Abstract
Formins are a conserved family of actin assembly-promoting factors with essential and diverse biological roles. Most of our biochemical understanding of formin effects on actin dynamics is derived from studies using formin fragments. In addition, all structural information on formins has been limited to fragments. This has left open key questions about the structure, activity and regulation of intact formin proteins. Here, we isolated full-length mouse mDia1 (mDia1-FL) and found that it forms tightly autoinhibited dimers that can only be partially activated by RhoA. We solved the structure of autoinhibited mDia1-FL using electron microscopy and single particle analysis. Docking of crystal structures into the 3D reconstruction revealed that the fork-shaped N-terminal DID-CC region hangs over the ring-shaped FH2 domain, suggesting that autoinhibition results from steric obstruction of actin binding. Deletion of the C-terminal DAD domain extended mDia1 structure and activated it for actin assembly. Using TIRF microscopy, we observed that RhoA-activated mDia1-FL persistently accelerated filament elongation in the presence of profilin similar to mDia1 FH1-FH2 fragment. These observations validate the known activities of FH1-FH2 fragments as reflecting those of the intact molecule. Our results further suggest that mDia1-FL does not readily snap back into the autoinhibited conformation and dissociate from growing filament ends, and thus additional factors may be required to displace formins and restrict filament length.
Keywords: actin, formin, autoinhibition, TIRF, diaphanous
INTRODUCTION
Formins comprise a large family of highly conserved proteins that stimulate both the nucleation and elongation phases of actin filament assembly and play critical roles in the formation of diverse actin-based structures in vivo, including stress fibers, actin cables, filopodia, and cytokinetic rings (Faix and Grosse 2006; Goode and Eck 2007). The C-terminal formin homology FH2 domain is required and sufficient for promoting actin assembly in vitro and associates with the barbed end of the filament (Pruyne et al. 2002; Sagot et al. 2002). The FH2 domain is dimeric, and mutations at conserved tryptophan residues disrupt dimerization and abolish actin assembly activity (Moseley et al. 2004). In addition, the adjacent diaphanous autoregulatory domain (DAD) binds to actin monomers and assists the FH2 domain in nucleating actin assembly (Gould et al. 2011). The FH2 domain has a donut-shaped structure, with two rigid halves connected by flexible ‘linker’ sequences (Xu et al. 2004), and this architecture enables the FH2 dimer to processively move with the growing end of the filament through sequential alternating contacts of its two halves in a ‘stair-stepping’ mechanism (Goode and Eck 2007; Xu et al. 2004). This processive attachment has been demonstrated visually by total internal reflection fluorescence (TIRF) microscopy (Kovar and Pollard 2004), and recently, the rotational movement of the FH2 along the actin filament helix, predicted by the stair-stepping mechanism, has been visualized in single molecule fluorescence polarization experiments (Mizuno et al. 2011). The processive capping abilities of the dimeric FH2 domain enable it to protect growing barbed ends of actin filaments from capping proteins (Moseley et al. 2004; Zigmond et al. 2003). Further, in combination with the FH1 domain and profilin, the FH2 can accelerate filament elongation by many fold over the rate of elongation at free filament ends (Kovar et al. 2006; Romero et al. 2004). This involves the unstructured FH1 domains recruiting profilin-actin complexes and delivering them to the FH2-capped filament end for insertional assembly (Paul and Pollard 2009a; Paul and Pollard 2009b). Thus, formins can influence at least three different aspects of actin assembly: de novo nucleation, rate of filament elongation, and duration of filament growth before capping occurs.
How the activities of formins are spatially and temporally controlled in vivo is only beginning to be understood (Chesarone et al. 2010). The first mode of formin regulation to be revealed was autoinhibition, which applies to a select subset of animal and fungal formins referred to as diaphanous-related formins (DRFs) (Castrillon and Wasserman 1994; Chalkia et al. 2008; Higgs and Peterson 2005; Rivero et al. 2005; Schonichen and Geyer 2010). DRFs contain a C-terminal DAD domain and an interacting N-terminal diaphanous inhibitory domain (DID) (Alberts 2001; Li and Higgs 2003; Li and Higgs 2005). DID-DAD binding is required for autoinhibition, and truncations of DID or DAD in vivo lead to excessive stress fibers, filopodia or actin cables (Evangelista et al. 1997; Tominaga et al. 2000; Watanabe et al. 1999). Further, overexpression of DAD peptides can activate endogenous formins to assemble excess stress fibers, and these effects depend on conserved Met and Leu residues in the DAD (Alberts 2001). Autoinhibition has been reconstituted in vitro using purified N-terminal DID-containing and C-terminal FH2-DAD-containing fragments that associate in trans (Alberts 2001; Lammers et al. 2005; Li and Higgs 2003; Li and Higgs 2005; Nezami et al. 2006). However, the structural basis for autoinhibition blocking actin assembly has been unclear. In most DRFs, there is a Rho-binding domain (RBD) adjacent to the DID. Based on RBD-truncation phenotypes in vivo, it was suggested that RhoGTP binding to RBD relieves autoinhibition (Evangelista et al. 1997; Imamura et al. 1997; Kohno et al. 1996; Watanabe et al. 1997), and indeed, purified RhoAGTP partially relieves the autoinhibition of purified RBD-DID-DD mDia1 interacting in trans with FH2-DAD, and competitively disrupts direct interactions between DID and DAD (Li and Higgs 2003; Li and Higgs 2005; Nezami et al. 2006; Otomo et al. 2005; Rose et al. 2005). However, an observation that has not yet been reconciled is that while RhoAGTP binds to RBD with high affinity (Kd=10–50 nM in mDia1), the concentrations of RhoAGTP required for activation of mDia1 from in trans autoinhibition are more than two orders of magnitude higher than the Kd (Li and Higgs 2003) and yield only partial (15–20%) activation. This has led to the suggestion that full-length mDia1 may be more readily activated by RhoAGTP than the fragments of formins interacting in trans (Li and Higgs 2005). A second hypothesis raised by in vivo studies is that additional cellular factors interacting with mDia1 are required for efficient RhoAGTP-dependent activation (Seth et al. 2006).
To date, the activities and structures of formin proteins have been defined primarily using fragments. Only a limited number of studies have begun to address the properties of intact formins. These studies have shown that full-length or nearly-full-length mDia1 and mDia2 expressed in E. coli are autoinhibited, that full-length mDia1 is activated to an unknown extent by RhoA (V14), and that full-length mDia1 is dimeric (Eisenmann et al., 2007; Ramalingam et al., 2010; Nezami et al., 2010; Otomo et al., 2010). However, many key questions remain open, including: (1) Do full-length formins have the same nucleation capabilities as C-terminal FH1-FH2 fragments? (2) Do they similarly accelerate filament growth in the presence of profilin? (3) Do they processively protect growing filament ends from capping proteins, and if so, how persistently? (4) Are full-length autoinhibited DRFs activated by RhoAGTP more efficiently than fragments interacting in trans? (5) What is the structure of intact formins, and what can the structure tell us about the molecular basis of their autoinhibition?
The limited analysis available on purified full-length formins has stemmed in part from their low endogenous expression levels, sensitivity to proteolysis, and insolubility in bacterial expression systems. We overcame these barriers with a GAL-inducible yeast expression system and developed a procedure for rapidly isolating full-length formin proteins (yeast or mammalian). Here, we report the activities and structure of full-length mouse mDia1.
RESULTS
Purified full-length mDia1 and mDia2 are autoinhibited
The purification of full-length formins has presented a number of technical challenges (above), which we overcame by developing a procedure to rapidly isolate full-length mammalian or yeast formins from S. cerevisiae. We expressed formins as N-terminal His6 fusion proteins under control of the galactose-inducible promoter in a protease-deficient yeast strain, and isolated them by sequential nickel affinity and gel filtration chromatography steps. This scheme yielded purified soluble formins that elute from gel filtration columns as mono-dispersed peaks. We used this system to isolate and characterize full-length (FL) mouse mDia1 and mDia2 formins (hereafter referred to as mDia1-FL and mDia2-FL), along with their corresponding C-terminal halves (C) consisting of the FH1-FH2-C domains (mDia1-C and mDia2-C) (see schematic, Fig. 1A).
Figure 1. Actin assembly activities of purified full-length mDia1 and mDia2.
(A) Schematic of full-length (FL) and C-terminal half (C) of mDia1/2, with domain boundaries corresponding to residue numbers in mDia1. FH, formin homology; G, Rho GTPase binding domain; DAD, diaphanous autoregulatory domain; DID, DAD interacting domain DD, dimerization domain; CC, coiled coil domain. (B,C) Comparison of pyrene-actin assembly activities for FL and C constructs of mDia1 (B) and mDia2 (C). Coomassie stained gels of the purified formins are shown alongside the assembly curves. Reactions contained 2 μM monomeric actin (5% pyrene-labeled) and the indicated concentrations (nM) of formin.
We first asked whether purified mDia1-FL and mDia2-FL (Fig. 1B and 1C) were autoinhibited by directly comparing activities of freshly purified FL and C polypeptides in pyrene-actin assembly assays (Fig. 1B and 1C). mDia1-C and mDia2-C each robustly stimulated actin assembly, as expected, whereas mDia1-FL and mDia2-FL exhibited minimal nucleation activity, consistent with the full-length formins being autoinhibited. mDia1-FL also gave much higher yields than mDia2-FL, and we chose to focus all remaining analyses on mDia1-FL. Results were indistinguishable for mDia1-FL (1-1255) and a slightly truncated mDia1-FL (55-1255). In all of the remaining analyses below, we designate which mDia1-FL polypeptide was used (1-1255 or 55-1255). However, yields were significantly higher for mDia1-FL (55-1255), and therefore it was the predominant construct used.
Purified full-length mDia1 forms dimers
Formins have two separate domains that dimerize (the DD and FH2), which has left open the possibility that native formins assemble into dimers, tetramers, or higher order oligomers. To determine the oligomeric state of purified mDia1-FL, we first employed multi-angle light scattering analysis (Fig. 2A), which revealed that native mDia1-FL (55-1255) has an estimated molecular weight of 267 kDa. This value is almost precisely two times the predicted molecular weight of mDia1-FL (55-1255), 134 kDa, strongly suggesting that mDia1-FL dimerizes. As a second test, we determined the hydrodynamic properties of purified mDia1-FL (1-1255), measuring the Stokes radius by analytical gel filtration and the sedimentation coefficient on sucrose gradients (Fig. 2B). From these values, we calculated the molecular weight of native mDia1-FL (1-1255) to be 282 kDa, again very close to two times the molecular weight of mDia1-FL (1-1255), 139 kDa. For comparative purposes, we determined the hydrodynamic properties of native mDia1-C, which was also found to be a dimer (Fig. 2B).
Figure 2. Biophysical properties of full-length mDia1.
A) Multi-angle light scattering (MALS) analysis for purified mDia1-FL (55-1255). B) Hydrodynamic properties of mDia1-FL (1-1255), mDia1 ΔDAD (1-1179), and mDia1-C (549-1255). Listed predicted molecular weights (MW) correspond to the monomeric form of the protein, including its N-terminal 6xHis tag. Stokes radii were determined by gel filtration analysis, and sedimentation coefficients were determined by velocity sedimentation analysis on sucrose gradients. Native MW was calculated from the Stokes radius and sedimentation coefficient as described (Schuyler and Pellman 2002). Molar stoichiometry was determined by dividing the native MW by the monomeric MW. ND, not determined.
Single particle electron microscopy of full-length mDia1
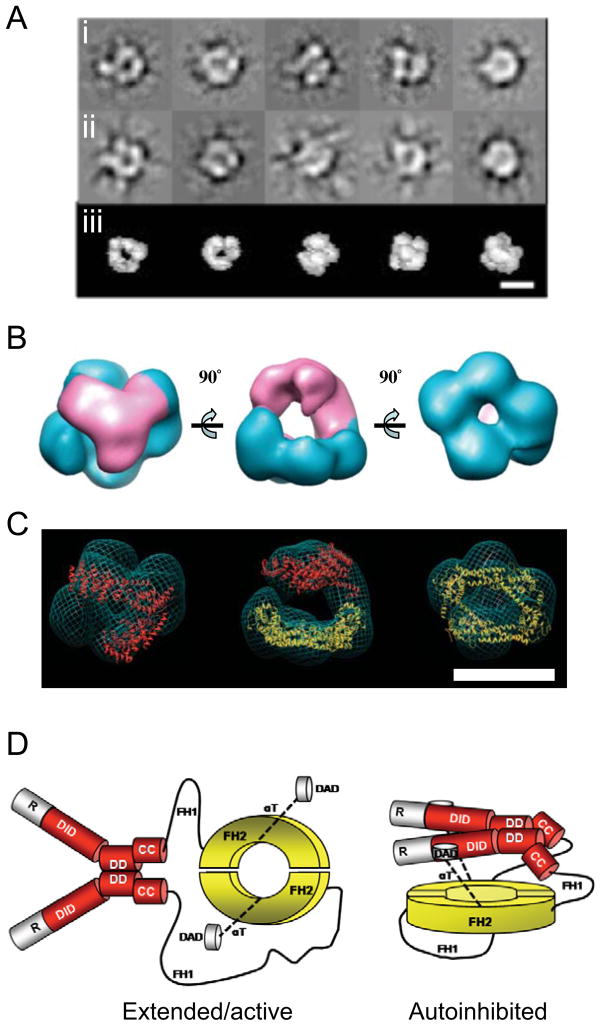
To better understand the conformation of full-length mDia1 (55-1255), we next employed single particle electron microscopy. We were not able to isolate mDia1-FL at high enough concentrations required for cryo-EM analysis, and therefore we used negative stain to enhance image contrast for optimal classification of single particles. 3590 particles were collected and categorized into 20 classes (representatives shown in Fig. 3A), which were then used to generate a 3D reconstruction (Fig. 3B). The resolution of the obtained structure was 24 Å, estimated by the Fourier shell correlation (FSC) between two reconstructions calculated from two half-sets of the data (Fig. S1B). Despite the fact that no symmetry was imposed on the final structure, it revealed a distinct two-fold symmetrical ‘fork’ on one layer (colored pink in Fig. 3B), and a four-fold symmetrical ring beneath (colored blue in Fig. 3B). This compact structure presumably represents mDia1-FL in an autoinhibited conformation.
Figure 3. Structure of autoinhibited full-length mDia1.
(A) Single particle electron microscopy analysis of purified mDia1-FL (55-1255). Row ‘i’, examples of particle classifications. Row ‘ii’, reprojections of the 3D structure before refinement matching the views in the row above. Row ‘iii’, corresponding views of the 3D structure without imposing symmetry. Scale bar, 10 nm. (B) CTF corrected and refined 3D structure of mDia1-FL (55-1255). The portion of the structure in which the N-terminal dimer (‘fork’) was docked is colored pink. Alternative views are shown at 90° angles. (C) Docking of crystal structures into our 3D electron density of mDia1-FL (55-1255). Crystal structures of mDia1 DID-DD-CC region (residues 131-516) (Otomo et al. 2005) are shaded red, and Bni1 FH2 domain (residues 1350-1760) (Xu et al. 2004) are shaded yellow. For clarity, in the top and bottom views only one of the two crystal structures is shown. Both structures are shown in the side view (middle panel). Scale bar, 10 nm. (D) Cartoons of mDia1-FL in the extended/activated (left) and closed/autoinhibited (right) conformations, with domains labeled as in Figure 1A and color-coded to match Figure 3C above: red (residues 131-516), yellow (residues 1350-1760).
To interpret our 3D structure of mDia1-FL, we docked the crystal structures of N- and C-terminal formin domains into the 3D reconstruction using an automated unbiased method (Fig. 3C). The crystal structure of the dimeric mDia1 N-terminus (consisting of DID, DD, and CC domains) (colored red) (Otomo et al. 2005) docked well into the density of the upper ‘fork’ layer, whereas the crystal structure of the Bni1 FH2 dimer (colored yellow) (Xu et al. 2004) docked well into the density of the donut-shaped layer. In this orientation, the actin-binding surfaces of the FH2 dimer point upward toward the N-terminal fork, which suggests that in this conformation the N-terminal half of the formin may obstruct actin binding to inhibit polymerization. In addition, our EM data further support the view that mDia1-FL is a dimer, since the 3D reconstructions cannot accommodate more than the one FH2 dimer and one N-terminal dimer.
Combining this information, we propose a molecular architecture for mDia1-FL in which mDia1 molecules in the dimer interact intermolecularly at two separate points (DD and FH2 domains), as depicted in the cartoons of mDia1-FL extended/activated and autoinhibited conformations (Fig. 3D).
Deletion of the DAD domain activates mDia1
We next tested the formin autoinhibition model by asking whether truncation of the DAD domain would activate and extend mDia1. To address this, we purified mDia1-ΔDAD (Fig. 4A and 4B) and assessed its activity in pyrene-actin assembly assays. We observed strong concentration-dependent nucleation activity, consistent with a release from autoinhibition (Fig. 4C). In addition, mDia1-ΔDAD had a larger Stokes radius compared to mDia1-FL (Fig. 2B), consistent with extension of its structure.
Figure 4. Effects of ΔDAD truncation on mDia1 activity and structure.
(A) Schematic of mDia1-ΔDAD polypeptide. (B) Coomassie stained gel of purified mDia1-ΔDAD. (C) Concentration-dependent effects of mDia1-ΔDAD on polymerization kinetics of 2 μM monomeric actin (5% pyrene-labeled). (D) Quantitative comparison of the physical dimensions of mDia1-ΔDAD and mDia1-FL particle classes expressed as length-to-width (X/Y) ratio. mDia1-ΔDAD (1-1179), n=25 classes; mDia1-FL (55-1255), n=20 classes; mDia1-FL (1-1255), n=30 classes.
We investigated the structural differences between mDia1-FL and mDia1ΔDAD using single particle analysis. Compared to mDia1-FL (55-1255) and mDia1-FL (1-1255), mDia1-ΔDAD showed substantial heterogeneity in particles, which precluded processing to obtain a 3D reconstruction. Nonetheless, mDia1-ΔDAD particles had distinct conformations, and from the class averages we were able to compare the physical dimensions of mDia1-ΔDAD to those of mDia1-FL. The degree of ‘compaction’ for particle class averages was calculated as a length to width ratio (X/Y), where X is the measured length of the particle class average along its longest axis, and Y is the measured length along its shortest axis. These values were averaged for all classes of mDia1-FL (1-1255 and 55-1255, separately) and mDia1-ΔDAD and plotted (Fig. 4D). This revealed that the average length to width ratio for mDia1-ΔDAD was almost double that of mDia1-FL.
Taken together, the data above indicate that truncation of DAD leads to a substantial lengthening of mDia1 structure that correlates with a release from autoinhibition.
RhoAGTP weakly activates mDia1-FL for actin assembly
Evidence from multiple studies has suggested that DRFs are activated by their interactions with GTP-bound Rho family GTPases (RhoGTP). However, this mechanism remains only partially understood. The binding affinity of DAD-DID interactions is in the sub-micromolar range (Kd~100–200 nM) (Nezami et al. 2006; Otomo et al. 2005; Rose et al. 2005). Non-prenylated RhoAGTP binds to the N-terminus of mDia1 with even higher affinity (Kd=6 nM) and forms a ternary complex with the RBD and DID domains (Otomo et al. 2005; Rose et al. 2005). Further, nanomolar concentrations of non-prenylated RhoAGTP are sufficient to competitively displace DAD-DID interactions (Otomo et al. 2005; Rose et al. 2005). However, it has been perplexing that the same concentrations of the same preparations of non-prenylated RhoAGTP are relatively ineffective in releasing mDia1 from in trans autoinhibition using N- and C-terminal fragments larger than the DAD and DID domains (Li and Higgs 2003; Otomo et al. 2005). Thus, for reasons that are still not well understood, micromolar concentrations of RhoAGTP are required to relieve autoinhibition, and even the highest concentrations of RhoAGTP tested yield only partial (15%) activation of mDia1. These observations have led to the suggestion that additional binding partners of mDia1 may be required for efficient activation in vivo, or possibly that RhoAGTP is more effective in activating intact full-length mDia1 compared to N- and C-terminal fragments that interact in trans (Li and Higgs 2003). With purified full-length mDia1, we were able to directly test the latter hypothesis. We first asked whether RhoAGTP activates mDia1-FL (55-1255) in pyrene-actin assembly assays (Fig. 5A). Similar to the above-mentioned studies, we observed that micromolar concentrations of RhoAGTP were required to observe any effect, and that activation was only partial. Thus, our data demonstrate that RhoAGTP alone does not efficiently activate mDia1-FL (see Discussion).
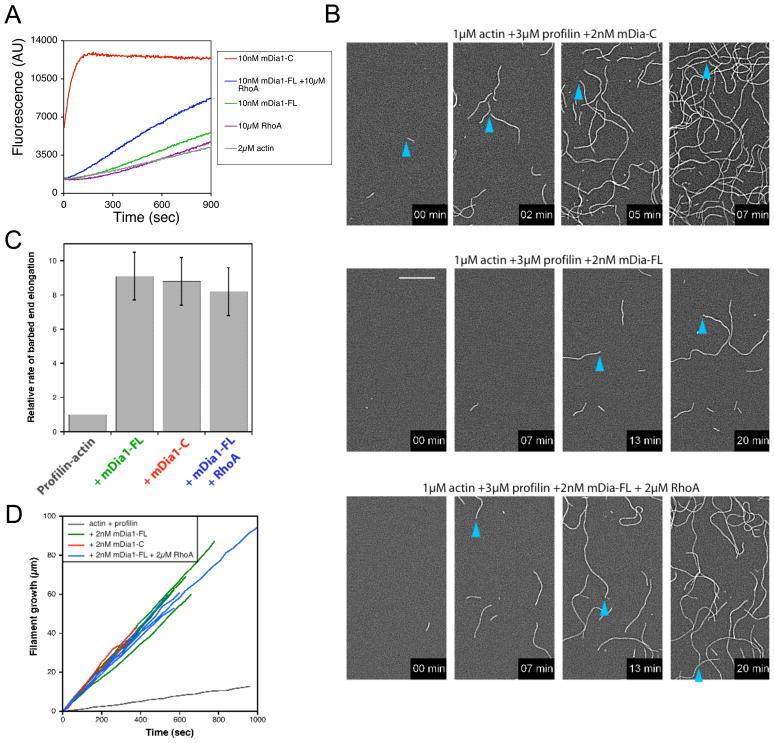
Figure 5. RhoA activation and TIRF microscopy analysis of mDia1-FL effects on actin filament nucleation and elongation.
(A) Effects of 10μM RhoA (GTP bound) on the ability of 10 nM mDia1-FL (55-1255) to stimulate polymerization of 2 μM monomeric actin (5% pyrene-labeled). (B) Time lapse TIRF microscopy comparing barbed end elongation rates of filaments in reactions containing 1.0 μM actin and 3 μM profilin +/- 2 nM mDia1-C or 2 nM mDia1-FL (55-1255) +/− 2μM RhoAGTP. Filaments were imaged at the indicated time points after initiation of polymerization. Time points are taken from movies (see Supplementary Data). Blue arrows trace the growth of single barbed ends. Scale bar, 15 μm. (C) Examples of individual filament measurements, where filament length increases over time. (D) Average rates of barbed end elongation (n = 10 filaments); color-coding as in C.
TIRF microscopy of actin elongation rates supported by mDia1-FL
It has been shown that FH1-FH2 fragments of various formins (including mDia1, mDia2, Bni1, and Cdc12) increase the rate of filament barbed end elongation to different extents, with mDia1 causing the greatest acceleration of elongation, and that these effects depend critically on profilin-FH1 interactions (Kovar et al. 2006). However, the effects of intact formins on filament elongation rates have remained unknown, as inclusion of the N-terminal half of the formin in principle could affect the rate of movement of the FH1-FH2 on the barbed end of the filament and/or the persistence of formin movement. If for instance, full-length formins snap back into an autoinhibited conformation at some frequency and fall off the ends of filaments, then by TIRF microscopy one might expect to see a major difference in the average filament elongation rates over time supported by mDia1-C versus mDia1-FL. On the other hand, if mDia1-FL supports persistent movement on growing ends like mDia1-C, then it might be expected to yield a similar average rate of filament elongation, or only slightly reduced due to steric hindrance resulting from inclusion of the N-terminal half.
To address this, we directly compared the effects of mDia1-FL (55-1255) and mDia1-C on rate of elongation in the presence of 1 μM actin and 3 μM profilin using TIRF microscopy. In the absence of formins, filament barbed ends grew at 4.9 subunits per second. Consistent with RhoA activation of mDia1-FL in pyrene-actin assembly assays above, we observed greatly increased numbers of filaments in reactions containing mDia1-FL + RhoA versus mDia1-FL alone (Fig. 5B). However, despite the very small number of filaments nucleated, we could observe that mDia1-FL accelerated barbed end elongation to the same extent (~8 fold) as mDia1-C or mDia1-FL + RhoA (~40 subunits per second) compared to free barbed ends. In addition, the accelerated elongation of individual filaments persisted for periods exceeding 10 minutes (Fig. 5C and 5D). These data suggest that mDia1-FL molecules accelerate filament elongation to rates indistinguishable from mDia1-C, and persist on growing ends for many minutes, which has important implications for their regulation in vivo, discussed below.
DISCUSSION
Until now, the biochemical activities and structures of formins have been defined primarily by studies using smaller fragments of these proteins. This has left many of the key properties of intact formin molecules a mystery. Here, we developed procedures for the rapid isolation of full-length mammalian formin proteins from a yeast expression system, and characterized the structure and activities of full-length mouse mDia1 (mDia1-FL). Our results shed new light on the autoinhibited structure and activities of full-length formins. Further, our work opens the door to characterizing the properties of other full-length formins from a wide range of organisms, which ultimately is required to understand their cellular functions and regulation.
Oligomeric state of full-length formins
It has long been suggested that formins self-associate into oligomers, but there have been mixed reports concerning the oligomerization state, i.e. dimers, tetramers or higher order concatemers (Zigmond et al., 2003). Until recently, only the oligomeric states of formin fragments had been determined, which indicated that the N- and C-terminal halves of formins independently dimerize (Otomo et al. 2005; Rose et al. 2005; Xu et al. 2004). Here, we examined the oligomeric state of purified mDia1-FL by multi-angle light scattering (MALS), hydrodynamic analysis, and single particle EM. By all three approaches, mDia1-FL formed dimers, which also agrees with other recent reports (Nezami et al. 2010; Otomo et al. 2010). Thus, despite containing two different dimerization domains (the N-terminal DD and the C-terminal FH2), mDia1-FL does not appear to form higher order oligomers.
Based on mDia1-FL being a dimer, and factoring in the geometry of the crystal structures available for N- and C-terminal formin domains, we propose the depicted molecular architectures for full-length mDia1 in the extended/activated and closed/autoinhibited states (Fig. 3D; autoinhibited and activated states). Our preliminary hydrodynamic analysis for three other purified full-length Bni1, Bnr1, and mDia2 further indicates that each of these formins is a dimer (C.G. and B.G., unpublished observations). Thus, in the absence of other factors, a number of formins from evolutionarily diverse species exist as dimers. In a number of in vivo systems, GFP-tagged formins have been shown to localize as faint puncta in the cytosol, and are then recruited to membranes where they stimulate actin assembly (Buttery et al. 2007; Higashida et al. 2004; Martin and Chang 2006). We suggest that these puncta may represent inactive dimers, which are subsequently recruited and activated at the membrane by Rho GTPases, lipids, and other formin ligands (Fig 6).
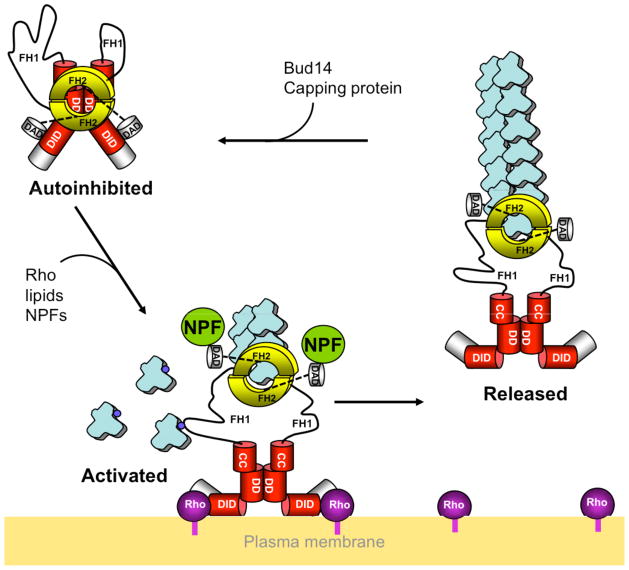
Figure 6. Model for the integrated regulation of mDia1 activity.
Autoinhibited mDia1-FL dimers in the cytoplasm are recruited to the plasma membrane through interactions with lipids, prenylated RhoAGTP, and possibly additional ligands, including DAD-binding nucleation promoting factors (NPFs). These binding events activate mDia1-FL for actin assembly. NPFs facilitate the nucleation phase of actin filament assembly (Gould et al. 2011; Okada et al. 2010; Vizcarra et al. 2011). Subsequently, the formin may be released from the membrane, possibly in response to Rho GTPase activity and/or changes in lipid composition. Our data suggest that activated full-length mDia1 dimers can remain processively attached to growing barbed ends until additional factors (e.g. Bud14 or capping protein) displace the formin, leading mDia1-FL to return to the autoinhibited state.
Structure of full-length mDia1 in the autoinhibited conformation
Using single particle electron microscopy, we solved the 3D structure of autoinhibited mDia1-FL. This revealed a two-layered structure with a forked domain sitting approximately 20–40 Å above a donut-shaped domain (Fig. 3B). An automated docking program assigned the crystal structure of the FH2 dimer into the lower domain, and the crystal structure of the N-terminal DID-DD-CC domain into the upper forked domain (Fig. 3C). In this conformation, the actin binding sites on the FH2 face upward toward the forked domain, suggesting that formin autoinhibition is achieved through steric obstruction of actin binding. This conformation does not rule out the possibility that the FH2 may still be capable of binding to one or two actin subunits in the autoinhibited state, as suggested by NMR data in another recent study (Otomo et al. 2010). However, our structure suggests that further polymerization of actin would be blocked, and that autoinhibited mDia1-FL would not be able to associate with the barbed ends of existing filaments. The fixed distance separating the FH2 and DID-DD-CC layers in the structure is presumably maintained by DAD-DID interactions and the long α-T helix extending from the FH2 that holds the DAD domain on its end (Xu et al., 2004; Otomo et al. 2010; Nezami et al. 2010). Consistent with this view, deletion of the DAD region activated mDia1 for actin assembly (Fig. 4C) and extended its conformation (Fig. 4D). As expected, this effect was accompanied by a modest decrease in actin nucleation activity due to loss of the C-terminal tail, which binds actin monomers and assists the FH2 in nucleating actin (Gould et al. 2011; Heimsath and Higgs, 2012). Neither the flexible FH1 sequences nor the αT helices (see schematic in Fig. 3D) were visible in our low-resolution structure. This is presumably because these elements lack tertiary structure, as predicted for the proline-rich FH1 (Paul and Pollard 2009a; Paul and Pollard 2009b) and verified for αT helices (Nezami et al. 2010; Otomo et al. 2010). In our model of the structure of autoinhibited mDia1-FL, the two αT helices (dotted lines, Fig. 3D) extend from the coiled coil subdomains of the FH2 halves and hold the DADs at the established DAD-binding surfaces on the DID domains (Lammers et al. 2005; Nezami et al. 2006) (Fig. 3C and 3D).
Our autoinhibited structure also provides new structural insights into the formin activation mechanism. Only one of the three crystal structures reported for the N-terminal half of mDia1 (mDia1-N) could be docked into our 3D reconstruction. In the Rho-bound and DAD peptide-bound structures of the mDia1-N dimer (Rose et al. 2005; Lammers et al. 2005), the two DID-containing halves flare out to form essentially a straight line. In contrast, in the unbound structure of mDia1-N (Otomo et al. 2005), the two halves are angled to form a ‘V’ shaped dimer. Only this V-shaped conformation could be docked into our 3D reconstruction of autoinhibited mDia1-FL. This raises the intriguing possibility that Rho-binding might alter the conformation of the N-terminal half of mDia1 as part of the activation mechanism. In addition, we note that the mDia1-N construct that forms a V-shape contains a short coiled coil segment (C-terminal to its DD domain) that is lacking in both the Rho- and DAD peptide-bound structures. Thus, it is possible that the presence of the coiled coil segment promotes the V-shaped autoinhibitory conformation of mDia1-N.
How does our EM structure of the autoinhibited mDia1-FL dimer relate to the recently reported tetrameric structures of interacting mDia1 N- and C-terminal fragments (Nezami et al. 2010; Otomo et al. 2010)? These studies obtained similar tetrameric structures for mDia1, consisting of two interlocking FH2-DAD dimers at the center and N-terminal dimers bound at each end, and proposed a number of working models for how autoinhibition might work in an intact formin dimer. Eck and co-workers essentially proposed two models. In the first, the DID would intimately contact the FH2 to directly block the actin binding sites, and in the second, the DID would sit well above the FH2, sterically obstructing actin polymerization. Our EM structure agrees with the second model, which is similar to one of four models proposed by Rosen and co-workers (Otomo et al. 2010). Thus, our EM structure has greatly clarified the spatial organization of DID and FH2 domains in the autoinhibited state.
While our structure generally supports these previously proposed models, one important difference is the orientation of the FH2. In our structure, the actin binding sites of the FH2 face in the direction of the DID-DD-CC domains, which offers a relatively simple explanation for how actin polymerization is blocked. In the tetrameric crystal structures of the studies above, the FH2 was oriented in the opposite direction, i.e. facing away from the DID-DD. This led to alternative explanations for how DAD interactions with DID-DD might block actin assembly. For instance, it was proposed that in the autoinhibited conformation there might be large changes in FH2 conformation, thereby increasing the distance separating the two functional halves of the FH2 (hemi-dimers or half-bridges) so that they could no longer efficiently stabilize an actin nucleus (dimer or trimer). Our structure suggests a far simpler mechanism at work, because the actin binding surfaces on the FH2 domain face the inhibitory domain, and thus conformational changes in the FH2 are not required for autoinhibition.
Activity and regulation of full-length mDia1
Another key question we addressed is whether activated full-length formins have the same or distinct effects in stimulating actin assembly compared to their C-terminal FH1-FH2-C fragments. We observed that RhoA-activated mDia1-FL molecules produced the same rapid and persistent actin filament elongation rates in the presence of profilin as mDia1-C (FH1-FH2-C). These results further validate the relevance of the many studies reporting actin assembly activities of FH1-FH2 fragments. Further, they offer new insights into formin mechanics and regulation. Specifically, we observed that inclusion of the relatively large and dimeric N-terminal half of the formin does not interfere with processive insertional assembly by the C-terminal half. Specifically, our data suggest that the N-terminus did not cause the formin to ‘reduce speed’ on the filament end, or rapidly ‘snap back’ into an autoinhibited conformation and fall off. Individual filaments were observed to elongate at the accelerated speed (~40 actin subunits/sec) for periods of > 10 min (Fig 5D, blue and green traces). These observations have at least two important implications for formin regulation in vivo. First, they suggest that once activated, full-length formins may persist on growing ends of filaments until additional factors displace the formin. Potential mechanisms include competition for the barbed end with capping protein and/or displacement by formin-binding partners such as Bud14 (Chesarone et al. 2009) (see model, Fig. 6). Second, if activated formins are released from membranes, they should be capable of persisting on growing ends of filaments and accelerating elongation, which might explain observed actin polymerization-dependent long-range movements of GFP-tagged formins in cells (Higashida et al. 2004).
Finally, we addressed whether Rho GTPase is sufficient to activate full-length formins for actin assembly. Previously it was shown that GTP-bound RhoA only partially activates mDia1 N- and C-terminal fragments interacting in trans (Li and Higgs 2003). This led to the suggestion that RhoA might be more effective in activating full-length mDia1. Here, we directly tested this possibility in bulk assays using mDia1-FL, and observed that concentrations of non-prenylated RhoA as high as 8 μM led to only partial activation (~15%), matching the results of Li and Higgs. Taken together, these results suggest that in the absence of other factors, non-prenylated RhoA is not sufficient to fully release/activate mDia1 from autoinhibition. To account for these observations, we propose that full and efficient activation may require a convergence of multiple inputs (Fig. 6). In this model, Rho binding plays a central role in formin activation but additional factors amplify Rho effects. These include (i) specific lipids in the membrane, which bind to N- and C-terminal domains of formins (Ramalingam et al. 2010) and may also orient prenylated RhoA to optimize activation, (ii) NPFs and other ligands of the formin C-terminus (e.g. Dishevelled, Bud6, Spire and APC) (Liu et al. 2008; Moseley and Goode 2005; Okada et al. 2010; Quinlan et al. 2007; Vizcarra et al. 2011), and (iii) kinases that phosphorylate formins (e.g. ROCK, Prk1, Aurora B, and CK2) (Cheng et al. 2011; Hannemann et al. 2008; Iskratsch et al. 2010; Staus et al. 2011; Wang et al. 2009). In this manner, formin activation at specific locations in vivo may require the integration of multiple signal inputs, akin to coincidence detection and activation of WASP-Arp2/3 complex-mediated actin assembly (Padrick and Rosen 2010).
MATERIALS AND METHODS
Plasmid construction
For yeast expression of mDia1-FL (amino acids 1-1255 and 55-1255), mDia1-C (549-1255), mDia1-ΔDAD (1-1179), mDia2-FL (1-1171) and mDia2-C (478-1171), the DNA sequences encoding these regions were PCR amplified and subcloned into pBG564 (URA3, 2μ, GAL1/10 promoter) (Moseley et al. 2006).
Protein expression and purification
Rabbit skeletal muscle was purified as described (Pollard 1984), and labeled with pyrenyliodoacetamide (Higgs and Pollard 1999) or Alexa-488 (Isambert et al. 1995). All formin constructs were purified from S. cerevisiae as described with minor modifications (Moseley et al. 2006). Briefly, plasmids were transformed into yeast strain BJ2168, and cells were grown at 25°C in 2 L cultures to OD600 = 0.8 in selective medium containing 2% raffinose. Galactose (2% final) along with peptone (10 g/L) and yeast extract (5 g/L) were added to induce expression at 25°C for 8 h. Cells were harvested by centrifugation, washed in water, resuspended in a 1:3 (v/w) ratio of water, lysed by mechanical perturbation in a coffee grinder with liquid N2, and stored as a dry powder at −80°C. To isolate His6-fusion proteins, 10 g of frozen powder was thawed in a 1:4 (v/w) ratio of 20 mM imidazole pH 8.0, 1.5x PBS (40 mM sodium phosphate buffer, 200 mM NaCl, pH 7.4), 0.5% IGEPAL (v/v), 0.2% Thesit, 1 mM DTT, and protease inhibitors (final: 1 mM phenylmethylsulfonyl fluoride and 0.5 μg/ml each of antipain, leupeptin, pepstatin A, chymostatin, and aprotinin). 50 ml of crude lysate was centrifuged at 80,000 rpm for 20 min in a TLA100.3 rotor (Beckman/Coulter, Fullerton, CA), and the supernatant was incubated for 1.5 h with Ni2+-NTA-agarose beads (Qiagen, Valencia, CA). Beads were washed three times with 20 mM imidazole (pH 8.0), 1x PBS, 1 mM DTT, 200 mM NaCl. Then, proteins were eluted with 0.5 ml buffer (300 mM imidazole pH 8.0, 50 mM Tris pH-8.0, 100 mM NaCl, 1 mM DTT, 5% glycerol), and purified further by gel filtration on a Superose 6 column (GE Healthcare, Piscataway) equilibrated in HEKG5 buffer (20 mM HEPES pH 7.5, 1 mM EDTA, 50 mM KCl, 5% glycerol). Peak fractions were pooled and stored on ice until use in kinetic assays. Activities remained constant for 1–2 weeks on ice. Human GST-RhoA (QL) was expressed in BL21 (DE3) E. coli as described with minor modifications (Nezami et al. 2006), then purified on glutathione agarose, released from GST by TEV protease digestion, and purified further by gel filtration on a Superose 12 column (GE Heathcare) equilibrated in 50 mM Tris pH 8.0, 100 mM NaCl, and 0.1 mM GTP. Peak fractions were pooled, supplemented with 1 mM GTP, and frozen. Human profilin was expressed in BL21 (DE3) E. coli and purified as described (Moseley et al. 2004).
Pyrene-actin assembly assays
Monomeric rabbit skeletal muscle actin was prepared by gel filtration on a Sephacryl S-200 column (GE healthcare) equilibrated in G-buffer (10 mM Tris pH 8.0, 0.2 mM ATP, 0.2 mM CaCl2, and 0.2 mM DTT). Final reaction volumes were 60 μl. Gel-filtered monomeric actin (final 2 μM, 5% pyrene-labeled) was converted to Mg-ATP-actin for 2 min immediately before each reaction as described (Kovar et al. 2003). To initiate assembly, Mg-ATP-actin was mixed with 15 μl HEKG5 buffer or proteins in HEKG5 and added to 3 μl of 20x initiation mix (40 mM MgCl2, 10 mM ATP, 1 M KCl). Pyrene fluorescence was monitored over time at 25 °C, excitation 365 nm and emission 407 nm, in a fluorescence spectrophotometer (Photon Technology International, Lawrenceville, NJ).
Total internal reflection fluorescence (TIRF) microscopy
Reactions contained 1 μM rabbit muscle actin monomers (30% Alexa-488 labeled) and 3 μM human profilin, with or without 2 nM mDia1-FL, 2 nM mDia1-C, and 2 μM RhoA (QL). Protein mixtures were diluted in freshly prepared fluorescence buffer containing 10 mM imidazole-HCl pH 7.8, 50 mM KCl, 1 mM MgCl2, 100 mM DTT, 3 mg/ml glucose, 20 mg/ml catalase, 100 mg/ml glucose oxidase, and 0.5% methylcellulose to induce polymerization, and imaged at 20 sec intervals on an objective-based TIRF microscope (Nikon TE2000E). Metamorph software (version.6.3r7; Universal Imaging, Media, PA) was used for image acquisition and analysis. Barbed end elongation rates were 4.9 μM−1 s−1 for Mg-ATP/ADP+Pi-actin with profilin.
Electron microscopy and single particle analysis
After gel filtration, purified mDia1-FL (two different constructs, 1-1255 and 55-1255), and mDia1-ΔDAD were diluted with one volume of HEK buffer, and 3 μl were applied to freshly glow-discharged 400 mesh carbon-coated copper grids. Grids were stained with a solution of 1% uranyl acetate for 30 sec and blotted to remove excess stain. Grids were examined on a Philips CM12 or a FEI Tecnai G12 transmission electron microscope at 120 kV under low-dose conditions. Images were captured at 60,000x magnification and 1.5–1.8 μm underfocus. The negatives were scanned on a Zeiss SCAI scanner, using a pixel size of 7 μm. The pixel size on the specimen was 3.5 Å/pixel after 3x3 pixel binning. Particles were collected manually and processed as described (Rodal et al. 2005) using IMAGIC software (van Heel et al. 1996) for the angular reconstitution method, and Spider software (Frank et al. 1996) for the random conical tilt method. For angular reconstitution, the data sets contained 3654 particles for mDia1-FL (55-1255), 2953 particles for mDia1-FL (1-1255) and 1690 particles for mDia1-ΔDAD. To generate the preliminary 3D reconstructions for mDia1-FL (both constructs), we used the angular reconstitution method. For mDia1-FL (55-1255), this produced 20 classes (from 3590 particles), and for mDia1-FL (1-1255) this produced 18 classes (from 2425 particles). Preliminary structures of mDia1-FL were highly similar, but the mDia1-FL (55-1255) data set had more particle orientations and its reconstruction had higher resolution. Therefore, we used mDia1-FL (55-1255) for the final 3D reconstruction shown in Figure 3B. Refinement and correction for contrast transfer function (CTF) was performed using Frealign software (Grigorieff 2007), and the results were visualized in the UCSF Chimera package (Pettersen et al. 2004). The resulting mDia1-FL structure has a resolution of 24 Å (Fig. S1B). To interpret this 3D map of mDia1-FL, we docked the crystal structures of mDia1 N-terminal DID-DD-CC region (residues 131-516) (pdb 2BNX, (Otomo et al. 2005)) and Bni1 FH2 domain (residues 1350-1760) (pdb 1UX5, (Xu et al. 2004)) automatically using the program Situs (Wriggers 2010). The correlation coefficients were 0.65 for FH2, and 0.59 for N-terminus.
To ensure that the 3D structure obtained by angular reconstitution above did not result from a model-dependent bias (imposed by an alignment based on cross-correlation), we independently determined the 3D structure by the random conical tilt method (Fig. S1A). We collected 3990 tilted pairs at 0 and 45° tilt angles, aligned them using a reference-free procedure, and categorized them by a hierarchical clustering procedure into 22 classes. We generated corresponding 3D reconstructions for the 10 most prevalent classes, each of which yielded a tiered structure with a donut-shaped layer and a fork-like layer above. These 10 classes (2394 particles in total) were combined to generate a 3D reconstruction that was further refined and corrected for CTF using Frealign (Fig. S1A). The resolution of this 3D structure was 33 Å (Fig. S1B).
Supplementary Material
(A) 3D structure of mDia1-FL (55-1255) calculated using RCT method, refined and CRF corrected (left panel). Docking of the crystal structures of mDia1 N-terminal DID-DD-CC region (residues 131-516) (Otomo et al. 2005) (blue) and Bni1 FH2 domain (residues 1350-1760) (Xu et al. 2004) (purple) into the 3D density of mDia1-FL (55-1255) (right panel). (B) Fourier shell correlation plot for the structure of mDia1-FL (55-1255), calculated using angular reconstitution method (AR) (solid line) and structure of same molecule, calculated using RCT, (dotted line), calculated between structures each containing one half of the data.
Acknowledgments
We are grateful to G. Gundersen for providing mDia1 and mDia2 clones, M. Eck for providing purified RhoA GTPase (QL), and J. Eskin, B. Graziano, and A. Rodal for editing the manuscript. This work was supported by grants from RFBR (08-04-91125) to O.S., a joint CRDF grant (RUB1-2918-MO-07) to B.G. and O.S, and the N.I.H. (GM083137) to B.G.
References
- Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276(4):2824–30. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- Buttery SM, Yoshida S, Pellman D. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol Biol Cell. 2007;18(5):1826–38. doi: 10.1091/mbc.E06-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon DH, Wasserman SA. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120(12):3367–77. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Chalkia D, Nikolaidis N, Makalowski W, Klein J, Nei M. Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol Biol Evol. 2008;25(12):2717–33. doi: 10.1093/molbev/msn215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhang J, Ahmad S, Rozier L, Yu H, Deng H, Mao Y. Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev Cell. 2011;20(3):342–52. doi: 10.1016/j.devcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M, Gould CJ, Moseley JB, Goode BL. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev Cell. 2009;16(2):292–302. doi: 10.1016/j.devcel.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, Dupage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010:1–13. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol. 2007;17(7):579–91. doi: 10.1016/j.cub.2007.03.024. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276(5309):118–22. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Faix J, Grosse R. Staying in shape with formins. Dev Cell. 2006;10(6):693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Gould CJ, Maiti S, Michelot A, Graziano BR, Blanchoin L, Goode BL. The Formin DAD Domain Plays Dual Roles in Autoinhibition and Actin Nucleation. Curr Biol. 2011;21(5):384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157(1):117–25. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Hannemann S, Madrid R, Stastna J, Kitzing T, Gasteier J, Schonichen A, Bouchet J, Jimenez A, Geyer M, Grosse R, et al. The Diaphanous-related Formin FHOD1 associates with ROCK1 and promotes Src-dependent plasma membrane blebbing. J Biol Chem. 2008;283(41):27891–903. doi: 10.1074/jbc.M801800200. [DOI] [PubMed] [Google Scholar]
- Heimsath EG, Higgs HN. The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J Biol Chem. 2012;287(5):3087–98. doi: 10.1074/jbc.M111.312207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashida C, Miyoshi T, Fujita A, Oceguera-Yanez F, Monypenny J, Andou Y, Narumiya S, Watanabe N. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303(5666):2007–10. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16(1):1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD. Regulation of actin polymerization by Arp2/3 complex and WASp/Scar proteins. J Biol Chem. 1999;274(46):32531–4. doi: 10.1074/jbc.274.46.32531. [DOI] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16(10):2745–55. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isambert H, Venier P, Maggs AC, Fattoum A, Kassab R, Pantaloni D, Carlier MF. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J Biol Chem. 1995;270(19):11437–44. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- Iskratsch T, Lange S, Dwyer J, Kho AL, dos Remedios C, Ehler E. Formin follows function: a muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J Cell Biol. 2010;191(6):1159–72. doi: 10.1083/jcb.201005060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, et al. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15(22):6060–8. [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124(2):423–35. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161(5):875–87. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101(41):14725–30. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers M, Rose R, Scrima A, Wittinghofer A. The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J. 2005;24(23):4176–87. doi: 10.1038/sj.emboj.7600879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Higgs HN. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13(15):1335–40. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Li F, Higgs HN. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem. 2005;280(8):6986–92. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- Liu W, Sato A, Khadka D, Bharti R, Diaz H, Runnels LW, Habas R. Mechanism of activation of the Formin protein Daam1. Proc Natl Acad Sci U S A. 2008;105(1):210–5. doi: 10.1073/pnas.0707277105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr Biol. 2006;16(12):1161–70. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Higashida C, Yuan Y, Ishizaki T, Narumiya S, Watanabe N. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011;331(6013):80–3. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem. 2005;280(30):28023–33. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Maiti S, Goode BL. Formin proteins: purification and measurement of effects on actin assembly. Meth Enzymol. 2006;406:215–34. doi: 10.1016/S0076-6879(06)06016-2. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15(2):896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami A, Poy F, Toms A, Zheng W, Eck MJ. Crystal structure of a complex between amino and carboxy terminal fragments of mDia1: insights into autoinhibition of diaphanous-related formins. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami AG, Poy F, Eck MJ. Structure of the autoinhibitory switch in formin mDia1. Structure. 2006;14(2):257–63. doi: 10.1016/j.str.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189(7):1087–96. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005;18(3):273–81. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Machius M, Rosen MK. Crystal structure of the Formin mDia1 in autoinhibited conformation. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Rosen MK. Physical mechanisms of signal integration by WASP family proteins. Annu Rev Biochem. 2010;79:707–35. doi: 10.1146/annurev.biochem.77.060407.135452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. J Biol Chem. 2009a;284(18):12533–40. doi: 10.1074/jbc.M808587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009b;66(8):606–17. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Polymerization of ADP-actin. J Cell Biol. 1984;99(3):769–77. doi: 10.1083/jcb.99.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297(5581):612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179(1):117–28. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam N, Zhao H, Breitsprecher D, Lappalainen P, Faix J, Schleicher M. Phospholipids regulate localization and activity of mDia1 formin. Eur J Cell Biol. 2010;89(10):723–32. doi: 10.1016/j.ejcb.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Rivero F, Muramoto T, Meyer A-K, Urushihara H, Uyeda TQP, Kitayama C. A comparative sequence analysis reveals a common GBD/FH3-FH1-FH2-DAD architecture in formins from Dictyostelium, fungi and metazoa. BMC Genomics. 2005;6:28. doi: 10.1186/1471-2164-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Sokolova O, Robins DB, Daugherty KM, Hippenmeyer S, Riezman H, Grigorieff N, Goode BL. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. 2005;12(1):26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier M-F. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119(3):419–29. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435(7041):513–8. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal A, Moseley J, Goode B, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4(8):626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Schonichen A, Geyer M. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim Biophys Acta. 2010;1803(2):152–63. doi: 10.1016/j.bbamcr.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Schuyler SC, Pellman D. Analysis of the size and shape of protein complexes from yeast. Methods Enzymol. 2002;351:150–68. doi: 10.1016/s0076-6879(02)51845-0. [DOI] [PubMed] [Google Scholar]
- Seth A, Otomo C, Rosen MK. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J Cell Biol. 2006;174(5):701–13. doi: 10.1083/jcb.200605006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staus DP, Blaker AL, Medlin MD, Taylor JM, Mack CP. Formin homology domain-containing protein 1 regulates smooth muscle cell phenotype. Arterioscler Thromb Vasc Biol. 2011;31(2):360–7. doi: 10.1161/ATVBAHA.110.212993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T, Sahai E, Chardin P, McCormick F, Courtneidge SA, Alberts AS. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell. 2000;5(1):13–25. doi: 10.1016/s1097-2765(00)80399-8. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116(1):17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Vizcarra CL, Kreutz B, Rodal AA, Toms AV, Lu J, Zheng W, Quinlan ME, Eck MJ. Structure and function of the interacting domains of Spire and Fmn-family formins. Proc Natl Acad Sci U S A. 2011;108(29):11884–9. doi: 10.1073/pnas.1105703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Neo SP, Cai M. Regulation of the yeast formin Bni1p by the actin-regulating kinase Prk1p. Traffic. 2009;10(5):528–35. doi: 10.1111/j.1600-0854.2009.00893.x. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1(3):136–43. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16(11):3044–56. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2(1):21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116(5):711–23. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13(20):1820–3. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) 3D structure of mDia1-FL (55-1255) calculated using RCT method, refined and CRF corrected (left panel). Docking of the crystal structures of mDia1 N-terminal DID-DD-CC region (residues 131-516) (Otomo et al. 2005) (blue) and Bni1 FH2 domain (residues 1350-1760) (Xu et al. 2004) (purple) into the 3D density of mDia1-FL (55-1255) (right panel). (B) Fourier shell correlation plot for the structure of mDia1-FL (55-1255), calculated using angular reconstitution method (AR) (solid line) and structure of same molecule, calculated using RCT, (dotted line), calculated between structures each containing one half of the data.