: 4610–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Santos F, et al. 2007. Metagenomic approach to the study of halophages: the environmental halophage 1. Environ. Microbiol. 9: 1711–1723 [DOI] [PubMed] [Google Scholar]
- 56. Santos F, et al. 2011. Metatranscriptomic analysis of extremely halophilic viral communities. ISME J. 5: 1621–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Santos F, Yarza P, Parro V, Briones C, Antón J. 2010. The metavirome of a hypersaline environment. Environ. Microbiol. 12: 2965–2976 [DOI] [PubMed] [Google Scholar]
- 58. Santos F, et al. 2012. Viruses from hypersaline environments: a culture-independent approach. Appl. Environ. Microbiol. 6: 1635–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schoenfeld T, et al. 2008. Assembly of viral metagenomes from Yellowstone hot springs. Appl. Environ. Microbiol. 74: 4164–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Short SM, Suttle CA. 2003. Temporal dynamics of natural communities of marine algal viruses and eukaryotes. Aquat. Microb. Ecol. 32: 107–119 [Google Scholar]
- 61. Sime-Ngando T, et al. 2010. Diversity of virus-host systems in hypersaline Lake Retba, Senegal. Environ. Microbiol. 8: 1956–1972 [DOI] [PubMed] [Google Scholar]
- 62. Simpson JT, et al. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19: 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Snyder JC, et al. 2007. Virus movement maintains local virus population diversity. Proc. Natl. Acad. Sci. U. S. A. 104: 19102–19107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sullivan MB, et al. 2010. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ. Microbiol. 12: 3035–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Suttle C, Fuhrman J. 2010. Enumeration of virus particles in aquatic or sediment samples by epifluorescence microscopy, p 145–153. In Wilhelm S, Weinbauer M, Suttle C. (ed), Manual of aquatic viral ecology. American Society of Limnology and Oceanography, Waco, TX [Google Scholar]
- 66. Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5: 801–812 [DOI] [PubMed] [Google Scholar]
- 67. Tang S-L, Nuttall S, Dyall-Smith M. 2004. Haloviruses HF1 and HF2: evidence for a recent and large recombination event. J. Bacteriol. 186: 2810–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tang S-L, et al. 2002. HF2: a double-stranded DNA tailed haloarchaeal virus with a mosaic genome. Mol. Microbiol. 44: 283–296 [DOI] [PubMed] [Google Scholar]
- 69. Thingstad TF. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45: 1320–1328 [Google Scholar]
- 70. Thompson LR, et al. 2011. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. U. S. A. 108: E757–E764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thurber RV. 2009. Current insights into phage biodiversity and biogeography. Curr. Opin. Microbiol. 12: 582–587 [DOI] [PubMed] [Google Scholar]
- 72. Tucker KP, Parsons R, Symonds EM, Breitbart M. 2011. Diversity and distribution of single-stranded DNA phages in the North Atlantic Ocean. ISME J. 5: 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tyson GW, Banfield JF. 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 10: 200–207 [DOI] [PubMed] [Google Scholar]
- 74. Tyson GW, et al. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43 [DOI] [PubMed] [Google Scholar]
- 75. Ugalde J, Podell S, Narasingarao P, Allen E. 2011. Xenorhodopsins, an enigmatic new class of microbial rhodopsins horizontally transferred between archaea and bacteria. Biol. Direct 6: 52 doi:10.1186/1745-6150-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Williams WD. 2001. Anthropogenic salinisation of inland waters. Hydrobiology 466: 329–337 [Google Scholar]
- 77. Williamson SJ, et al. 2008. The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One 3: e1456 doi:10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Winget DM, et al. 2011. Repeating patterns of virioplankton production within an estuarine ecosystem. Proc. Natl. Acad. Sci. U. S. A. 108: 11506–11511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wommack KE, Bhavsar J, Ravel J. 2008. Metagenomics: read length matters. Appl. Environ. Microbiol. 74: 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wommack KE, Ravel J, Hill RT, Chun J, Colwell RR. 1999. Population dynamics of Chesapeake Bay virioplankton: total-community analysis by pulsed-field gel electrophoresis Appl. Environ. Microbiol. 65: 231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.