Abstract
The RidA (YjgF/YER057c/UK114) family of proteins is broadly conserved in the three domains of life yet the functional understanding of these proteins is at an early stage. Physiological studies of ridA mutant strains of Salmonella enterica provided a framework to inform in vitro studies and led to the description of a conserved biochemical activity for this family. ridA mutant strains of S. enterica have characteristic phenotypes including new synthesis of thiamine biosynthetic intermediate phosphoribosylamine (PRA), inability to grow on pyruvate as a sole carbon and energy source or when serine is present in the minimal growth medium, and a decreased specific activity of transaminase B (IlvE). Secondary mutations restoring growth to a ridA mutant in the presence of serine were in dapA (encoding dihydrodipicolinate synthase) and thrA (encoding homoserine dehydrogenase). These mutations suppressed multiple ridA mutant phenotypes by increasing the synthesis of threonine. The ability of threonine to suppress the metabolic defects of a ridA mutant is discussed in the context of recent biochemical data and in vivo results presented here.
Introduction
The RidA (formerly YjgF/YER057c/UK114) family of proteins is well conserved throughout the three domains of life. Members of this protein family have been implicated in a diverse number of phenotypes in a variety of organisms [1]–[12]. However, a common mechanism to explain these phenotypes was not obvious. Strains of Salmonella enterica lacking RidA display several characteristic phenotypes, including: synthesis of thiamine biosynthetic intermediate phosphoribosylamine (PRA), inability to grow on pyruvate as a sole carbon and energy source or in the presence of serine [13], and a decreased specific activity of transaminase B (IlvE) [14]. Each of these phenotypes required the presence of a functional threonine dehydratase (IlvA; EC 4.3.1.19). These phenotypic analyses in Salmonella enterica led to a general model in which RidA eliminated reactive products that were generated in normal metabolic reactions involving IlvA [5].
In vitro studies, which were informed by the phenotypic analyses, identified a biochemical function for the RidA protein family. RidA deaminated reactive enamine/imine metabolites generated by IlvA [15]. These enamine/imine compounds were normal intermediates in the pyridoxal-5′-phosphate-dependent dehydration of both threonine and serine. Further, reconstitution of the PRA formation phenotype required a short-lived intermediate produced by IlvA from threonine. This molecule, presumed to be the 2-aminocrotonate enamine was utilized by anthranilate phosphoribosyltransferase (TrpD; EC 2.4.2.18) to generate PRA [16]. RidA inhibited the formation of PRA in vitro by this mechanism, which was consistent with the phenotype observed only in a ridA mutant.
Aside from the IlvA-, TrpD-dependent formation of PRA, the in vivo consequences of a ridA mutation are not understood in the context of the biochemical activity of RidA. The in vitro biochemical work characterizing RidA did not address the significance of the enamine deaminase activity in vivo or relate the previously observed phenotypes to the in vitro activity. Herein suppressor analyses dissected the basis of the other phenotypes caused by the loss of RidA in vivo. The data showed that threonine reversed many of the phenotypes of a ridA mutant of S. enterica. We propose that threonine outcompetes serine for the active site of threonine dehydratase (IlvA) thus preventing the formation of a deleterious serine-derived reactive intermediate that is normally removed by RidA.
Materials and Methods
Bacterial Strains, Media, and Chemicals
Strains used in this study are derivatives of S. enterica serovar Typhimurium LT2 and are listed with their respective genotypes in Table 1.
Table 1. Bacterial strains.
| Strain | Relevant Genotype* | Source |
| DM3480 | ridA3::MudJ | Lab collection |
| DM3871 | ridA3::MudJ purF2085 | Lab collection |
| DM6309 | ridA3::MudJ purF2085 thrA1371 | This study |
| DM7608 | ridA3::MudJ ilvA3211 | [4] |
| DM7610 | ridA3::MudJ ilvA3210 | [4] |
| DM9404 | Wild type (isogenic to DM3480) | Lab collection |
| DM9521 | ridA3::MudJ dapA356 zxx4116::Tn10d(Tc) | This study |
| DM10009 | ridA3::MudJ ilvY3212::Tn10d(Tc) ilvA3210 | [4] |
| DM10010 | ridA3::MudJ ilvY3212::Tn10d(Tc) | [4] |
| DM10331 | ilvY3212::Tn10d(Tc) ilvA3210 | [4] |
| DM10332 | ilvY3212::Tn10d(Tc) | [4] |
| DM10460 | dapA362::cat | This study |
| DM11412 | ridA3::MudJ purF2085 dapA356 | This study |
| DM11558 | ilvY3212::Tn10d(Tc) ilvA3211 | [4] |
| DM11609 | ridA3::MudJ thrA1371 stm0014-13::Tn10d(Tc) | [4] |
| DM11635 | ridA3::MudJ dapA357 | This study |
| DM11636 | ridA3::MudJ dapA358 | This study |
| DM11637 | ridA3::MudJ dapA356 | This study |
| DM11638 | ridA3::MudJ dapA361 | This study |
| DM11639 | ridA3::MudJ dapA359 | This study |
| DM11640 | ridA3::MudJ dapA360 | This study |
| DM11877 | ridA3::MudJ thrA1371 stm0014-13::Tn10d(Tc) | This study |
| DM11878 | ridA3::MudJ stm0014-13::Tn10d(Tc) | This study |
| λ3520 | ΔasdA1 zhf4::Tn10 | R. Curtiss III [36] |
No-carbon E medium (NCE), supplemented with 1 mM MgSO4 [17], trace minerals [18], and 11 mM glucose (or 50 mM pyruvate as indicated) was used as minimal medium. Difco nutrient broth (8 g/L) with NaCl (5 g/L) was used as rich (NB) medium. Luria broth was used for experiments involving plasmid isolation. Super Broth containing tryptone (32 g/L), yeast extract (20 g/L), NaCl (5 g/L), and NaOH (5 mM) was used to grow cultures for protein purification. Difco BiTek agar was added (15 g/L) for solid medium. When present in the culture medium the final concentrations of serine and isoleucine were 5 and 0.3 mM, respectively. The final concentrations of the antibiotics in rich and minimal medium, respectively, were: tetracycline, 20, 10 mg/L, chloramphenicol, 20, 5 mg/L, and ampicillin, 150, 15 mg/L. Unless otherwise noted, all chemicals were from Sigma-Aldrich. Aspartate 4-semialdehyde was custom synthesized commercially at the University of Canterbury by the Gerrard Laboratory.
Growth Quantification
Cells from overnight cultures in NB medium were pelleted and resuspended in an equal volume of saline (0.85% NaCl), and an aliquot (0.2 mL) was used to inoculate 5 mL of the appropriate minimal medium. Cell growth was monitored as optical density (OD) at 650 nm over time at 37°C with shaking. Growth rates (in h−1) were determined as µ = ln(X/X0)/T where X = OD at 650 nm and T = time in hours during logarithmic growth.
Genetic Techniques
Transductional crosses were performed using the high-frequency general transducing mutant of bacteriophage P22 (HT105/1, int-201) [19]. Methods for transductional crosses, purification from phage, and identification of phage-free transductants have been described elsewhere [20]. Multiply-mutant strains were constructed using standard genetic techniques. When necessary, genetic backcrosses were performed to confirm the presence of a respective allele.
To isolate mutants, independent cultures of ridA3::MudJ (DM3480) were grown overnight in NB, centrifuged, and resuspended in the same volume of saline. 107 cells were spread on solid minimal glucose medium with 5 mM serine. Spontaneously arising mutations (∼10−7) that allowed ridA mutants to grow on serine were isolated after 36 hours at 37°C. A transposon (Tn10d(Tc)) genetically linked to the causative mutation in one strain was isolated by standard genetic techniques and used to reconstruct the mutant for phenotypic confirmation. The chromosomal location of relevant insertions was determined by sequencing using a PCR-based protocol [21]. A DNA product was amplified with degenerate primers and primers derived from the Tn10d(Tc) insertion sequence and sequenced at the University of Wisconsin Biotechnology Center. Strains carrying suppressor mutations were reconstructed by transducing the relevant allele into dapA::cat (DM10460) and selecting for growth without diaminopimelic acid.
Molecular Techniques
The dapA genes from strains DM3480, DM7604, DM7606, and DM11019 were amplified by PCR using Herculase II Fusion DNA Polymerase (Stratagene) and primers 5′ DapANdeI (GGGGCATATGTTCACGGGAAGTATTC) and 3′DapAXhoI (GGGGCTCGAGTTACAGCAGGCCAGC) and cloned into the pET20b vector (Novagen) at NdeI and XhoI restriction sites. Sequence analysis of each clone confirmed the presence of the N-terminal hexahistidine tag and the relevant lesion. The construct carrying the wild-type allele (pLD-dapA) complemented a dapA mutant (DM10460), indicating that the gene was expressed in this construct (data not shown).
Protein Purification
The wild-type and variant DapA proteins were overexpressed in E. coli BL21(AI) according to the manufacturer’s protocol (Invitrogen). Cells from the resulting cultures were broken at 15,000 psi in a French Pressure cell at 4°C. Cell debris was removed by centrifugation (42,000×g) for 30 min at 4°C. Proteins were purified using a column containing Ni-NTA superflow resin (QIAGEN) according to manufacturer’s protocol. Fractions containing DapA were concentrated at 30 psi under Argon gas using a 10,000 Da molecular weight cut-off membrane (Amicon). The protein was dialyzed in 0.5 M NaCl, 20 mM Tris-HCl, 5 mM imidazole, pH 7.9 and stored at −80°C. DapB was purified according to standard protocol using a hexahistidine-tagged dapB clone from the ASKA collection [22]. IlvE was purified as a hexahistidine-tagged protein as has been described [14]. Protein concentration was estimated with bovine serum albumin as the standard using a Bradford assay [23].
Biochemical Assays
i) Dihydrodipicolinate synthase (DapA) assay
DapA activity was measured in a coupled assay with DapB (dihydrodipicolinate reductase; E.C. 1.3.1.26) following a published protocol [24]. A typical 1 mL reaction contained ∼2 µg DapB, 100 mM HEPES pH 8.0, 0.125 mM NADPH, 40 mM pyruvate, and 0.05–1.0 µg DapA (>95% pure) and was initiated by the addition of ASA at concentrations ranging from 0–2 mM. Enzyme-dependent oxidation of NADPH was quantified at 340 nm.
ii) Threonine dehydratase (IlvA) assay
IlvA was assayed as previously described [4], [25], or alternatively, by quantification of [14C]-2-ketobutyrate (2-KB) formed from [14C(U)]-L-threonine. 200 µL reactions containing 100 mM Tris pH 8.0, 50 µM pyridoxal-5′-phosphate, 20 mM ammonium chloride, 1 mM dithiothreitol (DTT), and 2 µg purified IlvA were initiated with a final concentration of 40 mM [14C(U)]-L-Threonine (12.5 µCi mmol−1), incubated for 12 minutes at 37°C, and stopped with 0.5 mL 0.1% 2,4-dinitrophenylhydrazine in 2 N HCl. Derivatized [14C]-2-KB was extracted with 0.5 mL toluene and radioactivity from 200 µL toluene phase, representing quantity of [14C]-2-KB generated, was counted in 5 mL scintillation fluid using a scintillation counter (Packard).
iii) Transaminase B (IlvE) assay
The transaminase B activity assay was based on previously described protocols [14], [26]. Cells were permeabilized by sonication. Known concentrations of product 2-keto-3-methylvalerate were subjected to the extraction procedure to generate a standard curve.
iv) Homoserine dehydrogenase (ThrA) assay
The homoserine dehydrogenase activity assay was adapted from a previously described protocol [27]. Cells were grown in 100 mL minimal medium to an OD650 nm of 0.8, pelleted, and resuspended in 0.5 mL 100 mM HEPES pH 8.0 with 0.125 mM DTT. Cells were disrupted by sonication, extract was clarified by centrifugation, and total protein concentration was estimated by the method of Bradford [23]. Assay mixtures contained 100 mM HEPES pH 8.0, 0.125 mM DTT, 200 mM potassium chloride, 0.3 mM NADP+, and ∼300 µg cell extract, in a final volume of 200 µL. Assays were initiated by the addition of 15 mM homoserine and activity was monitored by the increase in absorbance at 340 nm at 30°C, representing NADPH production. Inhibitor L-threonine was added to a final concentration of 0.5 mM when indicated.
Results
Alleles of dapA Restore Growth of a ridA Mutant Strain on Glucose Serine
A ridA null mutant (DM3480) cannot grow on minimal glucose medium in the presence of 5 mM serine [13]. Six independent mutant derivatives of ridA that grew in the presence of serine were isolated. Using Tn10d(Tc) insertions to map the location of the mutations, each of the causative mutations was subsequently found to affect the dapA locus, encoding dihydrodipicolinate synthase (EC 4.2.1.52). Table 2 summarizes the six lesions that allowed growth of the ridA mutant in the presence of serine. Four lesions generated variant DapA proteins (DapAA563G was isolated twice), one affected the Shine-Dalgarno sequence and one was in the dapA promoter. Strains with each of the mutant alleles were reconstructed (DM11635–40) and were analyzed in liquid media for growth in the presence of serine. ridA mutant strains containing alleles dapA356, dapA357, or dapA358 grew similar to a wild-type strain in the presence of serine and are represented by strain DM11637 in Figure 1. The parent ridA strain (DM3480) failed to grow after 12 hours as expected. The strain carrying a lesion 36 nucleotides upstream of dapA (DM11640) had limited growth with serine and was concluded to decrease transcription of the dapA gene. (The promoter of dapA from E. coli resides within a 70-base region upstream of dapA containing an extended −10 and −35 site [28].) Growth of the suppressor-containing strains, with the exception of strain ridA dapA359 (DM11639), was indistinguishable from the parental strain on minimal glucose medium (data not shown). The dapA359 allele encoded a variant with two deleted amino acid residues and despite growth on solid medium with serine, growth was not detected in liquid media after 24 hours in the absence of exogenous diaminopimelic acid (DAP).
Table 2. Suppressing DapA variants have decreased specific activities.
| Strain | Allele* | DNA change | Proteinchange | Specificactivity† |
| DM9404 | WT | – | – | 5.10±1.60 |
| DM11637 | dapA356 | A563G | D188G | 0.12±0.04 |
| DM11635 | dapA357 | C143T | S48F | 0.15±0.04 |
| DM11636 | dapA358 | A(−10)T | – | N.D.‡ |
| DM11637 | dapA359 | ΔG249–C254 | ΔE84–A85 | 0.02± <0.01 |
| DM11640 | dapA360 | T(−36)C | – | N.D. |
| DM11638 | dapA361 | A563G | D188G | N.D. |
A ridA strain carrying any of the listed alleles is able to grow in the presence of serine.
Specific activity of DapA in µmol NADPH oxidized/sec/mg of purified protein.
N.D. = not determined.
Figure 1. Mutations in dapA restore growth to ridA mutants in the presence of serine.
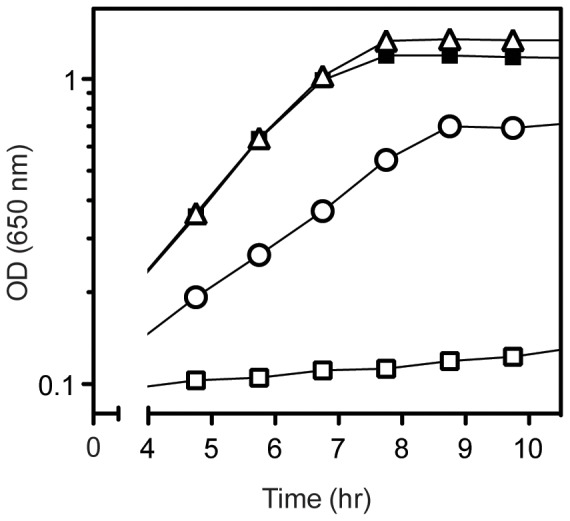
Growth was monitored over time as optical density at 650 nm. Strains were grown at 37°C in minimal glucose medium with no additions (closed symbols) or 5 mM serine (open symbols). Shown are strains ridA (DM3480), squares; ridA dapA356 (DM11637), triangles; and ridA dapA360 (DM11640), circles. Curves displayed were representative of 3 biological replicates.
Suppressor Alleles of dapA Encode Variants with Decreased Specific Activity
The wild-type gene and each of three suppressor alleles of dapA were cloned into the pET20b vector to generate C-terminal hexahistidine tagged proteins, creating pLD-dapA, pLD-dapAD188G, pLD-dapAS48F and pLD-dapAΔ84–85. The recombinant proteins were purified by affinity chromatography. Wild-type and variant proteins were assayed for dihydrodipicolinate synthase activity using a coupled assay [24]. The variant proteins all had more than a 30-fold decrease in specific activity when compared to the wild-type protein, as shown in Table 2.
A simple interpretation of the above results was that decreased activity of DapA allowed growth of a ridA mutant in the presence of serine. Complementation analysis eliminated the formal possibility that an altered function of DapA was responsible for allowing growth of a ridA mutant. When provided in trans, wild-type dapA eliminated growth of the ridA dapA356 mutant strain in the presence of serine and did not affect growth of a ridA mutant (data not shown).
Aspartate 4-semialdehyde Accumulation Mediated Phenotypic Suppression by the dapA Alleles
DapA functions in the synthesis of some aspartate-derived amino acids and uses aspartate 4-semialdehyde (ASA) as a substrate (Figure 2). In one scenario, a recessive lesion in dapA results in accumulation of ASA that allows a ridA mutant to grow in the presence of serine. ASA itself restored the growth of a ridA mutant in the presence of serine, supporting a role for this molecule in suppression of the ridA phenotype. As little as 0.5 mM ASA in the medium allowed a ridA mutant to reach full density in medium with 5 mM serine. Growth rate (µ) of ridA (DM3480) in the presence of serine (µ = 0.06±0.01) was restored by 1 mM ASA (µ = 0.55±0.03) and was the same as the growth rate of the same strain grown on minimal medium without serine (µ = 0.54±0.03). The nutritional requirements of an asd mutant (methionine, lysine, DAP, and threonine), which cannot make ASA, [29] were satisfied with ∼1.3 mM exogenous ASA, indicating the cells have the ability to transport and incorporate ASA into the biosynthetic pathways (data not shown).
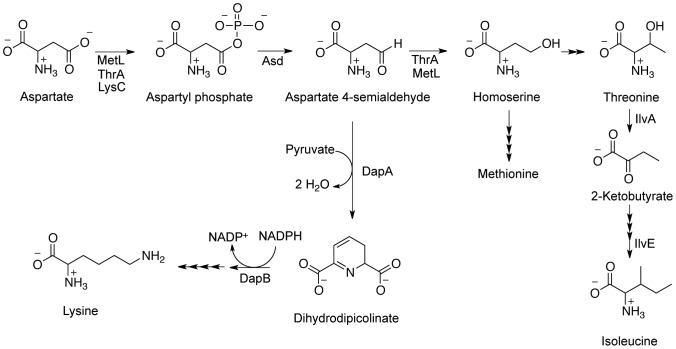
Figure 2. Pathway for synthesis of aspartate-derived amino acids.
Aspartate is a precursor to lysine, methionine, threonine, and isoleucine, as depicted here. Aspartate 4-semialdehyde (ASA) is a branchpoint metabolite controlled by the activities of DapA, ThrA, and MetL.
In addition to suppressing serine sensitivity, the dapA alleles restored IlvE activity in a ridA mutant. The IlvE activity in the ridA strain carrying the dapA356 allele (230±7 nmol/min/mg) was restored to an intermediate level between the wild-type (303±13 nmol/min/mg) and ridA mutant strain (140±7 nmol/min/mg). This result suggested intracellular accumulation of ASA could impact the activity of IlvE in a ridA mutant. No evidence of a direct role for ASA in mediating phenotypic suppression was found. The activity of purified IlvE was not significantly affected by 10 min incubation with 10 mM ASA (26.1±7 µmol/min/mg without ASA, 18.6±6 µmol/min/mg with ASA). Further, ASA had no detectable effect on the activity of threonine deaminase (IlvA) in vitro. While as little as 500 µM isoleucine inhibited IlvA, ASA failed to inhibit IlvA in vitro at a range of concentrations (0.1 µM –1.0 mM) (data not shown). These data showed that the effect of ASA was not due to mimicking the effect of isoleucine as a feedback inhibitor [14], and suggest that further metabolism of this molecule was required.
Analysis of a Second Suppressor Locus Provides Insight into Role of ASA
In addition to the alleles of dapA described above, a mutation in thrA (thrA1371), encoding aspartokinase I/homoserine dehydrogenase I, previously reported to suppress serine sensitivity of a ridA mutant [4] was sequenced and found to encode variant ThrAG403D. The homoserine dehydrogenase activity in a strain with the ThrAG403D variant was indistinguishable from the wild-type parental strain. The location of the G403D substitution suggested the variant could be altered in allosteric interaction properties [30]–[32]. Data in Table 3 showed that the homoserine dehydrogenase activity of the ThrAG403D variant was resistant to inhibition by threonine. Significantly, this effect was evident at a low of concentration of threonine, as would be expected under in vivo conditions where the threonine concentration was reported to be 0.2 mM [33]. Taken together, the data suggested the ThrAG403D variant could increase conversion of ASA to homoserine in vivo, consistent with the above conclusion that metabolism of ASA is required for suppression.
Table 3. The ThrAG403D variant is insensitive to feedback inhibition by threonine and serine.
| Homoserine dehydrogenaseactivity* | |||
| thrA allele | Protein variant | No inhibitor | + Thr (0.5 mM) |
| thrA WT | WT | 44±5 | 18±3 |
| thrA1371 | ThrAG403D | 37±5 | 38±6 |
Homoserine dehydrogenase activity was measured in crude extracts from isogenic strains DM11877 (ridA thrA1371) and DM11878 (ridA) by following reduction of NADP+ and was reported as ΔA420 nm/min/µg protein.
Threonine, not Isoleucine is the Metabolite Responsible for Suppression
ASA is a biosynthetic precursor to isoleucine, which is known to allow a ridA mutant to grow in the presence of serine [13], so it was a formal possibility that ASA was correcting growth by leading to increased levels of isoleucine. Two IlvA variants with decreased threonine dehydratase activity were used to constrict flux between ASA and isoleucine. Neither of the ilvA alleles caused a detectable growth defect on minimal glucose medium (Table 4). However, they each resulted in derepression of the ilv operon [4] indicating the strains were limited for isoleucine. Despite the constriction of flux between ASA and isoleucine, the double mutants ridA ilvA3210 (DM10009) and ridA ilvA3211 (DM11558) had the same growth rates as a ridA mutant (DM10010) (µ = 0.53±0.10, 0.54±0.04, and 0.56±0.01, respectively) when grown in a minimal medium containing 5 mM serine and 1 mM ASA. These data suggested that ASA did not correct growth by increasing intracellular isoleucine levels.
Table 4. IlvA variants have reduced activity.
| ilvA allele | Proteinvariant | Activity* | µ†(Glc) | µ†(Glc Ile) |
| ilvA WT | WT | 0.22±0.01 | 0.54±0.05 | 0.53±0.01 |
| ilvA3210 | IlvAA142T | B.D.‡ | 0.62±0.01 | 0.60±0.03 |
| ilvA3211 | IlvAG191S | 0.05±0.01 | 0.56±0.03 | 0.54±0.01 |
Threonine dehydratase (IlvA) activity measured in crude extracts from DM3480 (ridA), DM7610 (ridA ilvA3210) and DM7608 (ridA ilvA3211) and reported as ΔA540 nm/min/mg protein.
Growth rate (in h−1) (µ = ln(X/X0)/T where X = optical density at 650 nm and T = time in hours during logarithmic growth) for strains DM10332 (WT), DM10331 (ilvA3210), and DM11558 (ilvA3211) determined from growth in minimal medium with glucose (Glc) and glucose with isoleucine (Glc Ile).
Below Detection.
Other metabolites in the pathway from ASA to the branch chain amino acids were considered and tested for their ability to suppress growth of a ridA mutant with serine. Nutritional tests showed qualitative suppression of multiple phenotypes with both homoserine and threonine. Addition of exogenous threonine to the growth medium of a ridA mutant restored growth on serine (µ = 0.09±0.01 without threonine, 0.50±0.01 with threonine), growth on pyruvate (µ = 0.06±0.01 without threonine, 0.37±0.02 with threonine), and IlvE activity (160±31 nmol/min/mg in minimal medium without threonine versus 287±33 nmol/min/mg in minimal with threonine).
Threonine is a precursor in PRA formation in a ridA mutant [16]. This fact provided a means to directly test whether the suppressor mutations in dapA and thrA generated increased cellular threonine levels. If the dapA and thrA mutations acted by increasing flux to threonine, they would be expected to increase the PRA formed in a ridA mutant. A purF mutant strain background was used to detect PRA, as it requires PRA to make thiamine and allow growth. The data in Figure 3 showed that the thrA and dapA suppressors increased growth of a purF ridA strain, and exogenous threonine further increased growth. These results supported the conclusion that flux to threonine was increased by these mutations. Additionally, since isoleucine has been shown to have the opposite effect and inhibit PRA synthesis in a ridA mutant [13], these data were consistent with the interpretation that the dapA mutations were not increasing the synthesis of isoleucine. Considering the results of nutritional and suppressor analyses in total, threonine was identified as the metabolite that had a direct effect in suppressing the phenotypes caused by lack of RidA.
Figure 3. Suppressor mutations increase growth in purF ridA strain background.
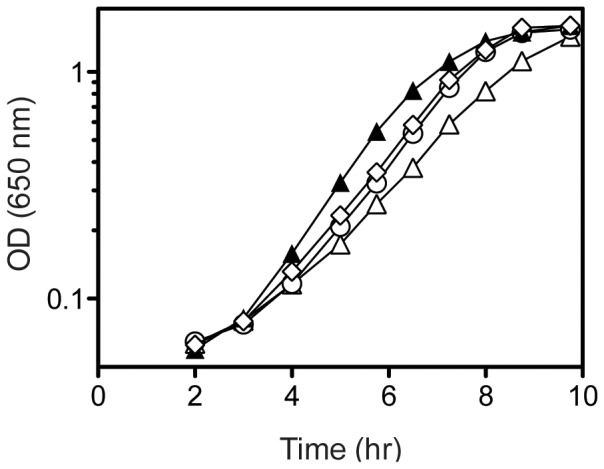
Strains were grown at 37°C in minimal glucose medium with adenine (open symbols) or further supplemented with 0.3 mM threonine (closed symbols). Growth was monitored over time as optical density at 650 nm. Shown are strains purF ridA (DM3871), triangles; purF ridA thrA1371 (DM6309), diamonds; and purF ridA dapA356 (DM11412), circles. Error bars represent standard deviations of three biological replicates.
Discussion
The RidA (YjgF/YER057c/UK114) family of proteins is highly conserved, but the diverse cellular defects caused by its absence are not understood [1]–[11]. Recently it was shown in vitro that RidA family members deaminate reactive enamine/imine intermediates generated by threonine dehydratases (e.g., IlvA) [15]. This study investigated the relationship between the characterized biochemical activity of RidA and the in vivo phenotypes observed in a ridA mutant in S. enterica. Suppressor analyses identified an important role for threonine in attenuating multiple phenotypes of a ridA strain, including sensitivity to exogenous serine, lack of growth on pyruvate, and a decreased specific activity of IlvE.
When considering the results of this study in combination with the biochemical activity of RidA, we proposed a mechanism by which threonine could suppress the mutant phenotypes. Our model predicted that threonine relieved the sensitivities of a ridA mutant by outcompeting serine in the IlvA active site. Threonine dehydratase (IlvA) was required for a number of ridA phenotypes [4], [13], [14], [16]. The fact that threonine reversed those phenotypes suggested the metabolic defects required IlvA to use a different substrate. To our knowledge, the only other reported physiological substrate of IlvA is serine, and IlvA has a much higher Km for serine than for threonine (90 mM versus 4.5 mM, respectively [34]). Threonine and serine use the same active site in IlvA [35], and the presence of additional threonine would preclude IlvA from binding and dehydrating serine instead. This model suggested that the intermediate derived from serine, but not threonine, was deleterious to the cell unless it was removed by RidA.
The significance of threonine as a key metabolite that can modulate the ridA serine-sensitivity phenotype was further emphasized by the saturation of the suppressor analyses. Repeated attempts to isolate serine-resistant mutants only produced the decreased activity dapA (dihydrodipicolinate synthase) alleles and the feedback-resistant thrA (homoserine dehydrogenase) allele described here. These mutants not only demonstrated that increased flux to threonine was key to reversing the serine-sensitivity of a ridA mutant, but they also suggested that the primary control of threonine levels in the cell occurs at the homoserine dehydrogenase step and can be affected by increasing substrate (ASA) or decreasing the allosteric control of ThrA. This finding has important implications for metabolic engineering and groups endeavoring to generate organisms that overproduce threonine or downstream metabolites.
The findings herein emphasized the central role of threonine in compensating for the lack of RidA. In combination with past results, these data refine a model to explain the phenotypes of ridA mutants. It has been shown that IlvA generates reactive enamine/imines that are removed by RidA [15]. We suggest that serine is used as a substrate by IlvA to generate a reactive intermediate that attacks cellular components if it is not quenched by RidA. This is in contrast to the reactive intermediate derived from threonine reported to serve as a substrate for an alternative mechanism of PRA synthesis [16]. Thus, the IlvA-generated intermediates that accumulate in vivo in the absence of RidA can have either deleterious or productive consequences, depending on the substrate used (e.g.,serine versus threonine). Together these results suggest a complex role for IlvA in the in vivo phenotypes of ridA mutants. Continued studies are needed to identify the diversity of both the reactive metabolites eliminated by RidA and the targets of these reactive intermediates to better understand the breadth of metabolic consequences that result from the lack of the conserved RidA protein.
Acknowledgments
We thank Dr. George Schmitz for isolating the ridA suppressor mutants in the presence of serine and for the initial characterization of the thrA1371 allele, Benjamin Bice for assaying IlvA variants, and Rebecca Schomer for performing the IlvE assay in the presence of threonine.
Funding Statement
This work was supported by competitive grants GM47296 and GM95837 from the National Institutes of Health (NIH) to DMD and a National Science Foundation Graduate Research Fellowship to MRC. JAL was supported as a trainee on the Molecular Biosciences Training Grant from the NIH (T32 GM07215). DD was the recipient of undergraduate research fellowships from the Department of Bacteriology, the Hilldale Competition, and Merck and Co. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schmiedeknecht G, Kerkhoff C, Orso E, Stohr J, Aslanidis C, et al. (1996) Isolation and characterization of a 14.5-kDa trichloroacetic-acid-soluble translational inhibitor protein from human monocytes that is upregulated upon cellular differentiation. Eur J Biochem 242: 339–351. [DOI] [PubMed] [Google Scholar]
- 2. Oxelmark E, Marchini A, Malanchi I, Magherini F, Jaquet L, et al. (2000) Mmf1p, a novel yeast mitochondrial protein conserved throughout evolution and involved in maintenance of the mitochondrial genome. Mol Cell Biol 20: 7784–7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim JM, Yoshikawa H, Shirahige K (2001) A member of the YER057c/YjgF/UK114 family links isoleucine biosynthesis and intact mitochondria maintenance in Saccharomyces cerevisiae . Genes Cells 6: 507–517. [DOI] [PubMed] [Google Scholar]
- 4. Christopherson MR, Schmitz GE, Downs DM (2008) YjgF is required for isoleucine biosynthesis when Salmonella enterica is grown on pyruvate medium. J Bacteriol 190: 3057–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Browne BA, Ramos AI, Downs DM (2006) PurF-independent phosphoribosyl amine formation in yjgF mutants of Salmonella enterica utilizes the tryptophan biosynthetic enzyme complex anthranilate synthase-phosphoribosyltransferase. J Bacteriol 188: 6786–6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goupil-Feuillerat N, Cocaign-Bousquet M, Godon JJ, Ehrlich SD, Renault P (1997) Dual role of alpha-acetolactate decarboxylase in Lactococcus lactis subsp. lactis. J Bacteriol 179: 6285–6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farkas A, Nardai G, Csermely P, Tompa P, Friedrich P (2004) DUK114, the Drosophila orthologue of bovine brain calpain activator protein, is a molecular chaperone. Biochem J 383: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leitner-Dagan Y, Ovadis M, Zuker A, Shklarman E, Ohad I, et al. (2006) CHRD, a plant member of the evolutionarily conserved YjgF family, influences photosynthesis and chromoplastogenesis. Planta 225: 89–102. [DOI] [PubMed] [Google Scholar]
- 9. Marchini A, Accardi R, Malanchi I, Schyr E, Oxelmark E, et al. (2002) Schizosaccharomyces pombe Pmf1p is structurally and functionally related to Mmf1p of Saccharomyces cerevisiae . Yeast 19: 703–711. [DOI] [PubMed] [Google Scholar]
- 10. Morishita R, Kawagoshi A, Sawasaki T, Madin K, Ogasawara T, et al. (1999) Ribonuclease activity of rat liver perchloric acid-soluble protein, a potent inhibitor of protein synthesis. J Biol Chem 274: 20688–20692. [DOI] [PubMed] [Google Scholar]
- 11. D’Inca R, Marteil G, Bazile F, Pascal A, Guitton N, et al. (2010) Proteomic screen for potential regulators of M-phase entry and quality of meiotic resumption in Xenopus laevis oocytes. J Proteomics 73: 1542–1550. [DOI] [PubMed] [Google Scholar]
- 12. Kim KS, Pelton JG, Inwood WB, Andersen U, Kustu S, et al. (2010) The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems. J Bacteriol 192: 4089–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Enos-Berlage JL, Langendorf MJ, Downs DM (1998) Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J Bacteriol 180: 6519–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmitz G, Downs DM (2004) Reduced transaminase B (IlvE) activity caused by the lack of yjgF is dependent on the status of threonine deaminase (IlvA) in Salmonella enterica serovar Typhimurium. J Bacteriol 186: 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lambrecht JA, Flynn JM, Downs DM (2012) Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5′-phosphate (PLP)-dependent enzyme reactions. J Biol Chem 287: 3454–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lambrecht JA, Browne BA, Downs DM (2010) Members of the YjgF/YER057c/UK114 family of proteins inhibit phosphoribosylamine synthesis in vitro . J Biol Chem 285: 34401–34407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vogel HJ, Bonner DM (1956) Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218: 97–106. [PubMed] [Google Scholar]
- 18. Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS (1979) Methanogens: reevaluation of a unique biological group. Microbiol Rev 43: 260–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmieger H (1972) Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet 119: 75–88. [DOI] [PubMed] [Google Scholar]
- 20. Downs DM, Petersen L (1994) apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium . J Bacteriol 176: 4858–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caetano-Anolles G (1993) Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl 3: 85–94. [DOI] [PubMed] [Google Scholar]
- 22. Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, et al. (2005) Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12: 291–299. [DOI] [PubMed] [Google Scholar]
- 23. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 24. Yugari Y, Gilvarg C (1965) The condensation step in diaminopimelate synthesis. J Biol Chem 240: 4710–4716. [PubMed] [Google Scholar]
- 25.Burns RO (1971) L-Threonine deaminase–biosynthetic (Salmonella typhimurium). Methods Enzymol. 555–560.
- 26. Duggan DE, Wechsler JA (1973) An assay for transaminase B enzyme activity in Escherichia coli K-12. Anal Biochem 51: 67–79. [DOI] [PubMed] [Google Scholar]
- 27. Angeles TS, Smanik PA, Borders C Jr, Viola RE (1989) Aspartokinase-homoserine dehydrogenase I from Escherichia coli: pH and chemical modification studies of the kinase activity. Biochemistry 28: 8771–8777. [DOI] [PubMed] [Google Scholar]
- 28. Acord J, Masters M (2004) Expression from the Escherichia coli dapA promoter is regulated by intracellular levels of diaminopimelic acid. FEMS Microbiol Lett 235: 131–137. [DOI] [PubMed] [Google Scholar]
- 29. Jagusztyn-Krynicka EK, Smorawinska M, Curtiss R 3rd (1982) Expression of Streptococcus mutans aspartate-semialdehyde dehydrogenase gene cloned into plasmid pBR322. J Gen Microbiol 128: 1135–1145. [DOI] [PubMed] [Google Scholar]
- 30. Szczesiul M, Wampler DE (1976) Regulation of a metabolic system in vitro: synthesis of threonine from aspartic acid. Biochemistry 15: 2236–2244. [DOI] [PubMed] [Google Scholar]
- 31. Omori K, Imai Y, Suzuki S, Komatsubara S (1993) Nucleotide sequence of the Serratia marcescens threonine operon and analysis of the threonine operon mutations which alter feedback inhibition of both aspartokinase I and homoserine dehydrogenase I. J Bacteriol. 175: 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paris S, Viemon C, Curien G, Dumas R (2003) Mechanism of control of Arabidopsis thaliana aspartate kinase-homoserine dehydrogenase by threonine. J Biol Chem 278: 5361–5366. [DOI] [PubMed] [Google Scholar]
- 33. Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, et al. (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli . Nat Chem Biol 5: 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burns RO, Hofler JG, Luginbuhl GH (1979) Threonine deaminase from Salmonella typhimurium. Substrate-specific patterns of inhibition in an activator site-deficient form of the enzyme. J Biol Chem 254: 1074–1079. [PubMed] [Google Scholar]
- 35. Hofler JG, Burns RO (1978) Threonine deaminase from Salmonella typhimurium. Effect of regulatory ligands on the binding of substrates and substrate analogues to the active sites and the differentiation of the activator and inhibitor sites from the active sites. J Biol Chem 253: 1245–1251. [PubMed] [Google Scholar]
- 36. Galan JE, Nakayama K, Curtiss R 3rd (1990) Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene 94: 29–35. [DOI] [PubMed] [Google Scholar]
- 37. Castilho BA, Olfson P, Casadaban MJ (1984) Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol 158: 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Way JC, Davis MA, Morisato D, Roberts DE, Kleckner N (1984) New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32: 369–379. [DOI] [PubMed] [Google Scholar]