Abstract
Screening for gene mutations in CDH23, which has many exons, has lagged even though it is likely to be an important cause for hearing loss patients. To assess the importance of CDH23 mutations in non-syndromic hearing loss, two-step screening was applied and clinical characteristics of the patients with CDH23 mutations were examined in this study. As a first screening, we performed Sanger sequencing using 304 probands compatible with recessive inheritance to find the pathologic mutations. Twenty-six possible mutations were detected to be pathologic in the first screening. For the second screening, using the probes for these 26 mutations, a large cohort of probands (n = 1396) was screened using Taqman amplification-based mutation analysis followed by Sanger sequencing. The hearing loss in a total of 52 families (10 homozygous, 13 compound heterogygous, and 29 heterozygous) was found to be caused by the CDH23 mutations. The majority of the patients showed congenital, high frequency involved, progressive hearing loss. Interestingly, some particular mutations cause late onset moderate hearing loss. The present study is the first to demonstrate the prevalence of CDH23 mutations among non-syndromic hearing loss patients and indicated that mutations of the CDH23 gene are an important cause of non-syndromic hearing loss.
Introduction
Mutations in the CDH23 (NM_22124) gene are known to be responsible for both Usher syndrome type ID (USH1D) and non-syndromic hearing loss (DFNB12) [1], [2]. Molecular confirmation of CDH23 mutations has become important in the diagnosis of these conditions.
This gene encodes cadherin 23, a protein of 3354 amino acids with 27 extracellular (EC) domains, a single transmembrane domain and a short cytoplasmic domain. Cadherin-specific amino acid motifs such as DRE, DXNDN, and DXD, that are highly conserved in sequence and spacing and required for cadherin dimerization and calcium binding were found in each extracelluar domain [3].
The cadherin 23 protein is known to be an important composition of the tip link that maintains the arrangement of streocilia [4].
More than 50 mutations have been reported for the Usher phenotype (USH1D) and 24 mutations reported for the non-syndromic hearing loss phenotype (DFNB12) [1], [2], [5]–[7]. As suggested by genotype–phenotype correlation study, Usher 1D, which has congenital profound hearing impairment, vestibular dysfunction, and retinitis pigmentosa, is usually associated with nonsense mutations, whereas DFNB12, which has a milder phenotype, is associated with missense mutations [1], [2], [5]–[8].
We previously reported that four pathologic mutations were identified in 5 out of 64 Japanese families compatible with autosomal recessive inheritance, suggesting that CDH23-caused deafness may be commonly found among non-syndromic hearing loss patients [6]. GJB2 has been shown to be a common gene involved in congenital hearing impairment. SLC26A4 is also frequently involved among those patients. GJB2 and SLC26A4 are comparatively small making Sanger sequencing relatively easy. The latter is also associated with the typical inner ear anomaly, enlarged vestibular aqueduct. Therefore, screening is relatively easy and many studies have focused on just these two genes. Clinical molecular diagnosis has been dramatically improved for these genes. However, screening strategy of other hearing loss genes is difficult and Sanger sequencing of the candidate genes, such as CDH23, with many exons is time consuming. Consequently, only a few reports are available for the mutation spectrum of CDH23.
In the present study, we performed Sanger sequencing using 304 patients whose pedigrees are compatible with recessive inheritance to find additional pathologic mutations. Also, to find the novel pathologic mutations and to clarify the frequency and clinical characteristics of patients with CDH23 mutations, a large cohort of probands from unrelated families (n = 1396) was screened using TaqMan amplification-based mutation analysis of the variants observed in the initial 304 patients.
Results
The first screening using 304 Japanese probands compatible with autosomal recessive inheritance identified 26 candidates for disease causing mutations. These include four previously reported pathologic mutations: p.P240L, p.R301Q, p.Q1716P, and p.R2029W, as well as 6 possible pathologic variants in the coding region of CDH23. All of the mutations were missense mutations. The following second screening based on TaqMan assay followed by Sanger sequencing confirmed 10 “possibly pathologic” mutations (Table 1) and 17 variants with uncertain pathogenicity (Table 2) in a large cohort of the patients. “Possible pathologic” mutations were defined as 1) mutations found to be homozygotes or compound heterozygotes (and determined by segregation study), 2) variants which were not found or were very few in 192 control subjects, 3) amino acids that were well-conserved among various species, 4) compatible with next generation sequencing database, and 5) compatible with the predicted effect of missense mutations on CDH23 protein function. Results of the compatibility of the next generation sequence database, the SIFT and PolyPhen2 score for prediction are shown in Tables 1 and 2.
Table 1. Possible pathologic variants found in this study.
| Amino acid change | Nucleotide change | EXON | Domain | Evolutionary conservation | The highly conserved calcium-binding elements | Number in probands (n = 1396) | Allele frequency in patients (in 2792 allele) | Allele frequency in control (in 384 allele) | Allele frequency in HL patients based on a Next generation sequencing database (in 432 allele) | Allele frequency in controls based on a Next generation sequencing database (in 144 allele) | PolyPhen 2 score* | SIFT Score* | Reference | ||
| homozygote | compound heterozygote | heterozygote | |||||||||||||
| p.P240L | c.719C>T | 7 | EC3 | 7 | - | 7 | 12 | 19 | 1.612 | 0.260 | 0.63 | 0.67 | 0.999 | 0.06 | Wagatsuma et al. |
| p.R301Q | c.902G>A | 9 | EC3 | 7 | DRE | - | 3 | - | 0.107 | 0.260 | 0 | 0 | 1.000 | 0 | Wagatsuma et al. |
| p.E956K | c.2866G>A | 25 | EC9 | 7 | DRE | - | 1 | 2 | 0.107 | 0 | 0.21 | 0 | 1.000 | 0.04 | this study |
| p.T1368M | c.4103C>T | 32 | EC13 | 7 | - | - | 1 | - | 0.036 | 0 | 0 | 0 | 1.000 | 0 | this study |
| p.R1417W | c.4249C>T | 35 | EC13 | 5 | - | 1 | - | 2 | 0.143 | 0 | 0.25 | 0 | 0.998 | 0.19 | Wagatsuma et al. |
| p.D1626A | c.4877A>C | 39 | EC15 | 7 | DXNDN | - | 1 | - | 0.036 | 0 | 0 | 0 | 0.999 | 0.01 | this study |
| p.Q1716P | c.5147A>C | 39 | EC16 | 7 | - | - | 3 | - | 0.107 | 0 | 0 | 0 | 0.957 | 0.3 | Wagatsuma et al. |
| p.R2029W | c.6085C>T | 46 | EC19 | 7 | DRE | 2 | 2 | 6 | 0.430 | 0 | 0 | 0 | 0.999 | 0.01 | Wagatsuma et al. |
| p.N2287K | c.6861T>G | 50 | EC21 | 7 | DXNDN | - | 2 | - | 0.072 | 0 | 0 | 0 | 0.971 | 0 | this study |
| p.E2438K | c.7312G>A | 52 | EC23 | 6 | - | - | 1 | - | 0.036 | 0 | 0 | 0 | 0.986 | 1 | this study |
Computer analysis to predict the effect of missense variants on CDH23 protein function was performed with Sorting Intolerant from Tolerant (SIFT; http://sift.jcvi.org/), and Polymorphism Phenotyping (PolyPhen2;http://genetics.bwh.harvard.edu/pph2/).
Table 2. Variants with uncertain pathogenicity found in this study.
| Amino acid change | Nucleotide change | EXON | Domain | Evolutionary conservation | The highly conserved calcium-binding elements | Number in probands (n = 1396) | Allele frequency in patients (in 2792 allele) | Allele frequency in control (in 384 allele) | Allele frequency in HL patients based on a Next generation sequencing database (in 432 allele) | Allele frequency in controls based on a Next generation sequencing database (in 144 allele) | PolyPhen 2 score*** | SIFT Score*** | Reference | ||
| homozygote | compound heterozygote | heterozygote | |||||||||||||
| p.D160N | c.478G>A | 4 | EC2 | 7 | DXD | - | - | 2 | 0.072 | 0.260 | 0 | 0 | 1.000 | 0 | this study |
| p.V803I | c.2407G>A | 23 | EC8 | 7 | - | - | - | 3 | 0.107 | 0 | 0 | 0 | 0.761 | 0.41 | this study |
| p.S1415I | c.4244G>T | 35 | EC13 | 7 | - | - | - | 1 | 0.036 | 0 | 0 | 0 | 0.840 | 0.06 | this study |
| p.A1443G * | c.4328C>G | 35 | EC14 | 7 | - | 1* | - | 2 | 0.143 | 0 | 0.2 | 0 | 0.944 | 0.06 | this study |
| p.R1588W ** | c.4762C>T | 38 | EC15 | 7 | - | 4** | - | 18 | 0.931 | 0.260 | 2.22 | 0 | 1.000 | 0.01 | Wagatsuma et al. |
| p.V1711I | c.5131G>A | 40 | EC16 | 7 | - | - | - | 2 | 0.072 | 0 | 0 | 0 | 0.970 | 0.12 | Wagatsuma et al. |
| p.V1807M | c.5419G>A | 42 | EC17 | 5 | - | - | 1 | - | N/A | 0.260 | 0 | 0 | 0.054 | 0.22 | this study |
| p.S1876N | c.5627G>A | 43 | EC18 | 5 | - | - | - | 6 | 0.215 | 0 | 0 | 0 | 0.981 | 0.26 | Wagatsuma et al. |
| p.V1908I | c.5722G>A | 44 | EC9 | 5 | - | - | - | 12 | 0.430 | 0.260 | 1.09 | 0.53 | 0.948 | 1 | Wagatsuma et al. |
| p.A2130V | c.6389C>T | 48 | EC20 | 6 | - | - | - | 1 | 0.036 | 0 | 0 | 0 | 0.999 | 0.24 | this study |
| p.R2171C | c.6511C>T | 48 | EC20 | 7 | DXNDNR | - | - | 1 | 0.036 | 0.521 | 0 | 0 | 0.999 | 0.11 | Wagatsuma et al. |
| p.Q2227P | c.6680A>C | 48 | EC21 | 6 | - | - | - | 1 | 0.036 | 0.260 | 0 | 0 | 0.930 | 0.2 | Wagatsuma et al. |
| p.L2473P | c.7418T>C | 53 | EC23 | 7 | - | - | - | 1 | 0.036 | 0 | 0 | 0 | 0.999 | 0 | Wagatsuma et al. |
| p.I2669V | c.8005A>G | 56 | EC25 | 5 | - | - | - | 1 | 0.036 | 0 | 0 | 0 | 0.134 | 0.7 | Wagatsuma et al. |
| p.F2801V | c.8401T>G | 59 | EC26 | 5 | - | - | - | 1 | 0.036 | 0.781 | 1.52 | 1.27 | 0.800 | 0.01 | Wagatsuma et al. |
| p.G2912S | c.8734G>A | 61 | EC27 | 7 | - | - | - | 1 | 0.036 | 0 | 0.23 | 0 | 0.996 | 0 | this study |
| p.R3175C | c.9523C>T | 68 | CYTO | 7 | - | - | - | 1 | 0.036 | 0.260 | 0 | 0 | 0.886 | 0.01 | Wagatsuma et al. |
not confirmed by segregation study.
one normal hearing subject with homozygotes.
Computer analysis to predict the effect of missense variants on CDH23 protein function was performed with Sorting Intolerant from Tolerant (SIFT; http://sift.jcvi.org/), and Polymorphism Phenotyping (PolyPhen2;http://genetics.bwh.harvard.edu/pph2/).
N/A: TaqMan probe not available.
The 17 variants found as heterozygous and therefore with uncertain pathogenicity did not fulfill all the above criteria. For example, p.A1443G was uncertain because DNA samples from family members were not available and we could not confirm its pathogenicity by segregation study. p.R1588W was found to be homozygous in 4 patients and heterozygous in 16 patients, but only 1 was found in 384 control alleles. However, a member of the patient's family (#2841) showed normal hearing instead of being homozygous. Also p.V803I, p.V1807M and p.I2669V are obscure from the functional prediction analysis.
In one family (#4685), three mutation were found in proband and two of them were found in same allele p.[D16126A;V1807M] confirmed by segregation analysis.
As p.V1807M predicted to have no effect on CDH23 structure, p.D1626A might be a pathogenic mutation.
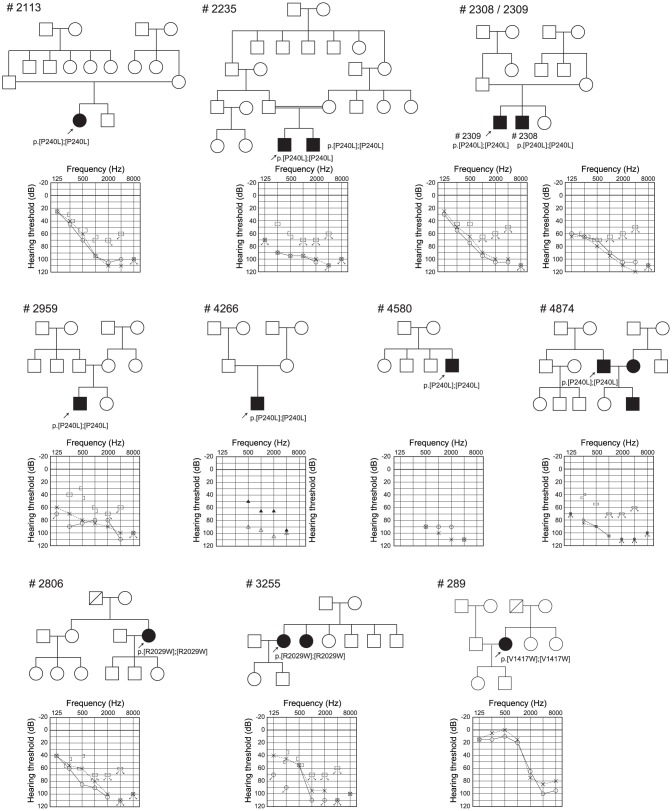
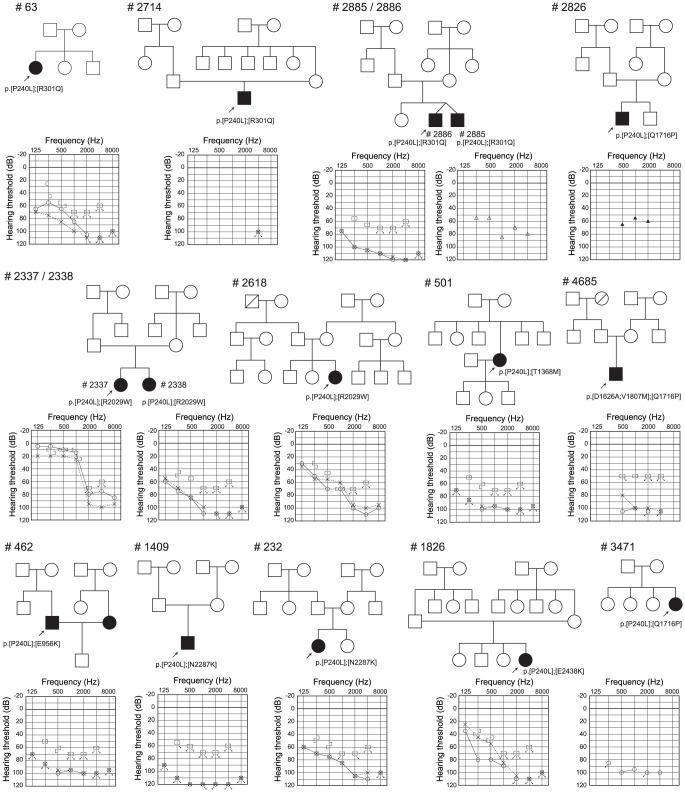
For 10 possible pathologic mutations, amino acids were well-conserved among various species, including Homo sapiens, P. troglodytes, B. traurus, M. musculus, R. norvegicus, G. gallus, and D. rario. Many mutations (5 out of 10 possible pathologic mutations, 2 out of 17 uncertain variants) were found in DRE, DXNDN, and DXD motif (Table 1 and 2). Ten possible pathologic mutations were found to be either homozygotes (n = 11, Table 3, Fig. 1) or compound heterozygotes (n = 15) (Table 4, Fig. 2). Twenty-nine patients were found to be heterozygous without a second mutation (Table 5).
Table 3. Details of phenotype and genotype of 11 patients in 10 families with homozygous CDH23 mutation.
| Sample No | relationship | Amino acid Change | Hereditary form | Threshold* (Rt)(dB) | Threshold* (Lt)(dB) | severity | Residual hearing in the lower frequencies** (dB) | Hearing in the higher frequencies*** (dB) | Age | Age of awareness | Progressiveness | Hearing aid/cochlear implant | Vertigo | Tinnitus |
| #2113 | p.[P240L];[P240L] | sporadic | 91.3 | 90 | severe | 44.2 | 104.2 | 12 | 6 | + | HA | − | − | |
| #2235 | p.[P240L];[P240L] | AR | 97.5 | 96.3 | profound | 85.0 | 104.2 | 22 | 0 | − | HA | − | − | |
| #2308 | p.[P240L];[P240L] | AR | 88.8 | 95 | severe | 67.5 | 110.0 | 11 | 0**** | − | HA | − | − | |
| #2309 | sibling of #2308 | p.[P240L];[P240L] | AR | 92.5 | 86.3 | severe | 50.0 | 105.0 | 9 | 0**** | − | HA | − | − |
| #2959 | p.[P240L];[P240L] | sporadic | 81.3 | 85 | severe | 75.8 | 96.7 | 8 | 0**** | − | HA | − | − | |
| #4266 | p.[P240L];[P240L] | sporadic | 96.3 | 96.3 | severe | 70.0 | 91.3 | 3 | 0**** | + | CI | − | − | |
| #4580 | p.[P240L];[P240L] | sporadic | 102.5 | 97.5 | profound | 88.3 | 106.7 | 1 | 0**** | − | CI | − | N/A | |
| #4874 | p.[P240L];[P240L] | sporadic | 102.5 | 102.5 | profound | 80.8 | 106.7 | 38 | 2 | + | HA | − | − | |
| #2806 | p.[R2029W];[R2029W] | sporadic | 92.5 | 80 | severe | 56.7 | 104.2 | 53 | 48 | + | HA | − | + | |
| #3255 | p.[R2029W];[R2029W] | AR | 96.3 | 85 | severe | 59.2 | 104.2 | 71 | 60 | + | HA | − | + | |
| #289 | p.[V1417W];[V1417W] | sporadic | 31.3 | 26.3 | mild | 10.0 | 85.0 | 34 | 14 | + | HA | − | − |
average of 500, 1000, 2000 and 4000 Hz.
average of 125, 250, and 500 Hz.
average of 2000, 4000, and 8000 Hz.
found by newborn hearing screening.
Figure 1. Pedigrees, mutations, and audiograms of the patients with homozygous CDH23 mutations.
Table 4. Details of phenotype and genotype of 15 patients in 13 families with compound heterozygous CDH23 mutation.
| Sample No | relationship | Amino acid Change | Hereditary form | Threshold* (Rt)(dB) | Threshold* (Lt)(dB) | severity | Residual hearing in the lower frequencies** (dB) | Hearing in the higher frequencies*** (dB) | Age | Age of awareness | Progressiveness | Hearing aid/cochlear implant | Vertigo | Tinnitus |
| #63 | p.[P240L];[R301Q] | sporadic | 85 | 98.8 | severe | 69.2 | 105.8 | 27 | 0 | − | HA | − | + | |
| #2714 | p.[P240L];[R301Q] | sporadic | 97.5 | 97.5 | profound | 71.7 | 105.0 | 2 | 0**** | + | HA | − | − | |
| #2885 | p.[P240L];[R301Q] | AR | 90 | 108.7 | profound | 55.0 | 75.0 | 13 | 3 | + | CI | − | − | |
| #2886 | sibling of #2885 | p.[P240L];[R301Q] | AR | 115 | 110 | profound | 93.3 | 115.8 | 13 | 2 | + | CI | − | − |
| #2337 | p.[P240L];[R2029W] | AR | 30 | 41.3 | mild | 13.3 | 88.3 | 13 | 11 | + | HA | − | + | |
| #2338 | sibling of #2337 | p.[P240L];[R2029W] | AR | 103.8 | 98.8 | profound | 71.7 | 106.7 | 8 | 2 | + | HA | − | − |
| #2618 | p.[P240L];[R2029W] | sporadic | 77.5 | 67.5 | moderate | 49.2 | 100.0 | 8 | 3 | + | CI | − | − | |
| #2826 | p.[P240L];[Q1716P] | sporadic | 91.3 | 95 | profound | 66.7 | 112.5 | 6 | 0 | + | HA | − | − | |
| #3471 | p.[P240L];[Q1716P] | sporadic | 97.5 | 97.5 | profound | 92.5 | 100.0 | 4 | 0 | − | CI | − | − | |
| #462 | p.[P240L];[E956K] | sporadic | 97.5 | 97.3 | profound | 84.2 | 98.3 | 38 | 10 | − | HA | − | − | |
| #501 | p.[P240L];[T1368M] | sporadic | >90 | >90 | profound | N/A | N/A | 68 | 44 | + | HA | + | + | |
| #1409 | p.[P240L];[N2287K] | sporadic | 120 | 120 | profound | 107.5 | 123.3 | 17 | 0 | + | HA | − | − | |
| #232 | p.[P240L];[N2287K] | sporadic | 87.5 | 86.3 | severe | 67.5 | 104.2 | 15 | 0 | − | HA | − | + | |
| #1826 | p.[P240L];[E2438K] | sporadic | 91.3 | 106.3 | severe | 70.8 | 105.8 | 11 | 3 | + | HA | − | − | |
| #4685 | p.[D1626A;V1807M];[Q1716P] | sporadic | 97.5 | 103.8 | severe | 96.3 | 105.0 | 1 | 0* | − | CI | − | N/A |
average of 500, 1000, 2000 and 4000 Hz.
average of 125, 250, and 500 Hz.
average of 2000, 4000, and 8000 Hz.
found by newborn hearing screening.
Figure 2. Pedigrees, mutations, and audiograms of the patients with compound heterozygous CDH23 mutations.
Table 5. Details of phenotype and genotype of 29 patients with heterozygous CDH23 mutation.
| Sample No | relationship | Amino acid Change | Hereditary form | Threshold* (Rt)(dB) | Threshold* (Lt)(dB) | severity | Residual hearing in the lower frequencies** (dB) | Hearing in the higher frequencies*** (dB) | Age | Age of awareness | Progressiveness | Hearing aid/cochlear implant | Vertigo | Tinnitus |
| #334 | p.[P240L];[-] | AD | 96.25 | 83.75 | severe | 63.3 | 96.7 | 23 | 0 | + | HA | N/A | + | |
| #340 | p.[P240L];[-] | sporadic | >90 | >90 | profound | N/A | N/A | 54 | 14 | + | HA | N/A | N/A | |
| #569 | p.[P240L];[-] | sporadic | 86.25 | 90 | severe | 75.0 | 98.3 | 26 | 3 | + | HA | − | − | |
| #653 | p.[P240L];[-] | sporadic | 53.75 | 57.5 | moderate | 44.2 | 71.7 | 36 | 33 | + | HA | − | + | |
| #754 | p.[P240L];[-] | sporadic | 110 | 101.25 | profound | 87.5 | 104.2 | 57 | 0 | + | HA | N/A | N/A | |
| #1039 | p.[P240L];[-] | sporadic | 48.75 | 56.25 | moderate | 33.3 | 74.2 | 76 | 76 | − | HA | + | − | |
| #1598 | p.[P240L];[-] | sporadic | 56.25 | 10 | unilateral | 34.2 | 41.7 | 60 | 49 | − | − | + | + | |
| #1807 | p.[P240L];[-] | sporadic | 110 | 8.75 | unilateral | 50.8 | 60.0 | 50 | 9 | − | − | − | − | |
| #1846 | p.[P240L];[-] | AD | 100 | 96.25 | profound | 83.3 | 98.3 | 62 | 6 | + | HA | + | + | |
| #2159 | p.[P240L];[-] | AR | 67.5 | 66.25 | moderate | 60.0 | 69.2 | 10 | 65 | + | HA | − | − | |
| #2374 | p.[P240L];[-] | AR | 86.25 | 90 | severe | 78.3 | 78.3 | 5 | 0 | − | HA | − | − | |
| #2835 | p.[P240L];[-] | sporadic | 85 | 91.25 | severe | 65.8 | 101.7 | 12 | 3 | + | HA | + | − | |
| #3492 | p.[P240L];[-] | AD | 103.75 | 103.75 | profound | 88.8 | 107.5 | 1 | 0 | − | HA | − | − | |
| #3499 | p.[P240L];[-] | AD | 96.25 | 110 | severe | 84.2 | 105.8 | 57 | 50 | − | CI | − | + | |
| #3761 | p.[P240L];[-] | AR | 32.5 | 40 | mild | 43.3 | 75.8 | 71 | 0 | − | − | − | + | |
| #4040 | p.[P240L];[-] | AR | S/O | S/O | profound | S/O | S/O | 2 | 0 | + | HA | − | − | |
| #4159 | p.[P240L];[-] | AR | 97.5 | 71.25 | severe | 71.7 | 95.0 | 38 | 38 | + | HA | + | + | |
| #4313 | p.[P240L];[-] | AD/Mit | 130 | 102.5 | profound | 107.5 | 116.7 | 6 | 0 | − | CI | − | − | |
| #4615 | p.[P240L];[-] | sporadic | 90 | 90 | profound | 90.0 | 90.0 | 0 | 0**** | − | CI | − | − | |
| #265 | p.[E956K];[-] | sporadic | 110 | 6.25 | unilateral | 57.5 | 59.2 | 16 | 0 | − | − | − | − | |
| #3116 | p.[E956K];[-] | AD | 47.5 | 53.75 | moderate | 58.3 | 40.8 | 63 | N/A | + | HA | − | + | |
| #280 | p.[R1417W];[-] | sporadic | 110 | 6.25 | unilateral | 50.0 | 55.8 | 8 | 3 | − | − | N/A | N/A | |
| #2649 | p.[R1417W];[-] | sporadic | 95 | 110 | profound | 87.5 | 105.0 | 11 | 0 | + | CI | − | N/A | |
| #1131 | p.[R2029W];[-] | sporadic | 73.75 | 72.5 | severe | 55.0 | 93.3 | 24 | 17 | + | HA | − | − | |
| #1539 | p.[R2029W];[-] | AD | 53.75 | 110 | moderate | 70.0 | 83.3 | 71 | 60 | + | HA | − | + | |
| #1618 | p.[R2029W];[-] | sporadic | 26.25 | 61.25 | mild | 31.7 | 60.8 | 67 | N/A | − | − | − | + | |
| #1919 | p.[R2029W];[-] | AD | 38.75 | 36.25 | mild | 20.8 | 75.0 | 25 | 3 | + | − | N/A | N/A | |
| #2271 | p.[R2029W];[-] | AD | 58.75 | 62.5 | moderate | 41.7 | 50.0 | 6 | N/A | N/A | HA | N/A | N/A | |
| #4138 | p.[R2029W];[-] | AR | 71.25 | 53.75 | moderate | 50.8 | 65.8 | 10 | 3 | + | HA | + | − |
average of 500, 1000, 2000 and 4000 Hz.
average of 125, 250, and 500 Hz.
average of 2000, 4000, and 8000 Hz.
found by newborn hearing screening.
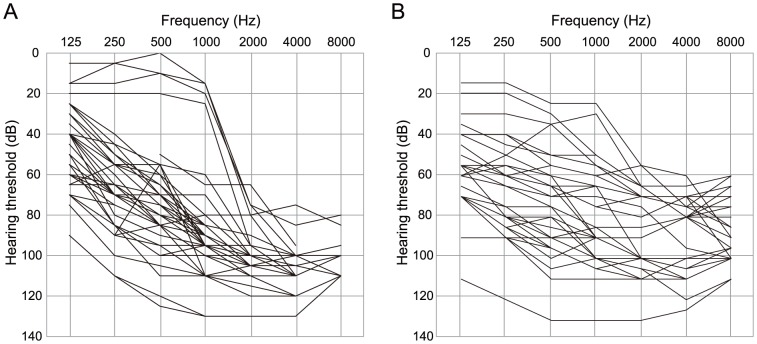
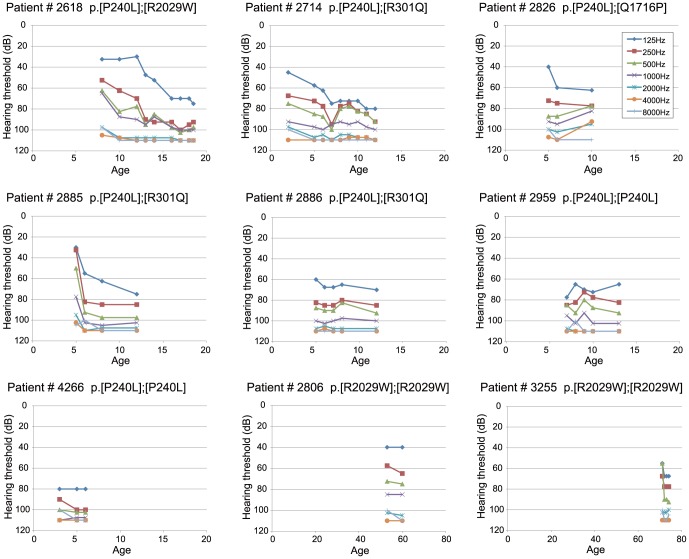
Tables 3 and 4 summarize 23 families with hearing loss caused by the CDH23 mutations (homozygous or compound heterozygous cases) and Table 5 summarizes 29 families with hearing loss potentially caused by the CDH23 mutations (heterozygous cases). The frequency was 1.6% (23/1396) or 2.1% (29/1396) of the overall hearing loss population. When restricted to patients compatible with recessive inheritance, the frequency was increased to 2.5% (23/919) or 3.2% (29/919). Table 3, 4 and 5 also summarize clinical characteristics including hereditary form, hearing threshold, severity, residual hearing in the lower frequencies, hearing in the higher frequencies, onset age (age of awareness), progressiveness of hearing loss, use of hearing aid/cochlear implantation, visual impairment, and vestibular symptoms. The ages of these patients were from 1 to 71 years. Age of onset (awareness of hearing loss) ranged from congenital to 60 years old, though the majority was congenital or early onset. There were some correlations between genotype and phenotype (onset age). The patients associated with p.P240L showed congenital and severe hearing loss regardless of whether associated with one more mutation, whereas the patients with p.R2029W or p.T1368M showed late-onset moderate hearing loss (Tables 3 and 4). Concerning type of hearing loss, the majority of the patients had some residual hearing in the lower frequencies, and overlapping audiograms showed characteristic high frequency involved hearing loss (Fig. 3). The majority of the patients showed progressive nature of hearing loss evaluated by serial audiogram (Fig. 4). No patients had associated visual impairment or vestibular symptoms (Tables 3, 4 and 5). Seven patients received cochlear implantation due to the insufficient amplification of hearing aids (Tables 3, 4 and 5).
Figure 3. Overlapping audiograms of the patients with CDH 23 mutations.
A: patients with hearing loss caused by the CDH23 mutations (homozygous or compound heterozygous cases), B: patients potentially caused by the CDH23 mutations (heterozygous cases).
Figure 4. Hearing progression of the patients with CDH23 mutations.
Note that the high frequency portion was already worsened, and the low frequency portion was deteriorated by ages.
Discussion
Mutations in the CDH23 gene are known to be responsible for both Usher syndrome type ID (USH1D) as well as non-syndromic hearing loss (DFNB12), and molecular confirmation of CDH23 mutations is clinically important for diagnosis of these conditions. However, clinical application of the detection of CDH23 mutations has lagged because of the size of the gene. Especially for DFNB12, which is not associated with visual impairment, screening is comparatively difficult, and therefore, little is known about frequencies among the hearing loss population as well as clinical characteristics.
In this study, we have applied two-step screening and identified a significant number of novel pathologic mutations of CDH23 responsible for non-syndromic hearing loss in a large cohort of patients. All of the possible pathologic mutations identified in this study (Table 1) were missense mutations, being consistent with previous reports that DFNB12 patients associated with missense mutations have milder hearing impairment than in USH1D, which is associated with nonsense, splice-site, or frameshift mutations [2], [5]–[7]. None had visual impairment, also supporting this rule. That the majority was found in the EC domain with only one exception found in the cytoplasmic domain, was also in line with the previous reports on DFNB12 [2], [5]–[7]. Of these 26 mutations, five out of 10 possible pathologic mutations were found in DRE, DXNDN, and DXD motifs, which are thought to be important for calcium binding property. These highly conserved EC calcium binding motifs are thought to be essential for linearization, rigidification, and dimerization of the cadherin molecules [9], [10]. And the results of computer analysis to predict the impact of amino acid change, all of 10 possible pathologic mutations predicted to cause a severe damage for protein function of CDH23.
As a result, 26 patients (from 23 families) had two mutations (in a homozygous or compound heterozygous state), and met criteria for recessive inheritance. A hallmark of recessive mutations is the detection of two mutations in the paternal and maternal alleles and the parents having normal hearing. As seen in previous mutation screening reports, including those for CDH23 [6], [7] as well as GJB2 and SLC26A4 [11], [12], we encountered a significant number of heterozygous cases without a second mutation even after direct sequencing of the coding region of the gene. Possible explanations are: 1) the existence of a second mutation in the intron or regulatory region of CDH23, which has not been explored, 2) the observed mutations are rare polymorphisms, 3) the screening method fails to detect the second mutation, and 4) an additional modulatory gene may contribute to hearing loss (for example, PCDH15). Although we have not reached the final conclusion, it is most likely that these heterozygous cases are also related to CDH23 mutations because: 1) allele frequencies are found to be higher in the hearing loss group (Table 2), and 2) the phenotype is similar to that of the patients with two mutations. As shown in Fig. 3, overlapping audiograms of the patients with only one mutation was similar to that with the patients with two mutations (high frequency involved sensorineural hearing loss with residual hearing at the lower frequencies).
Based on the frequencies of 3.7% (including heterozygous cases) of the hearing loss population and 5.7% (including heterozygous cases) of the recessive inherited cases in this study, we confirmed that mutations of CDH23 are an important cause for non-syndromic hearing loss and should be borne in mind next to GJB2 or SLC26A4 screening. This study revealed that p.P240L account for nearly 43.3%(45/104) of all CDH23 mutated families in Japan. Common mutations, such as c.35delG or c.235delC in GJB2 or p.H723R in the SLC26A4 gene, have been reported in many recessive deafness genes, and usually they are population-specific [12]–[14]. It is an interesting question whether p.P240L is frequent because of a founder effect or mutational hot spot, but the existence of such a common mutation makes mutation screening easier. Additional frequent mutations found in this study together with TaqMan procedures will facilitate genetic testing for deafness patients.
Concerning mutation spectrum, as in our previous report [6], the CDH23 mutation spectrum in Japanese is very different from that found in Caucasians and may be representative of those in Eastern Asian populations. Its elucidation is expected to facilitate the molecular diagnosis of DFNB12 and USH1D. It has also been known that prevalent GJB2 mutations are highly ethnic-specific (see The connexin-deafness homepage; http://davinci.crg.es/deafness/): c.35delG is common in the Caucasoid population, c.167delT was reported as prevalent in Ashkenazi Jews, p.R143W in a restricted village in Africa, and c.235delC in East Asian populations. A series of studies proved a founder effect for these frequent mutations [11], [15].
In the present study, using a large cohort of patients, clinical characteristics (onset age, progression, audiograms) of patients with CDH23 mutations were clarified.
Concerning genotype/phenotype correlations, hearing of the patients with p.[P240L];[P240L] is worse than in those with the other mutations, and tends to be congenital and severe. In contrast, the patients with p.[R2029W];[R2029W] showed a milder phenotype of middle age onset. Overlapping audiograms showed typical high frequency involved sensorineural hearing loss with residual hearing at the lower frequencies.
Concerning age of onset (awareness of hearing loss), the majority was congenital or early onset. But rather later-onset was seen in three patients (#2806, 3255, 501), and they were associated with some particular mutations (p.R2029W and p.T1368M). Their phenotype was rather mild and gradually progressive. It is interesting to note that their phenotype was similar to presbycusis. Actually, CDH23 mutations have been reported as responsible for age-related hearing loss in mice [16], [17].
Progressive nature of hearing loss and the presence of residual hearing are particular phenotypic features of the patients with CDH23 mutations. Our previous genetic analysis for the patients with high frequency involved hearing loss successfully identified CDH23 mutations [18]. Seven patients received cochlear implantation and showed good performance after implantation. For the patients with residual hearing, newly developed cochlear implantation; EAS (Electric Acoustic Stimulation) is a good therapeutic option and therefore much attention should be paid to the etiology when considering individual intervention, i.e., regular cochlear implantation or EAS. Genetic testing will be very important prognostic information together with various hearing tests.
In conclusion, a large cohort study using Taqman amplification-based mutation analysis indicated that mutations of the CDH23 gene are important causes of non-syndromic hearing loss. A mutation screening strategy using TaqMan assay based on the ethnic-specific frequent mutations is a powerful and effective method for such a large gene. Clinical characteristics of patients with CDH23 mutations is that hearing loss is progressive, high frequency involved sensorineural hearing loss with residual hearing in the lower frequencies. Most cases are congenital but care is needed because some patients show presbycusis-like hearing loss. Cochlear implantation (including EAS) is a good therapeutic intervention for the patients with CDH23 mutations.
Materials and Methods
To identify additional pathologic CDH23 mutations, two-step screening was applied in this study. Subjects from independent families were collected from 33 ENT departments nationwide in Japan. All subjects gave prior informed consent for participation in the project, which was approved by the ethical committee of each hospital. Genomic DNA was isolated from peripheral blood by DNeasy Blood and Tissue Kit (QIAGEN, Düsseldorf, Germany) according to the manufacturer's procedure.
First screening (Direct sequencing)
First, we sequenced the CDH23 gene in 304 Japanese non-syndromic sensorineural hearing loss probands (including our previously reported 64 samples [6]) compatible with autosomal recessive inheritance or sporadic cases. None of the subjects had any other associated neurological signs, vestibular or visual dysfunction. Sanger sequencing was applied to these samples to find mutations responsible for deafness. Detailed procedures were described in our previous report [6]. 26 candidates for disease causing mutations were collected according to the following criteria; 1) non-synonymous variants, and 2) allele carrier rates were less than 2% in control subjects.
Second screening (TaqMan genotyping assay based screening and Direct sequencing)
For the second screening, probes of these 26 mutations selected in the first screening was applied for a custom TaqMan® SNP Genotyping Assays (Applied Biosystems, Foster City, CA) [19]. 1396 probands of sensorineural hearing loss patients including 304 probands used in the first screening were used for the second assay. Of them, 1347 had bilateral sensorineural hearing loss and 49 had unilateral sensorineural hearing loss. The inheritance composition of the subjects was as follows: 298 subjects from autosomal dominant or maternally inherited families (two or more generations affected); 919 subjects from autosomal recessive families (parents with normal hearing and two or more affected siblings) or subjects with sporadic deafness (compatible with recessive inheritance or non-genetic hearing loss); the rest had unknown inheritance mode. After TaqMan assay, Sanger sequencing was performed: 1) to confirm these mutations found in TaqMan genotyping assays, 2) to confirm whether mutations were homozygotes or heterozygote, and 3) in cases found in heterozygous state, direct sequencing of the coding region of the CDH23 was performed.
Controls
The control group consisted of 192 unrelated Japanese individuals without any noticeable hearing loss evaluated by auditory testing.
Next generation sequencing and computer analysis
To elucidate the allele frequency of 26 mutations, comparison was made between allele frequency found in 216 deafness patients and 72 controls based on a next generation sequencing database that is currently being established at Shinshu University (unpublished). In brief, exome sequencing was performed with SureSelect target DNA enrichment (Agilent Technologies, Santa Clara, CA) and Illumina GAIIx sequencing (Illumina, San Diego, CA) according to the manufacturers' procedures. In the SureSelect library, 76 already reported genes responsible for sensorineural hearing loss and syndromic hearing loss were contained. After base calling, sequence results were aligned with a bowtie program [20] and allele frequencies of each CDH23 mutation in patients and the control population were calculated. Computer analysis to predict the effect of missense variants on CDH23 protein function was performed with Sorting Intolerant from Tolerant (SIFT; http://sift.jcvi.org/), and Polymorphism Phenotyping (PolyPhen2; http://genetics.bwh.harvard.edu/pph2/) [21], [22].
Acknowledgments
We thank Dr. William J Kimberling for helpful comments. We would also like to thank A. C. Apple-Mathews for help in preparing the manuscript.
Funding Statement
This study was supported by a Health and Labour Sciences Research Grant for Comprehensive Research on Disability Health and Welfare from the Ministry of Health, Labour and Welfare of Japan (http://www.mhlw.go.jp/english/) (SU), by the Acute Profound Deafness Research Committee of the Ministry of Health, Labour and Welfare of Japan (http://www.mhlw.go.jp/english/) (SU), by a Health and Labour Sciences Research Grant for Research on Specific Diseases (Vestibular Disorders) from the Japanese Ministry of Health, Labour and Welfare (http://www.mhlw.go.jp/english/) (SU), and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan (http://www.mext.go.jp/english/) (SU). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bolz H, von Brederlow B, Ramírez A, Bryda EC, Kutsche K, et al. (2001) Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet 27: 108–112. [DOI] [PubMed] [Google Scholar]
- 2. Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, et al. (2001) Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23 . Am J Hum Genet 68: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rowlands TM, Symonds JM, Farookhi R, Blaschuk OW (2000) Cadherins: crucial regulators of structure and function in reproductive tissues. Rev Reprod 5: 53–61. [DOI] [PubMed] [Google Scholar]
- 4. Muller U (2008) Cadherins and mechanotransduction by hair cells. Curr Opin Cell Biol 20: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Astuto LM, Bork JM, Weston MD, Askew JW, Fields RR, et al. (2002) CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am J Hum Genet 71: 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wagatsuma M, Kitoh R, Suzuki H, Fukuoka H, Takumi Y, et al. (2007) Distribution and frequencies of CDH23 mutations in Japanese patients with non-syndromic hearing loss. Clin Genet 72: 339–344. [DOI] [PubMed] [Google Scholar]
- 7. Oshima A, Jaijo T, Aller E, Millan JM, Carney C, et al. (2008) Mutation profile of the CDH23 gene in 56 probands with Usher syndrome type I. Hum Mutat 29: E37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McHugh RK, Friedman RA (2006) Genetics of hearing loss: Allelism and modifier genes produce a phenotypic continuum. Anat Rec A Discov Mol Cell Evol Biol 288: 370–381. [DOI] [PubMed] [Google Scholar]
- 9. Nagar B, Overduin M, Ikura M, Rini JM (1996) Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature 380: 360–364. [DOI] [PubMed] [Google Scholar]
- 10. Angst BD, Marcozzi C, Magee AI (2001) The cadherin superfamily: diversity in form and function. J Cell Sci 114: 629–641. [DOI] [PubMed] [Google Scholar]
- 11. Tsukada K, Nishio S, Usami S (2010) A large cohort study of GJB2 mutations in Japanese hearing loss patients. Clin Genet 78: 464–470. [DOI] [PubMed] [Google Scholar]
- 12. Tsukamoto K, Suzuki H, Harada D, Namba A, Abe S, et al. (2003) Distribution and frequencies of PDS (SLC26A4) mutations in Pendred syndrome and nonsyndromic hearing loss associated with enlarged vestibular aqueduct: a unique spectrum of mutations in Japanese. Eur J Hum Genet 11: 916–922. [DOI] [PubMed] [Google Scholar]
- 13. Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, et al. (2003) Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet 40: 242–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Usami S, Wagatsuma M, Fukuoka H, Suzuki H, Tsukada K, et al. (2008) The responsible genes in Japanese deafness patients and clinical application using Invader assay. Acta Otolaryngol 128: 446–454. [DOI] [PubMed] [Google Scholar]
- 15. Van Laer L, Coucke P, Mueller RF, Caethoven G, Flothmann K, et al. (2001) A common founder for the 35delG GJB2 gene mutation in connexin 26 hearing impairment. J Med Genet 38: 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson KR, Erway LC, Cook SA, Willott JF, Zheng QY (1997) A major gene affecting age-related hearing loss in C57BL/6J mice. Hear Res 114: 83–92. [DOI] [PubMed] [Google Scholar]
- 17. Zheng QY, Yan D, Ouyang XM, Du LL, Yu H, et al. (2005) Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum Mol Genet 14: 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Usami S, Miyagawa M, Suzuki N, Moteki H, Nishio S, et al. (2010) Genetic background of candidates for EAS (Electric-Acoustic Stimulation). Audiologidal Medicine 8: 28–32.20497527 [Google Scholar]
- 19. de Kok JB, Wiegerinck ET, Giesendorf BA, Swinkels DW (2002) Rapid genotyping of single nucleotide polymorphisms using novel minor groove binding DNA oligonucleotides (MGB probes). Hum Mutat 19: 554–559. [DOI] [PubMed] [Google Scholar]
- 20. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 22. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]