Abstract
Gaf1 is the first GATA family zinc-finger transcription factor identified in Schizosaccharomyces pombe. Here, we report that Gaf1 functions as a negatively acting transcription factor of ste11+, delaying the entrance of cells exposed to transient nitrogen starvation into the meiotic cycle. gaf1Δ strains exhibited accelerated G1-arrest upon nitrogen starvation. Moreover, gaf1Δ mutation caused increased mating and sporulation frequency under both nitrogen-starved and unstarved conditions, while overexpression of gaf1+ led to a significant impairment of sporulation. By microarray analysis, we found that approximately 63% (116 genes) of the 183 genes up-regulated in unstarved gaf1Δ cells were nitrogen starvation-responsive genes, and furthermore that 25 genes among the genes up-regulated by gaf1Δ mutation are Ste11 targets (e.g., gpa1 +, ste4 +, spk1 +, ste11 +, and mei2 +). The phenotype caused by gaf1Δ mutation was masked by ste11Δ mutation, indicating that ste11+ is epistatic to gaf1+ with respect to sporulation efficiency, and accordingly that gaf1+ functions upstream of ste11+ in the signaling pathway governing sexual development. gaf1Δ strains showed accelerated ste11+ expression under nitrogen starvation and increased ste11+ expression even under normal conditions. Electrophoretic mobility shift assay analysis demonstrated that Gaf1 specifically binds to the canonical GATA motif (5′-HGATAR-3′) spanning from −371 to −366 in ste11+ promoter. Consequently, Gaf1 provides the prime example for negative regulation of ste11+ transcription through direct binding to a cis-acting motif of its promoter.
Introduction
The fission yeast Schizosaccharomyces pombe reproduces asexually by mitosis under favorable conditions. When haploid cells are starved of nutrients, particularly of nitrogen, they arrest the cell cycle at G1 and undergo sexual differentiation [1]. Cells of opposite mating types, h+ and h−, fuse to form a diploid zygote, which undergoes meiosis to give four haploid ascospores that remain dormant until they encounter favorable growth conditions [2]. The transition from the mitotic cell cycle into meiosis is tightly regulated by a network of positive and negative factors that are controlled at various levels of gene expression, from transcription initiation [3]–[6] to protein modification [7]–[11].
One important regulatory component of S. pombe sexual development is Ste11. Ste11 positively regulates transcription of the mating type genes, matP and matM, and the mei2+ gene, which is essential for commitment to meiosis, by binding to an upstream cis-acting element under conditions of nitrogen starvation [12]. ste11Δ mutants are completely defective in mating and sporulation, while ectopic expression of ste11+ leads to sexual differentiation irrespective of nutritional conditions. The activity of Ste11 is regulated by two antagonistic protein kinases, Pat1 and Spk1, via the pheromone signaling pathway [13] and by the TOR protein kinase, Tor2, which is activated in the presence of nitrogen and represses sexual differentiation by directly interfering with the function of Ste11 and Mei2 [11]. Expression of ste11+ is regulated by at least three different signal transduction pathways: mating pheromone signaling (RAS/MAPK pathway), cyclic AMP (cAMP)-dependent protein kinase A (PKA), and stress-activated protein kinase (SAPK) in conjunction with MAPKKKs (Wis4 and Win1), MAPKK (Wis1), and MAPK (Sty1/Spc1/Phh1) [14], [15]. So far, only positive regulatory factors of ste11+ expression, such as Atf1 [6], [16], Pcr1 [5], Rst2 [4], Prr1 [17], and Ste11 itself [4] have been reported. Furthermore, only Rst2 and Ste11 are transcription factors that directly activate ste11+ expression. No transcription factor that directly represses the expression of ste11+ has been identified.
Here, we explore the role of Gaf1, the first GATA transcription factor identified in S. pombe [18], [19], in the expression of ste11+. GATA family transcription factors have a wide range of functions, from terminal differentiation in vertebrates [20]–[22] to nitrogen metabolism, siderophore biosynthesis, photoinduction, and mating type switching in fungi [23]. In S. pombe, Ams2 is a cell cycle-regulated GATA factor that is required for centromere function [24] and Fep1/Gaf2 occupies a central role in iron homeostasis by coordinating the reductive and non-reductive iron transport systems [25], [26]. GATA factors recognize a six base-pair consensus sequence, 5′-HGATAR-3′ (where H can be A/C/T and R can be A/G), contained in the promoters of their target genes. Although the C-terminal fragment of Gaf1 (Gaf1565–855) has been shown to bind specifically to the GATA motif DAL7 UAS, a canonical GATA motif of Saccharomyces cerevisiae [18], little is known about the function of Gaf1 in S. pombe. We present evidence that Gaf1 down-regulates the transcription of ste11+ via direct binding to its promoter and consequently delay the shift of nitrogen-starved cells from the vegetative cycle to the meiotic cycle.
Materials and Methods
S. pombe strains, media, and general procedures
S. pombe strains used in this study are listed in Table 1. Cells were maintained on complete medium (YES) containing 0.5% yeast extract, 3% glucose, 2% Bacto agar, adenine (225 µg ml−1), leucine (225 µg ml−1), and uracil (225 µg ml−1). Edinburgh minimal medium (EMM2) [27]–[29] was used as a minimal selective medium. EMM-N (EMM2 without NH4Cl) was used for starvation of nitrogen source, and EMM-G (EMM2 containing 0.5% instead of 2% glucose) for glucose-restriction experiments. All the minimal media were supplemented with required auxotrophic nutrients (adenine, leucine, and uracil) at the concentrations of 225 µg ml−1 each, which led to the presence of a starved amount of organic nitrogen source in EMM-N. Thiamine was added to the medium at a final concentration of 20 µM to repress expression from the thiamine-repressible nmt42+ (no message in thiamine) promoter. Transformation was performed by the lithium acetate procedure [27]. Standard techniques were used for genetic manipulation and analysis [29].
Table 1. S. pombe strains used in this study.
| Strain | Relevant genotype | Source or reference |
| 972 | h− | Lab stock |
| ED005 | h− ade6-M210 leu1–32 | Lab stock |
| ED665 | h− ade6-M210 leu1–32 ura4-D18 | Lab stock |
| ED668 | h+ ade6-M216 leu1–32 ura4-D18 | Lab stock |
| JY4 | h90 ade6-M216 leu1–32 ura4-D18 | Lab stock |
| JZ396 | h90 ade6-M216 leu1–32 ura4-D18 ste11::ura4+ | [12] |
| KL210 | h− ade6-M210 leu1–32 ura4-D18 gaf1::kanMX | This work |
| KL211 | h90 ade6-M216 leu1–32 ura4-D18 gaf1::kanMX | This work |
| KL213 | h90 ade6-M216 leu1–32 ura4-D18 gaf1::kanMX ste11::ura4+ | This work |
| KL216 | h+ ade6-M216 leu1–32 ura4-D18 gaf1::kanMX | This work |
| KL230 | h− gaf1::kanMX | This work |
| KL240 | h− ade6-M210 leu1–32 ura4-D18 gaf1::hphMX | This work |
| KL416 | h+ ade6-M216 leu1–32 ura4-D18 ste11::kanMX | This work |
Construction of plasmids
To construct pREP-Gaf1 with a full-length open reading frame (ORF) of gaf1+ downstream of the nmt42+ promoter, a 2.6-kb fragment was amplified by polymerase chain reaction (PCR) with the following primers: P1 (5′AACCCGGGCCATGGATCTAAAGTTTTCC3′) and P2 (5′AACCCGGGCATAACGCTATACCAATC3′) in which underlines designate SmaI sites. The resulting PCR product was cloned into pGEM-T (Promega) to yield pGEM-Gaf1. After confirming the absence of PCR artifact by sequence analysis, the gaf1+ ORF was excised by digestion with SmaI and ligated to SmaI-digested middle-copy expression vector pREP42 [30] to yield pREP-Gaf1. For construction of pGEX4T3-Gaf1 encoding the glutathione S-transferase (GST)-tagged Gaf1 (GST-Gaf1), the 2.6-kb gaf1+ ORF was excised from pGEM-Gaf1 by SmaI-digestion and cloned in frame into the SmaI site of pGEX4T3 (Amersham).
A lacZ-based reporter plasmid used for analysis of ste11+ promoter was constructed as follows. First, the replication origin of S. pombe, ars1+, was amplified from pESP1 (Stratagene) by PCR using primers P3 (5′CCGAATTCAGGCCTGAGTCTAACTCCTTAACCACT3′; underline designates StuI site) and P4 (5′CCGATATCCAACCTTCCAATTCATTAAATC3′; underline designates EcoRV site). The 1.2-kb DNA fragment was cloned into pGEM-T Easy vector to yield pGEM-ars1. A 1.5-kb fragment containing the kanMX gene was amplified from pFA6a-kanMX6 [31] using PCR primers P5 (5′CCGATATCGGGTTAATTAAGGCGCGCCAGA3′; underline designates EcoRV site) and P6 (5′CCGCATGCCACTGGATGACGGCGTTAGTAT3′; underline designates SphI site) and inserted into the pGEM-T vector to yield pGEM-kanMX6. The 1.5-kb kanMX fragment was then excised by EcoRV-SphI double digestion and subcloned into EcoRV-SphI-digested pGEM-ars1. From the resulting pGEM-ars1-kanMX6 vector, a 2.7-kb fragment containing both ars1 and kanMX was excised with SphI, Klenow enzyme, and StuI. The resulting 2.7-kb SphI(blunted)-StuI fragment was then ligated with the 6.5-kb SnaBI-StuI fragment of YEp353 plasmid to yield pJLC-LacZ. Then a 1.4-kb BamHI fragment containing the promoter region of ste11+ from −834 to +575 (the major transcription start site is assigned as position +1 for nucleotide numbering) [4] was PCR-amplified using primers P7 (5′GGATCCGCATGCCATCTCCAGGGAT3′) and P8 (5′GGATCCCAAAAGAACGTAGAGGCAA3′) in which underlines designate BamHI sites. The reporter plasmid pJLC-Ste11(p)-LacZ was constructed by subcloning the PCR-amplified 1.4-kb BamHI fragment into the BamHI site of pJLC-LacZ upstream of lacZ in the correct orientation.
Gene disruption
Construction of gaf1Δ strains was performed by direct chromosomal integration as described previously [28]. The 2.6-kb genomic regions corresponding to the entire gaf1+ ORF (855 amino acids) of the wild-type strains, 972, ED665, JY4, and ED668, carrying different auxotrophic markers or mating type were replaced with the 1.5-kb PCR-amplified gaf1::kanMX deletion cassette derived from pFA6a-kanMX4 [32]. Stable transformants were selected by resistance to G418, and the disruptions were confirmed by PCR with appropriate primers (data not shown) yielding the gaf1Δ strains, KL230, KL210, KL211, and KL216, respectively. ste11Δ strain JZ396 [kindly donated by Dr. Yamamoto [12]] was transformed with the gaf1::kanMX deletion cassette to yield the gaf1Δ ste11Δ double-deletion mutant (KL213). To construct a gaf1Δ strain carrying the hphMX marker, the gaf1::kanMX allele of KL210 strain was replaced with the 1.6-kb gaf1::hphMX deletion cassette amplified from pFA6a-hphMX6 [33]. Stable transformants were selected by resistance to hygromycin B and sensitivity to G418 (indicating loss of kanMX), and the disruptions were confirmed by PCR (data not shown) to yield strain KL240.
Growth tests
To analyze growth by plate assays, S. pombe cells grown to mid-exponential phase in EMM2 for 18 h were washed and resuspended in EMM2-N (for nitrogen starvation) or EMM-G (for glucose restriction) to a concentration of 5×106 cells ml−1. The cell suspensions were serially diluted in 5-fold steps, and then a 5 µl aliquot of each dilution was spotted onto EMM, EMM-N, and EMM-G plates. The plates were incubated at 30°C and photographed after 3 d.
Preparation of total RNA
For preparation of total RNA, wild-type (ED668) and gaf1Δ (KL216) strains were grown to mid-log phase in EMM2 for 18 h at 30°C. Cells were harvested from the mid-log phase cultures and used as unstarved cell preparations (+N). To prepare nitrogen-starved cell samples (−N), cells harvested from the mid-log phase cultures in EMM2 were washed with distilled water and shifted to EMM-N. Nitrogen-starved cells used for microarray analysis were harvested 4-h cultivation in EMM-N. For Northern blot analysis, aliquots of the nitrogen-starved culture were removed at intervals, i.e., at 0, 3, 6, 9, and 18 h after shift. The cells were washed twice with distilled water and frozen immediately at −70°C for total RNA preparation. Total RNA samples were extracted from approximately 2×108 cells using a bead beater as described previously [12].
Microarray analysis
Thirty-microgram total RNA of each sample was further purified using RNeasy (QIAGEN) columns and submitted for microarray analysis by the SeouLin Bioscience (Korea). Probes were generated and hybridized to the GeneChip Yeast Genome 2.0 Array (Affymetrix), and the data were analyzed using GeneSpring GX software (Agilent).
Northern blot analysis
Approximately 20 µg of total RNA was fractionated on a 1% agarose gel containing 18% formaldehyde, transferred onto a Hybond-N+ membrane (Amersham), and fixed by UV cross-linking (Stratagene). DNA probes for ste11+, gaf1+, and actin (act1 +) genes were prepared using the PCR fragments amplified with the following primer pairs: P11 (5′GACCTGCGATCCAGATGATT3′) and P12 (5′CCAACAGCACTCTTGACGAA3′) for ste11+; P13 (5′TTACAACTTGCGTCCAGCA3′) and P14 (5′TGAATTCAGGAGCACCTTCC3′) for gaf1+; and P15 (5′GAAGCACACCATGACGCTTA3′) and P16 (5′CCTTGATCTCACCACAAGCA3′) for act1 +. The DNA probes were labeled with [α-32p]-dCTP using a Random Primed DNA Labeling Kit (Amersham). Hybridization was carried out at 42°C overnight in Rapid-hyb solution (Amersham). The signal was visualized by exposing the membrane to X-ray film and the relative signal intensity was quantified using a shareware program (Scion Image Beta 4.0.2).
Flow cytometric analysis
For flow cytometry, cells grown to mid-log phase (5×106 cells ml−1) were harvested and fixed in 70% ethanol containing 50 mM sodium citrate overnight at 4°C. After brief centrifugation, cell pellets were washed twice with 1 ml of 50 mM sodium citrate buffer (pH 7) and treated with RNase A (10 µg ml−1) at 37°C for 2 h. The cells were stained with propidium iodide (16 µg ml−1) and analyzed using a BD FACScalibur Flow Cytometer as described previously [28]. Data were analyzed using WinMDI software, version 2.8.
Mating and sporulation assay
In order to monitor the efficiency of sporulation, S. pombe cells grown to mid-log phase were prepared by two successive transfers of young colonies on EMM2 agar plates. The cells were then collected and washed three-times with distilled water. Suspensions of homothallic haploid cells (h90) or mixtures of mating pairs (h+ and h−) were spotted in 10 µl aliquots (1.0×109 cells) onto EMM2 and EMM-N agar plates. Cultures were grown at 30°C, and the cells were observed at intervals by DIC microscopy to determine sporulation frequencies. At least 400 cells from three independent experiments were evaluated, and mating and sporulation frequencies (FM) were calculated using the following equation [34]: FM (%) = (2Z+2A+0.5S)×100/(H+2Z+2A+0.5S), where Z stands for the number of zygotes, A for the number of asci, S for the number of free spores, and H for the number of cells that failed to mate. When necessary, sporulation was visualized by iodine vapor staining.
β-Galactosidase reporter assay
S. pombe cells containing β-galactosidase reporter plasmids were pre-grown to mid-log phase in liquid EMM2 medium for 18 h and shifted to EMM2 or EMM-N medium. Cells were harvested at intervals by centrifugation, washed, and resuspended at a concentration of 5×106 cells ml−1. After the cells were permeabilized with 0.1% sodium lauroylsarcosinate, β-galactosidase activity was determined by measuring hydrolysis of the chromogenic substrate, o-nitrophenyl-β-D-galactoside, as described previously [35].
Electrophoretic mobility shift assay
To prepare GST and GST-Gaf1 fusion proteins for electrophoretic mobility shift assay (EMSA), cells of Escherichia coli BL21 strains carrying pGEX4T3 or pGEX4T3-Gaf1 were cultured in LB medium with 50 µg ml−1 of ampicillin at 30°C to A 600 of 0.5. Isopropyl β-D-thiogalactopyranoside was added to a final concentration of 1 mM and the cells were grown for 4 h at 25°C. Harvested cells were resuspended in Buffer A (20 mM Tris, pH 7.6; 137 mM NaCl) containing 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride, and 1 µg ml−1 lysozyme. Resuspended cells were lysed by sonication, and the lysate was cleared by centrifugation at 24,000×g for 20 min at 4°C. The protein extracts were purified through a Glutathione Sepharose 4B column (Amersham).
To prepare EMSA probes and cold competitors, the upstream 0.60-kb SphI-NdeI (PA) and downstream 0.25-kb NdeI-EcoRV (PB) segments of the partial ste11+ promoter region [4] were amplified by PCR from the genomic DNA of S. pombe 972 using the following primers: P17 (5′GCATGCCATCTCCAGGGA3′) and P18 (5′ACATATGATGCGAAAGCATT3′) for PA; and P19 (5′CATATGTTACTTTAACCCCT3′) and P20 (5′GGATATCCTTTTAATATATGCT3′) for PB. Specific double-stranded oligonucleotide probes for wild-type (PW) and mutant (PM) GATA motifs corresponding to nucleotides −385 to −352 of the ste11+ promoter were prepared by annealing complementary pairs of the following single-stranded oligonucleotides: P21 (5′CATTTTGCCTTGCGCTATCTCCCTCAACGAAAAG3′) and P22 (5′CTTTTCGTTGAGGGAGATAGCGCAAGGCAAAATG3′) for PW; and P10 and P9 for PM. The duplex DNAs were end-labeled with [γ-32P] ATP by T4-polynucleotide kinase and purified using a G-50 or G-25 column (Amersham). All binding reactions were carried out in 20 µl binding buffer (10 mM Tris pH 7.5, 2 mM MgCl2, 50 mM NaCl, 1 mM DTT, 5% glycerol) containing 2 µg of poly (dI·dC) as a non-specific competitor, 0.1–1 µg of recombinant GST-Gaf1 protein, and 5 ρmol 32P-end-labeled double-stranded probe, at room temperature for 20 min. For competition experiments, DNA binding reactions were allowed to reach equilibrium and a 50- or 100-fold excess of unlabeled specific competitor DNA was added to the binding reaction mixture. For detection of a specific DNA-protein complex, samples were loaded onto a 6% non-denaturation polyacrylamide gel in 0.5×Tris-glycine buffer and electrophoresed at 10 V cm−1 at room temperature. The gels were run for 2 h, dried on a gel dryer and autoradiographed at −70°C using Fuji X-ray film. Putative binding sites for transcription factors were searched by using TRANSFAC program (http://gene-regulation.com/pub/databases.html).
Results
Deletion of gaf1+ causes accelerated G1-arrest under nitrogen-starved conditions
Previously, we suggested that gaf1+ might function as a transcriptional activator, although its biological relevance was unclear as a deletion mutant showed no significant defects [18]. However, recent genome analysis revealed that the gaf1+ sequence previously reported (Accession No. AAC35593) contains only a partial ORF corresponding to the C-terminal 290-amino acid segment of Gaf1, and that the complete gaf1+ sequence comprises a 2,565-bp ORF encoding an 855-amino acid protein (Accession No. Q10280) [36].
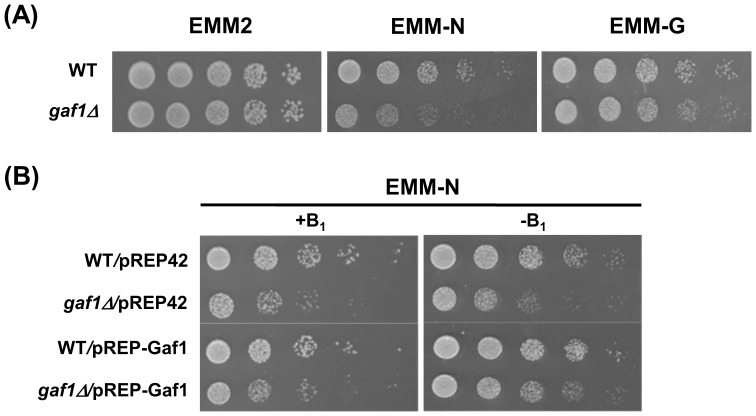
In the present study, we deleted the entire gaf1+ ORF from the genome of S. pombe (Table 1) and evaluated the response of gaf1Δ cells to nitrogen starvation and glucose restriction by plate assay. Cells of the gaf1Δ strain did not show any growth defect on EMM2 or EMM-G plate, indicating that the gaf1+ gene is dispensable for mitotic growth under normal and glucose-restricted conditions (Figure 1A). However, gaf1Δ cells showed significantly reduced growth on EMM-N which lacked the inorganic nitrogen source, NH4Cl, compared to wild-type cells that did grow to a limited extent by utilizing the supplementary auxotrophic nutrients, adenine, leucine, and uracil, as organic nitrogen sources.
Figure 1. Deletion of gaf1+ causes reduced growth on nitrogen-starved medium.
Cells grown to mid-log phase in EMM2 for 18 h were serially diluted 5-fold and spotted on agar plates. The plates were incubated at 30°C for 3 d. (A) Cells of WT (972) and gaf1Δ (KL230) strains were spotted on EMM, EMM-N, and EMM-G agar plates. (B) Cells of WT(ED665)/pREP42, gaf1Δ(KL210)/pREP42, WT(ED665)/pREP42-Gaf1, and gaf1Δ(KL210)/pREP-Gaf1 strains were spotted onto EMM-N plates with (+B1) or without (−B1) 20 µM thiamine. WT denotes the wild-type (gaf1+).
To confirm that the sensitivity of the gaf1Δ mutant to nitrogen starvation is due to the loss of Gaf1 activity rather than acquisition of abnormal activity, we constructed a system in which the production of Gaf1 could be shut off artificially using the thiamine-repressible nmt42+ promoter. In the absence of thiamine, the gaf1Δ/pREP-Gaf1 cells carrying ectopic copies of gaf1+ under the control of the nmt42+ promoter exhibited growth similar to the wild-type strains (WT/pREP42 and WT/pREP-Gaf1) on EMM-N plates (Figure 1B). In the presence of thiamine, gaf1Δ/pREP-Gaf1 cells exhibited as poor growth on EMM-N medium as gaf1Δ/pREP42 cells. These results suggest that the gaf1+ gene is dispensable for mitotic growth under normal conditions, but apparently plays a significant role in sustaining growth, though to a limited extent, under nitrogen-starved conditions.
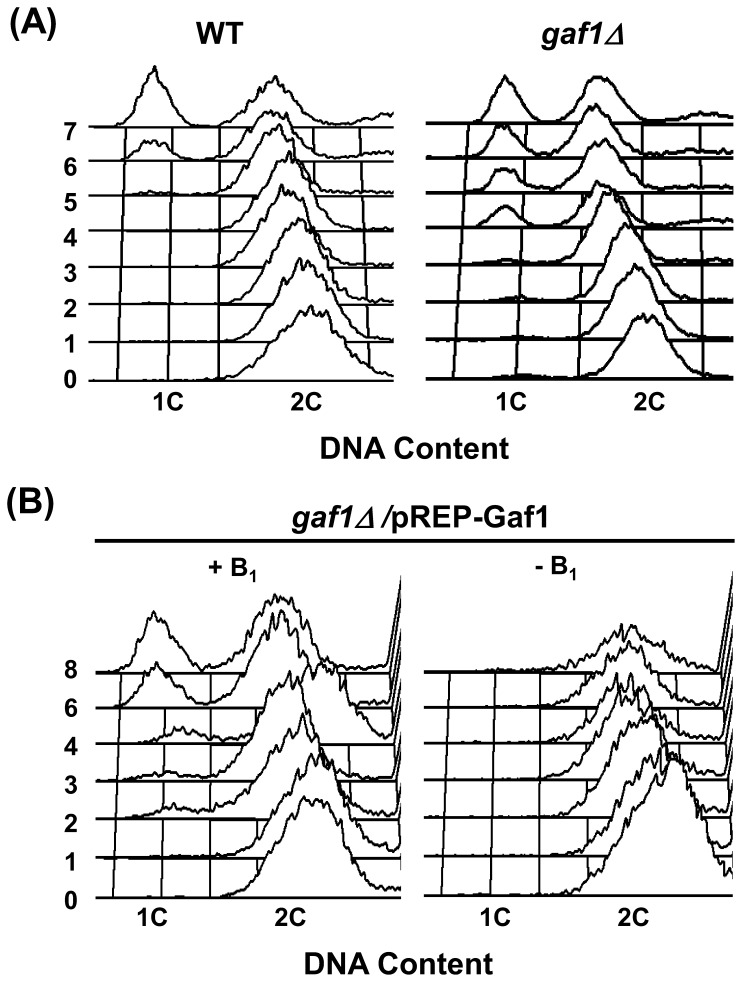
In S. pombe, as key nutrients become limited, cells exit the mitotic cycle and enter either G0 stationary phase or a program of sexual differentiation [37]. During early nitrogen starvation, S. pombe cells undergo several rounds of rapid cell division and then arrest at the G1 phase [8], [13]. In this study, the gaf1Δ strain accumulated G1-arrested cells after 4 h of nitrogen starvation, but the wild-type strain did not accumulate any detectable amount of G1-arrested cells until 6 h after the nitrogen shift (Figure 2A). Correspondingly, the homothallic haploid strain h90 gaf1Δ/pREP-Gaf1 began to accumulate G1-arrested cells after 2 h of nitrogen starvation in the presence of thiamine, however, it did not show any signs of G1-arrest even after 8 h of nitrogen starvation in the absence of thiamine (Figure 2B). Therefore, the deletion of gaf1+ causes accelerated entrance into G1 under nitrogen-starved conditions. Accordingly, the function of gaf1+ might be to delay the shift of nitrogen-starved cells from the vegetative cycle to the meiotic cycle, helping to sustain the vegetative cycle upon transient nitrogen starvation.
Figure 2. Deletion of gaf1+ causes accelerated G1-arrest under nitrogen-starved conditions.
Cells grown to mid-log phase in EMM2 for 18 h were shifted to EMM-N medium, and their DNA contents were monitored by FACS analysis at intervals. The FACS data represent typical examples of three independent experiments. (A) Cells of heterothallic (h−) WT (972) and gaf1Δ (KL230) strains were shifted to EMM-N medium. (B) Cells of homothallic (h90) gaf1Δ(KL211)/pREP-Gaf1 strain were shifted to EMM-N medium with (+B1) or without (−B1) 20 µM thiamine. WT denotes the wild-type (gaf1+).
Sporulation efficiency is enhanced by gaf1Δ mutation and reduced by gaf1+ overexpression
To evaluate the function of gaf1+ in sporulation under nitrogen-starved conditions, we spotted suspensions of pre-grown homothallic haploid cells (h90) and mixtures of mating pairs (h+ and h−) with or without gaf1Δ on EMM2 (+N) and EMM-N (−N) agar plates and estimated their mating and sporulation efficiencies (FM) after 3-d cultivation. The homothallic haploid gaf1Δ strain showed a significantly higher FM value (24%) than the wild-type strain (7%) on EMM2 plates (Table 2). In addition, the FM value of the h+ gaf1Δ×h− gaf1Δ mating mixture (63%) was approximately 5-fold higher than the h+×h− mating pair of wild-type (13%) on EMM2 plates. The homothallic gaf1Δ strain (85%) and h+ gaf1Δ×h− gaf1Δ mating mixture (80%) showed approximately 10–15% higher FM values than the homothallic wild-type strain (69%) and h+×h− mating mixture of wild-type strains (71%) on EMM-N plates. These results indicate that the gaf1Δ mutation elevates mating and sporulation efficiency by making cells more sensitive to nitrogen starvation.
Table 2. Mating and sporulation frequency of S. pombe strains.
| Mating and sporulation frequency (%)b | ||
| Strain genotypea | +N | −N |
| h90 | 6.5±2.1 | 69.2±6.1 |
| h90 gaf1Δ | 24.0±1.2 | 85.3±4.4 |
| h+×h− | 12.7±2.5 | 71.1±7.4 |
| h+ gaf1Δ×h− gaf1Δ | 62.5±2.9 | 80.0±9.6 |
| h+ ste11Δ×h− gaf1Δ | <0.01 | <0.01 |
| h90 ste11Δ | <0.01 | <0.01 |
| h90 gaf1Δ ste11Δ | <0.01 | <0.01 |
For analysis of homothallic strains, pre-grown cultures of h90 WT (JY4), h90 gaf1Δ (KL211), h90 ste11Δ (JZ396), and h90 gaf1Δ ste11Δ (KL213) were spotted onto EMM2 (+N) and EMM-N (−N) plates, and the cells were observed by DIC microscopy to determine sporulation frequencies after 3-d incubation at 30°C. For analysis of heterothallic strains, 1∶1 mixtures of the pre-grown mating pairs, h+ (ED668)×h − (ED665), h+ gaf1Δ (KL216)×h − gaf1Δ (KL210), and h+ ste11Δ (KL416)×h− gaf1Δ (KL240) were spotted onto EMM2 and EMM-N plates.
The values represent the average ± the standard deviation of at least three independent assays carried out in triplicate.
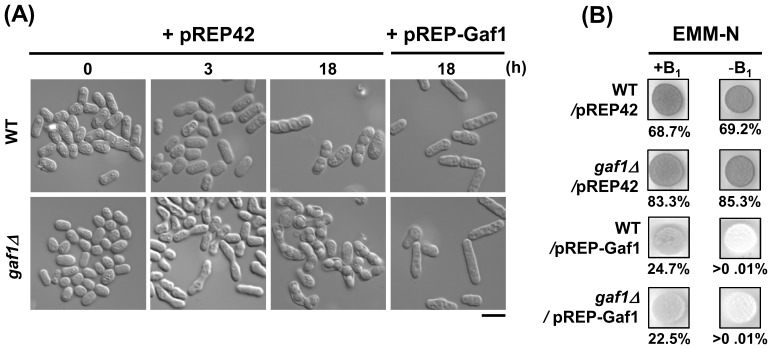
We also evaluated the role of gaf1+ in sporulation using an overexpression system of gaf1+ under the control of the nmt42+ promoter in homothallic strains. In liquid EMM-N medium, no sporulation was observed among h90 gaf1Δ/pREP-Gaf1 and h90 WT/pREP-Gaf1 cells even after 18-h exposure to nitrogen-starved conditions, while a considerable portion of the h90 gaf1Δ/pREP42 and h90 WT/pREP42 cells showed mating behavior after 3- and 18-h exposure, respectively (Figure 3A). On EMM-N plates, the h90 WT/pREP-Gaf1 and h90 gaf1Δ/pREP-Gaf1 strains exhibited negligible levels of iodine staining and FM value (<0.01%) in the absence of thiamine, but moderate levels of iodine staining and FM values (23–25%) in the presence of thiamine (Figure 3B). On the contrary, the spots of h90 WT/pREP42 and h90 gaf1Δ/pREP42 strains were stained dark brown with iodine vapor and exhibited high FM values (69–85%) after 3-d cultivation regardless of the presence of thiamine, indicating luxuriant sporulation under nitrogen-starved conditions. Together, these results demonstrate that overexpression of gaf1+ leads to a significant reduction of sporulation efficiency.
Figure 3. Overexpression of gaf1+ results in reduction of sporulation efficiency.
Cells and colonies of the homothallic (h90) strains, WT(JY4)/pREP42, gaf1Δ(KL211)/pREP42, WT(JY4)/pREP-Gaf1, and gaf1Δ(KL211)/pREP-Gaf1, were analyzed for sporulation by DIC microscopy and iodine staining, respectively. (A) Cells were grown to mid-log phase in EMM2 for 18 h and shifted to EMM-N medium. Samples were taken from the cultures at intervals, and the morphological characteristics of cells were observed by DIC microscopy. Bar, 10 µm. (B) Cells were grown to mid-log phase in EMM2 for 18 h and spotted onto EMM-N plates with (+B1) or without (−B1) 20 µM thiamine. Sporulation was monitored by iodine vapor staining of colonies after 3-d incubation at 30°C. The FM value presented under each panel was determined by observing the cells under a DIC microscope. The values represent the average of at least three independent assays carried out in triplicate. WT denotes the wild-type (gaf1+).
Gaf1 controls both the nitrogen starvation- and pheromone-responsive genes
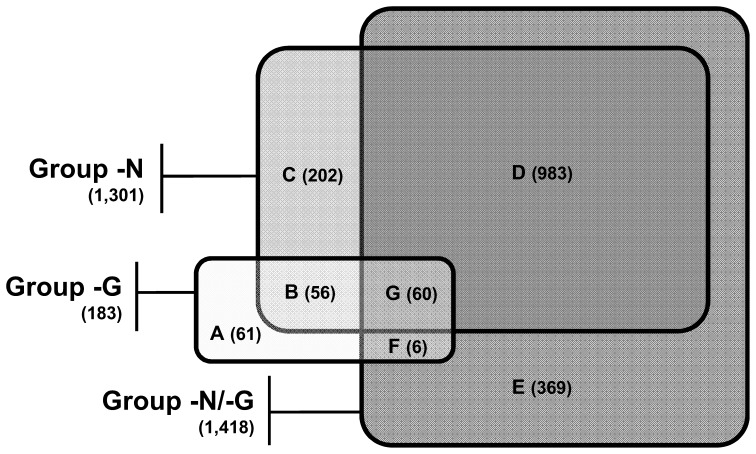
To search for a set of genes whose expression is specifically altered in response to the loss of gaf1+, microarray-based transcriptome analysis was performed using the RNA samples from the nitrogen-starved (−N) and unstarved (+N) cells of wild-type (gaf1+, ED668) and gaf1Δ (KL216) strains. A Venn diagram (Figure 4) was constructed from the groups of the genes up-regulated (≥1.5-fold, p<0.05) in unstarved gaf1Δ cells (Group −G), nitrogen-starved wild-type cells (Group −N), and nitrogen-starved gaf1Δ cells (Group −N/−G). One hundred and eighty-three genes were up-regulated in unstarved gaf1Δ cells (Group −G, Table S1). A total of 1,301 and 1,418 genes were up-regulated by nitrogen starvation in wild-type (Group −N, Table S2), and gaf1Δ cells (Group −N/−G, Table S3).
Figure 4. Effect of gaf1Δ mutation and nitrogen starvation on the global gene expression profiles of S. pombe.
RNA samples from the nitrogen-starved (−N) and unstarved (+N) cells of wild-type (gaf1+, ED668) and gaf1Δ (KL216) strains were used for the transcriptome analysis with the GeneChip Yeast Genome 2.0 Array. A Venn diagram is constructed from the lists of the genes up-regulated (≥1.5-fold, p<0.05) in unstarved gaf1Δ cells (Group −G, Table S1), nitrogen-starved wild-type cells (Group −N, Table S2), and nitrogen-starved gaf1Δ cells (Group −N/−G, Table S3). The overlapping and non-overlapping portions of the three groups (Group −G, −N, and −N/−G) are designated as Subgroup A to G. The lists of the genes included in Subgroup A–G are provided in Tables S4, S5, S6, S7, S8 S9, and S10, respectively, in the Supporting Information.
The overlapping and non-overlapping portions of the three groups (Group −G, −N, and −N/−G) are designated as Subgroup A to G (Figure 4), which enables more detailed clustering of the genes of interest. Approximately 63% (116 genes) of the members of Group −G were up-regulated by both gaf1Δ mutation and nitrogen starvation (Subgroup B+G, Table S5 and S10), while most of the remainders (61 genes) were up-regulated solely by gaf1Δ mutation (Subgroup A, Table S4). Therefore, it is suggested that Gaf1 down-regulates the basal expression levels of the nitrogen starvation-responsive genes included in Subgroup B+G. Although majorities of the members in Group −N and −N/−G were shown to be up-regulated by nitrogen starvation in both gaf1Δ and wild-type cells (1,043 genes, Subgroup D+G, Table S7 and S10), a considerable portions of them were up-regulated by nitrogen starvation in either gaf1Δ (375 genes, Subgroup E+F, Table S8 and S9) or wild-type cells (262 genes, Subgroup B+C, Table S5 and S6).
A considerable number of the genes of Subgroup B+G are up-regulated during mating and sporulation (the Sanger Centre Database; http://www.genedb.org/genedb/pombe) [38]: genes involved in nitrogen and/or pheromone response such as mei2 +, ste11+, spk1 +, ppk33 +, ste4 +, gpa1 +, mfm1 +, map1 +, map2 + and map3 +; genes function in double-strand break (DSB) formation, meiotic recombination, and/or nuclear segregation such as mug8 +, mug14 +, mug24 +, mug55 +, mug112 +, mug133 +, rec24 +, bqt2 +, and moa1 + [39]; genes encoding permeases (5 genes) and transporters (15 genes) for amino acids, sugars, urea, and other nutrients; and 5 wtf genes belonging to Tf transposon-containing sequences that are transcribed during meiosis. Taken together, it is suggested that deletion of gaf1+ may result in cellular physiology similar to one induced by nitrogen starvation and, accordingly, that Gaf1 plays an important role in both nitrogen starvation and mating response.
Furthermore, by comparing our data with the result of microarray analysis using the wild-type, Ste11-overexpressing, and ste11Δ strains [40], we found that 25 genes among those induced by gaf1Δ mutation are included in the list of the 61 Ste11 target genes (Table 3). Among the genes exhibiting increased expression in gaf1Δ cells, 10 genes including mei2+, spk1+, and ste11+ were up-regulated even under unstarved conditions (Subgroup G), while the remainders (15 genes) including ran1+, tht1+ and ste6+ were induced only under nitrogen-starved conditions (Subgroup D+E). This result supports the speculation that ste11+ is possibly a direct target of Gaf1 that negatively regulates its expression at the transcriptional level.
Table 3. List of the Ste11 target genes up-regulated by gaf1Δ mutation.
| Expression ratioa | ||||||
| Subgroup | Systematic No. | Gene | Description (GeneDB) |
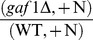
|
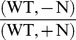
|
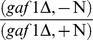
|
| (G)b | SPAC27D7.03c | mei2 + | RNA-binding protein involved in meiosis | 4.07 | 43.74 | 9.30 |
| SPAC32A11.01 | mug8 + | conserved fungal protein | 2.27 | 5.82 | 4.25 | |
| SPAC31G5.09c | spk1 + | MAP kinase | 1.95 | 36.46 | 23.90 | |
| SPBC32C12.02 | ste11 + | transcription factor | 1.76 | 5.87 | 3.10 | |
| SPBC359.06 | mug14 + | adducing | 1.73 | 82.76 | 45.98 | |
| SPAC11E3.06 | map1 + | MADS-box transcription factor | 1.72 | 11.77 | 6.14 | |
| SPBC19C2.04c | ubp11 + | ubiquitin C-terminal hydrolase | 1.72 | 5.07 | 3.84 | |
| SPBC24C6.06 | gpa1 + | G-protein alpha subunit | 1.70 | 11.54 | 7.32 | |
| SPAC1565.04c | ste4 + | adaptor protein | 1.69 | 10.87 | 5.41 | |
| SPCC162.10 | ppk33 + | serine/threonine protein kinase | 1.53 | 46.35 | 30.03 | |
| (D+E)c | SPAC1F5.09c | shk2 + | PAK-related kinase | 1.27 | 20.72 | 53.30 |
| SPCC1442.01 | ste6 + | guanyl-nucleotide exchange factor | 0.98 | 7.40 | 15.76 | |
| SPAC1093.06c | dhc1 + | dynein heavy chain | 0.30 | 1.47 | 15.66 | |
| SPAC23E2.03c | ste7 + | meiotic suppressor protein | 1.19 | 11.05 | 15.28 | |
| SPBC19C2.05 | ran1 + | serine/threonine protein kinase | 1.18 | 5.38 | 6.88 | |
| SPCC1393.07c | mug4 + | sequence orphan | 1.17 | 1.97 | 5.57 | |
| SPAC31G5.07 | conjugation protein (predicted) | 0.89 | 1.52 | 4.45 | ||
| SPBC354.08c | DUF221 family protein | 0.36 | 0.92 | 4.07 | ||
| SPBC2D10.06 | rep1 + | MBF transcription factor complex subunit | 0.92 | 2.00 | 3.36 | |
| SPAC13C5.03 | tht1 + | nuclear membrane protein involved in karyogamy | 1.04 | 0.62 | 3.01 | |
| SPAC1F5.08c | yam8 + | calcium transport protein | 1.07 | 1.55 | 2.65 | |
| SPAC27E2.07 | pvg2 + | galactose residue biosynthesis protein | 1.21 | 1.59 | 2.48 | |
| SPAPB2B4.03 | cig2 + | cyclin | 0.80 | 1.09 | 2.39 | |
| SPAC31G5.10 | eta2 + | Myb family protein | 1.04 | 1.41 | 2.22 | |
| SPBP4H10.11c | lcf2 + | long-chain-fatty-acid-CoA ligase | 0.75 | 1.02 | 2.08 | |
Relative expression of the genes was measured by microarray assay in the nitrogen-starved (−N) and unstarved (+N) cells of wild-type (ED668) and gaf1Δ (KL216) strains. Unstarved (+N) cells were prepared from the mid-log phase cultures in EMM2. Nitrogen-starved (−N) cells were prepared by shifting the mid-log phase cells to EMM-N for 4 h as described in Materials and Methods.
Genes up-regulated by gaf1Δ mutation under both nitrogen-starved and unstarved conditions (Subgroup G, p<0.05).
Genes up-regulated by gaf1Δ mutation only under nitrogen starved conditions (Subgroup D+E, p<0.05).
ste11+ is epistatic to gaf1+
Epistasis analysis was performed to determine whether Gaf1 functions in the same pathway as Ste11, a key transcription factor for sexual development [12]. Pre-grown cells of the homothallic haploid strains, h90 gaf1Δ, h90 ste11Δ, and h90 gaf1Δ ste11Δ, were spotted on both EMM2 and EMM-N plates and the sporulation efficiencies were monitored after 3 d. The FM value of h90 gaf1Δ reached approximately 85% on EMM-N and 24% on EMM2, but h90 ste11Δ and h90 gaf1Δ ste11Δ cells, and the h+ ste11Δ×h− gaf1Δ mating mixture exhibited negligible levels of sporulation efficiencies even on EMM-N (Table 2). Therefore, the phenotype of the gaf1Δ mutation, i.e., accelerated initiation of sporulation and elevated sporulation efficiency, is masked by the ste11Δ mutation, causing impaired sporulation. Thus, ste11+ is epistatic to gaf1+ with respect to sporulation efficiency, and furthermore, gaf1+ functions upstream of ste11+ in the signaling pathway governing sexual development.
Expression of ste11+ is accelerated and increased in gaf1Δ cells
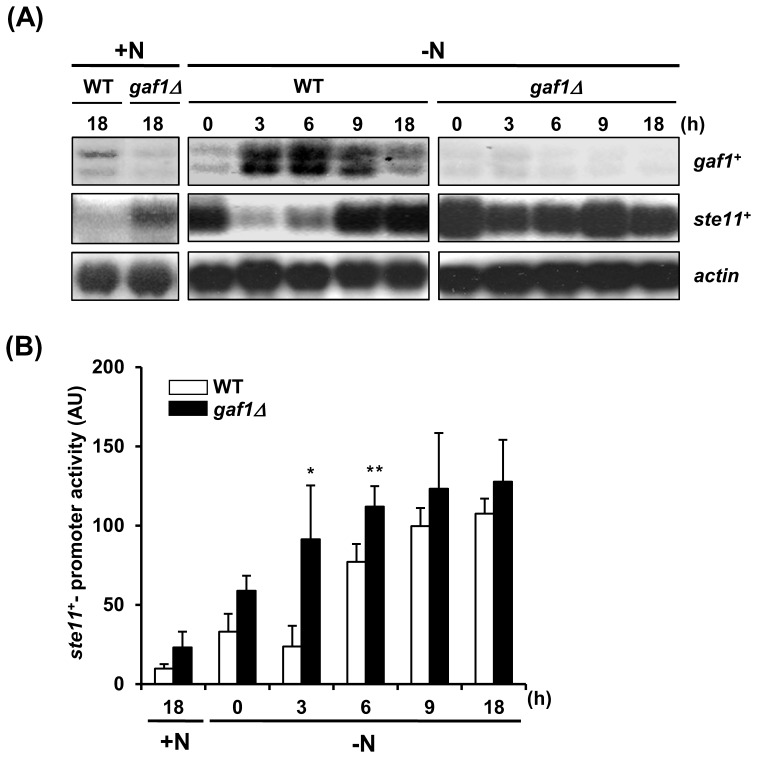
To test whether gaf1+ is responsible for the transcriptional regulation of ste11+, we analyzed the expression of ste11+ under both nitrogen-starved and normal conditions in gaf1Δ and wild-type strains by Northern blot analysis. Under normal conditions, gaf1Δ cells showed increased level of ste11+ compared to wild-type cells (Figure 5A). When the wild-type cells were exposed to nitrogen starvation, the level of gaf1+ transcript increased considerably during the first 6 h of nitrogen starvation, followed by a subsequent decline. Unexpectedly, the levels of ste11+ transcript in both the wild-type and gaf1Δ cells harvested from the nitrogen-starved cultures at the time point of 0 h were considerably higher than those in the corresponding cells from the unstarved culture, which might be due to the very short exposure of the cells to EMM-N medium followed by washing with distilled water. This result is similar to that observed in the previous study conducted by other research group [41]. Therefore, we adopted the data from the unstarved cells of corresponding strains cultivated in EMM2 for 18 h, rather than those from the nitrogen-starved cells sampled at the time point of 0 h, as references. As shown in Figure 5A, the amount of ste11+ transcript increased at a much slower rate than that of gaf1+ transcript, and it did not reach its highest level until at least 9 h after the transfer. The level of ste11+ transcript in gaf1Δ cells, however, increased steeply up to the plateau within 3 h and was maintained during the following 6 h. The expression of ste11+ was also monitored by assaying the ectopic expression of Ste11(p)-lacZ hybrid gene in wild-type and gaf1Δ strains carrying the pJLC-Ste11(p)-LacZ plasmid. In accordance with the result of Northern blotting, the expression of Ste11(p)-lacZ was higher in gaf1Δ cells than in wild-type cells under unstarved conditions (Figure 5B). When cells were subjected to nitrogen starvation, the expression of Ste11(p)-lacZ was induced more rapidly in gaf1Δ cells than in wild-type cells. These results suggest that deletion of gaf1+ causes accelerated induction of ste11+ expression under nitrogen-starved conditions and increased ste11+ expression even under unstarved conditions.
Figure 5. Deletion of gaf1+ results in accelerated induction of ste11+ expression under nitrogen-starved conditions.
(A) Northern blot analysis of ste11+ and gaf1+ mRNA from wild-type and gaf1Δ cells exposed to nitrogen starvation. Cells of wild-type (972) and gaf1Δ (KL230) strains pre-grown in EMM2 (+N) were shifted to EMM-N (−N) and cultured with constant shaking. At indicated time points, cells were harvested and washed twice with distilled water, and total RNAs were extracted from the cells. RNA blots were hybridized with 32P-labeled PCR-amplified gaf1+ and ste11+ probes. For internal control, all blots were stripped and subsequently rehybridized with 32P-labeled actin-specific probe (act1+). (B) β-Galactosidase reporter assay for analysis of ste11+ expression in wild-type and gaf1Δ cells subjected to nitrogen starvation. Cells of wild-type (ED665) and gaf1Δ (KL240) strains carrying pJLC-Ste11(p)-LacZ were cultivated to mid-log phase in EMM2 (+N) and shifted to EMM-N (−N). At indicated time points, cells were harvested and washed twice with distilled water, and the level of ste11 + expression was estimated by measuring the activity of β-galactosidase in each sample. Values are the mean ± standard error of three independent experiments carried out in triplicate, n = 9. *, p<0.01; **, p<0.05 (two-tailed Student's t-test, versus wild-type). WT denotes the wild-type (gaf1+).
Gaf1 protein binds to the promoter region of ste11+
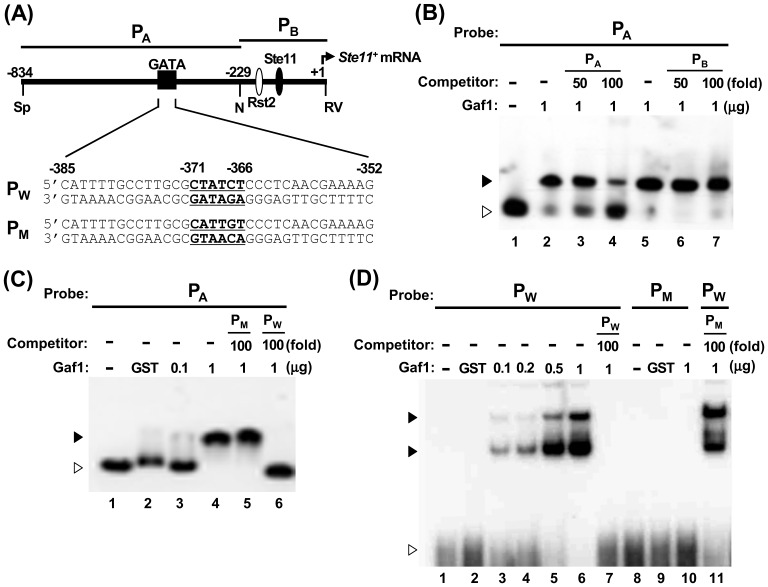
We performed EMSA to address whether Gaf1 can directly bind to the promoter region of ste11+. The upstream probe (PA), encompassing the region from −835 to −227 of the ste11+ promoter, was amplified by PCR and end-labeled with T4 polynucleotide kinase (Figure 6A). A DNA-protein complex was observed in the reaction mixture containing the 32P-labeled PA probe and the recombinant GST-Gaf1 (Figure 6B, lanes 2 and 5). The DNA-protein complex was specifically reduced in the presence of 50- and 100-fold molar excess of cold PA competitor probe (Figure 6B, lanes 3 and 4), but not in the presence of 50- or 100-fold molar excess of cold PB competitor (Figure 6B, lanes 6 and 7). This suggests that the Gaf1 protein specifically binds to a cis-element contained in the upstream region (from −828 to −227) of the ste11+ promoter.
Figure 6. Gaf1 protein specifically binds to the GATA binding motif of ste11+ promoter in vitro.
(A) Schematic diagram of the promoter region of ste11+. The open- and closed-ovals represent binding sites for Rst2 (UASst) and Ste11 (TR1 and TR2), respectively. The major transcription start site (position number +1) is designated by a hinged arrow. The regions contained in the upstream probe PA (−834∼−225) and the downstream probe PB (−231∼+10) are shown as bars. The dark square represents the GATA motif spanning from −385 to −352 for which wild-type (PW) and mutant (PM) double-stranded oligonucleotide probes were designed. Restriction sites: Sp, SphI; N, NdeI; RV, EcoRV. (B) Search for Gaf1-binding region in ste11+ promoter by EMSA. GST-Gaf1 protein (1 µg) was incubated with labeled PA probe containing the 0.6-kb upstream region of ste11+ promoter in the absence (lanes 2 and 5) or presence of excess cold competitor, PA (lane 3, 50-fold; lane 4, 100-fold) or PB (lane 6, 50-fold; lane 7, 100-fold). (C) Analysis of Gaf1 binding to the GATA motif of ste11+ promoter by EMSA. GST-Gaf1 protein (lane 3, 0.1 µg; lanes 4–6, 1 µg) was incubated with labeled PA probe in the absence (lanes 3 and 4) or presence of 100-fold excess of cold competitor PM (lanes 5) or PW (lanes 6). (D) Mutational dissection of the Gaf1-binding GATA motif of ste11+ promoter by EMSA. Varying amounts of GST-Gaf1 protein (lane 3, 0.1 µg; lane 4, 0.2 µg; lane 5, 0.5 µg; lanes 6, 7, 11, and 12, 1 µg) were incubated with labeled PW oligonucleotide probe in the absence (lanes 3 and 4) or presence of 100-fold excess of cold competitor PM (lanes 5) or PW (lanes 6). Mock reaction mixtures without GST-Gaf1 or with GST protein were used as negative controls in (B)–(D). The closed and open arrow heads in (B)–(D) represent shifted bands and free probes, respectively.
We also performed competition experiments using cold oligonucleotide probes spanning base pairs −385 to −351: PW containing a canonical GATA motif and PM containing mutations in the GATA motif (Figure 6A). The DNA-protein complex between the 32P-labeled PA probe and GST-Gaf1 protein was diminished by the addition of 100-fold molar excess cold PW (Figure 6C, lanes 4 and 6), but not by a similar amount of cold PM (Figure 6C, lanes 4 and 5). Accordingly, the amount of PW-GST-Gaf1 complex increased in proportion to the amount of GST-Gaf1 (Figure 6D, lanes 3–6) and was diminished to the basal level by addition of 100-fold molar excess of cold PW (Figure 6C, lane 6). The PM probe did not produce any detectable amount of DNA-protein complex with GST-Gaf1 (Figure 6D, lane 10) or exhibit competition against the PW probe to GST-Gaf1 even at a 100-fold molar excess (Figure 6D, lane 11). These results reflect that the canonical GATA motif from −371 to −366 in the promoter of ste11+ (Figure 6A) is the target sequence of the Gaf1 protein. It is thus suggested that the GATA motif in ste11+ promoter functions as a cis element to delay and attenuate the induction of ste11+ expression via the interaction with Gaf1.
Discussion
The S. pombe protein Ste11, which activates a number of genes required for mating and meiosis, is a pivotal regulator of sexual differentiation induced by nutrient starvation or environmental stress [12]. In the present study, we identified Gaf1, an S. pombe GATA factor, as a negative regulator of ste11+ expression.
Deletion of gaf1+ caused no growth defects under normal conditions. However, under nitrogen-starvation, it led to reduced mitotic growth (Figure 1), accelerated entrance into G1 (Figure 2), and elevated mating and sporulation efficiency (Table 2). Overexpression of gaf1+ resulted in a remarkable reduction of sporulation efficiency under nitrogen-starved conditions (Figure 3). It seems likely that Gaf1 functions as a modulator of the mitosis-meiosis transition, delaying the entrance of growing cells into the meiotic cycle during the initial stages of nitrogen starvation and signaling the optimal time for promoting sexual development. The delay in G1-arrest and subsequent sporulation may provide a safety mechanism allowing cells to revert to vegetative growth when nutrient availability again becomes favorable [6], [16], [42], [43]. In accordance with the present result, overexpression of tor2+ encoding the TOR protein kinase Tor2 strongly represses mating, meiosis and sporulation efficiency, whereas its inactivation has the opposite effect, leading to cell differentiation regardless of nutritional conditions [11]. In S. cerevisiae, it has been shown that Tor kinase and GATA transcription factor are involved in nitrogen catabolite repression (NCR), a regulatory event in which transcription of certain genes is down-regulated by a good nitrogen source such as glutamine but up-regulated by a poor nitrogen source such as proline [44]. Therefore, the involvement of TOR kinase and GATA transcription factor in nitrogen signaling may be a widely conserved phenomenon among various organisms.
Microarray analysis of the global gene expression profiles in gaf1Δ and wild-type cells under nitrogen-starved and unstarved conditions enabled us to search for a cluster of genes controlled by the action of Gaf1. Approximately 63% of the genes induced by the deletion of gaf1+ under normal conditions (Subgroup B+G, Figure 4) overlap with those induced by nitrogen starvation in wild-type cells. In addition, many of the Subgroup B+G genes are identified to be induced during mating and sporulation [38]. Thus, it is likely that Gaf1 plays an important role not only in nitrogen signaling pathway but also in mating response. In accordance with the present result, recent microarray analysis using temperature-sensitive tor2 mutants revealed that a total of 151 of 194 genes induced by the loss of Tor2 function are included in the list of roughly 1,000 genes found to be induced by nitrogen starvation in S. pombe [45]. This result pointed to an important role that Tor2 plays in nitrogen starvation and mating response. Interestingly, we also found that 13 genes among those induced by the deletion of gaf1+ overlap with those induced by the loss of Tor2 function, suggesting that the two genes might be involve in the same signaling pathway activated by nitrogen starvation. Thus, it would be interesting to determine the relationship between gaf1+ and tor2+ genes.
Comparison of our microarray data with the genome-wide view of Ste11 target genes reported previously [40] enabled us to speculate that ste11 + is a strong candidate for a direct transcriptional target of Gaf1 (Table 3). Especially, it is noteworthy that the genes involved in Ras/MAPK signaling pathway stimulated by pheromone such as gpa1 +, ste4 +, spk1 +, ste11 +, and mei2 + are transcriptionally induced in response to the loss of gaf1 + function under normal conditions. In addition, this finding is in agreement with the observation that deletion of gaf1+ causes accelerated entrance of cells into meiotic cell cycle (Figure 2) and elevated mating and sporulation efficiency on exposure to nitrogen starvation (Table 2).
In accordant with the result of microarray analysis, cells of the gaf1Δ ste11Δ strain were completely defective in mating and sporulation (Table 2), indicating that ste11 + is epistatic to gaf1+. Deletion of gaf1+ not only increases the expression of ste11+ in unstarved cells but also accelerates the induction of ste11+ transcription in nitrogen-starved cells (Figure 5). In addition, the result of EMSA provides compelling evidence that Gaf1 binds to the canonical GATA motif (5′-HGATAR-3′) spanning from −371 to −366 in the promoter of ste11+ to attenuate and delay its expression (Figure 5). Thus, it becomes evident that Gaf1 functions as a negative regulator of ste11+ transcription, via direct interaction with the GATA motif in the ste11+ promoter. The expression of ste11+ is regulated directly by two transcription factors, Rst2 and Ste11, that bind to the upstream activating sequence (UASst; 5′-CCCCTC-3′) and the T-rich box (TR box; 5′-TTCTTTGTTY-3′) in the ste11+ promoter, respectively [4]. No other proteins that either activate or repress the transcription of ste11+ through direct binding to its promoter had been identified. Our research indicates that Gaf1 provides the prime example for negative regulation of ste11+ transcription through direct binding to a cis-acting motif of its promoter.
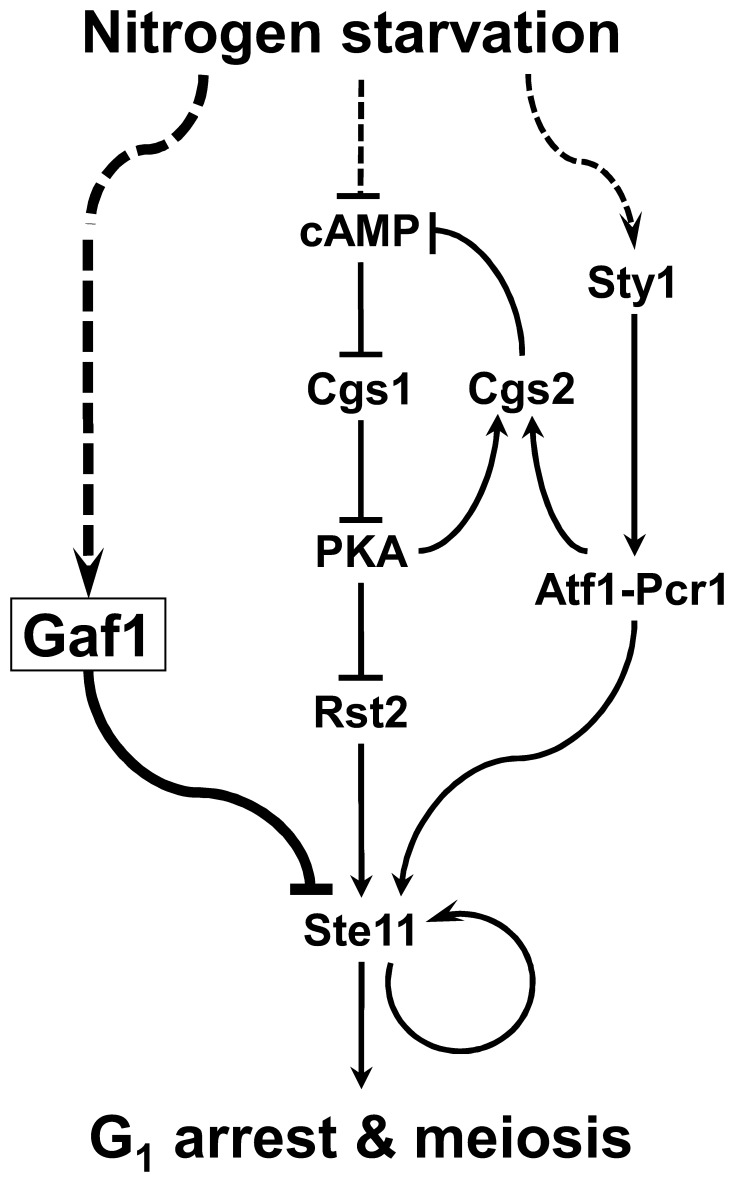
Figure 7 shows a simplified view of the proposed role of Gaf1 in the nitrogen-signaling pathways governing the expression of ste11+ and consequent sexual differentiation in S. pombe together with the cAMP-dependent PKA and stress-activated MAPK pathways determined previously. We found in the present study that nitrogen starvation causes induction of gaf1+ expression, and Gaf1, in turn, represses the expression of ste11+ via direct interaction with its promoter. Starvation of carbon or nitrogen source leads to decrease in the level of cAMP and subsequent drop of PKA activity, which consequently induces expression of ste11+ through the activation of Rst2 that can bind to the promoter region of ste11+ [4], [7], [46]. Nitrogen starvation also causes activation of the stress-activated MAPK pathway including Wis4, Win1, Wis1, and Sty1 [14], [15], leading to activation of Atf1 by phosphorylation [16], [47], [48]. Activated Atf1, in turn, forms a complex with another cAMP response element-binding protein Pcr1 to yield an Atf1-Pcr1 heterodimeric transcription factor, which is also required for expression of ste11+ [5], [6], [49]. It has not yet been determined whether the Atf1-Pcr1 complex directly regulates the expression of ste11+ or not. However, it has been reported that the Atf1-Pcr1 heterodimer directly activates the expression of cgs2+ encoding a phosphodiesterase that has a major role in regulating the single cAMP-dependent PKA pathway [3]. In addition, phosphodiesterase is most likely stimulated by PKA activity to create a feedback mechanism [50]. Thus there exists a direct connection between the MAPK and PKA pathways mediated by the action of Atf1-Pcr1 complex. It remains to be determined whether the expression of gaf1+ is subject to the control of either the cAMP-dependent PKA or the stress-activated MAPK pathway.
Figure 7. Schematic diagram showing the proposed function of Gaf1 in the nitrogen-signaling pathways in S. pombe.
Nitrogen starvation causes induction of gaf1+ expression, and Gaf1, in turn, represses the expression of ste11+ via direct interaction with its promoter. It has been previously known that nitrogen starvation leads to the activation of Rst2 via the cAMP-dependent PKA pathway [14], [15] as well as Atf1-Pcr1 via the Sty1 MAPK pathway [3], [5], [6], [14]–[16], [47]–[49], consequently resulting in induction of ste11+ expression. In addition, phosphodiesterase is most likely stimulated by PKA activity to create a feedback mechanism [50]. The pathway addressed in this study is shown in thick lines, and other paths previously determined are shown in thin lines. Activation and inhibition are indicated by arrows and crossing bars, respectively. Dotted lines indicate pathways remained to be fully determined.
Supporting Information
List of the genes up-regulated in unstarved (+N) gaf1 Δ cells (Group −G).
(PDF)
List of the genes up-regulated in nitrogen-starved wild-type ( gaf1 +) cells (Group −N).
(PDF)
List of the genes up-regulated in nitrogen-starved (−N) gaf1 Δ cells (Group −N/−G).
(PDF)
List of the genes in Subgroup A.
(PDF)
List of the genes in Subgroup B.
(PDF)
List of the genes in Subgroup C.
(PDF)
List of the genes in Subgroup D.
(PDF)
List of the genes in Subgroup E.
(PDF)
List of the genes in Subgroup F.
(PDF)
List of the genes in Subgroup G.
(PDF)
Funding Statement
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education, Science and Technology (MEST) (No. 313-2008-2-C00778). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Egel R (1973) Genes involved in mating type expression of fission yeast. Mol Gen Genet 122: 339–343. [DOI] [PubMed] [Google Scholar]
- 2. Davey J (1998) Fusion of a fission yeast. Yeast 14: 1529–1566. [DOI] [PubMed] [Google Scholar]
- 3. Davidson MK, Shandilya HK, Hirota K, Ohta K, Wahls WP (2004) Atf1-Pcr1-M26 complex links stress-activated MAPK and cAMP-dependent protein kinase pathways via chromatin remodeling of cgs2+. J Biol Chem 279: 50857–50863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kunitomo H, Higuchi T, Iino Y, Yamamoto M (2000) A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11+ gene, which encodes a pivotal transcription factor for sexual development. Mol Biol Cell 11: 3205–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Watanabe Y, Yamamoto M (1996) Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol Cell Biol 16: 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kanoh J, Watanabe Y, Ohsugi M, Iino Y, Yamamoto M (1996) Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells 1: 391–408. [DOI] [PubMed] [Google Scholar]
- 7. Higuchi T, Watanabe Y, Yamamoto M (2002) Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol Cell Biol 22: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamamoto M (1996) Regulation of meiosis in fission yeast. Cell Struct Funct 21: 431–436. [DOI] [PubMed] [Google Scholar]
- 9. Yamada H, Ohmiya R, Yamamoto E, Aiba H, Mizuno T (1997) Characterization of multicopy suppressor genes that complement a defect in the Wis1-Sty1 MAP kinase cascade involved in stress responses in Schizosaccharomyces pombe. J Gen Appl Microbiol 43: 209–215. [DOI] [PubMed] [Google Scholar]
- 10. Yamamoto TG, Chikashige Y, Ozoe F, Kawamukai M, Hiraoka Y (2004) Activation of the pheromone-responsive MAP kinase drives haploid cells to undergo ectopic meiosis with normal telomere clustering and sister chromatid segregation in fission yeast. J Cell Sci 117: 3875–3886. [DOI] [PubMed] [Google Scholar]
- 11. Alvarez B, Moreno S (2006) Fission yeast Tor2 promotes cell growth and represses cell differentiation. J Cell Sci 119: 4475–4485. [DOI] [PubMed] [Google Scholar]
- 12. Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M (1991) Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev 5: 1990–1999. [DOI] [PubMed] [Google Scholar]
- 13. Kjaerulff S, Lautrup-Larsen I, Truelsen S, Pedersen M, Nielsen O (2005) Constitutive activation of the fission yeast pheromone-responsive pathway induces ectopic meiosis and reveals ste11 as a mitogen-activated protein kinase target. Mol Cell Biol 25: 2045–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shiozaki K, Shiozaki M, Russell P (1997) Mcs4 mitotic catastrophe suppressor regulates the fission yeast cell cycle through the Wik1-Wis1-Spc1 kinase cascade. Mol Biol Cell 8: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shieh JC, Martin H, Millar JB (1998) Evidence for a novel MAPKKK-independent pathway controlling the stress activated Sty1/Spc1 MAP kinase in fission yeast. J Cell Sci 111 Pt 18:2799–2807. [DOI] [PubMed] [Google Scholar]
- 16. Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, et al. (1995) Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. Embo J 14: 6193–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohmiya R, Yamada H, Kato C, Aiba H, Mizuno T (2000) The Prr1 response regulator is essential for transcription of ste11+ and for sexual development in fission yeast. Mol Gen Genet 264: 441–451. [DOI] [PubMed] [Google Scholar]
- 18. Hoe KL, Won MS, Chung KS, Park SK, Kim DU, et al. (1998) Molecular cloning of gaf1, a Schizosaccharomyces pombe GATA factor, which can function as a transcriptional activator. Gene 215: 319–328. [DOI] [PubMed] [Google Scholar]
- 19. Won M, Hoe KL, Cho YS, Song KB, Yoo HS (1999) DNA-induced conformational change of Gaf1, a novel GATA factor in Schizosaccharomyces pombe. Biochem Cell Biol 77: 127–132. [DOI] [PubMed] [Google Scholar]
- 20. Charron F, Paradis P, Bronchain O, Nemer G, Nemer M (1999) Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol Cell Biol 19: 4355–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, et al. (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90: 109–119. [DOI] [PubMed] [Google Scholar]
- 22. Perlman H, Suzuki E, Simonson M, Smith RC, Walsh K (1998) GATA-6 induces p21(Cip1) expression and G1 cell cycle arrest. J Biol Chem 273: 13713–13718. [DOI] [PubMed] [Google Scholar]
- 23. Scazzocchio C (2000) The fungal GATA factors. Curr Opin Microbiol 3: 126–131. [DOI] [PubMed] [Google Scholar]
- 24. Chen ES, Saitoh S, Yanagida M, Takahashi K (2003) A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell 11: 175–187. [DOI] [PubMed] [Google Scholar]
- 25. Pelletier B, Beaudoin J, Philpott CC, Labbe S (2003) Fep1 represses expression of the fission yeast Schizosaccharomyces pombe siderophore-iron transport system. Nucleic Acids Res 31: 4332–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pelletier B, Trott A, Morano KA, Labbe S (2005) Functional characterization of the iron-regulatory transcription factor Fep1 from Schizosaccharomyces pombe. J Biol Chem 280: 25146–25161. [DOI] [PubMed] [Google Scholar]
- 27.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E (1993) Experiments with fission yeast: A laboratory course manual. Plainview, NY: Cold Spring Harbor Laboratory Press.
- 28. Forsburg SL, Rhind N (2006) Basic methods for fission yeast. Yeast 23: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823. [DOI] [PubMed] [Google Scholar]
- 30. Maundrell K (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130. [DOI] [PubMed] [Google Scholar]
- 31. Siam R, Dolan WP, Forsburg SL (2004) Choosing and using Schizosaccharomyces pombe plasmids. Methods 33: 189–198. [DOI] [PubMed] [Google Scholar]
- 32. Pearson BM, Hernando Y, Schweizer M (1998) Construction of PCR-ligated long flanking homology cassettes for use in the functional analysis of six unknown open reading frames from the left and right arms of Saccharomyces cerevisiae chromosome XV. Yeast 14: 391–399. [DOI] [PubMed] [Google Scholar]
- 33. Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM (2005) Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 22: 1013–1019. [DOI] [PubMed] [Google Scholar]
- 34. Kunitomo H, Sugimoto A, Wilkinson CR, Yamamoto M (1995) Schizosaccharomyces pombe pac2+ controls the onset of sexual development via a pathway independent of the cAMP cascade. Curr Genet 28: 32–38. [DOI] [PubMed] [Google Scholar]
- 35. Guarente L (1983) Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol 101: 181–191. [DOI] [PubMed] [Google Scholar]
- 36. Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- 37. Li P, McLeod M (1996) Molecular mimicry in development: identification of ste11+ as a substrate and mei3+ as a pseudosubstrate inhibitor of ran1+ kinase. Cell 87: 869–880. [DOI] [PubMed] [Google Scholar]
- 38. Mata J, Lyne R, Burns G, Bahler J (2002) The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147. [DOI] [PubMed] [Google Scholar]
- 39. Martin-Castellanos C, Blanco M, Rozalen AE, Perez-Hidalgo L, Garcia AI, et al. (2005) A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol 15: 2056–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mata J, Bahler J (2006) Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc Natl Acad Sci U S A 103: 15517–15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakashima A, Hasegawa T, Mori S, Ueno M, Tanaka S, et al. (2006) A starvation-specific serine protease gene, isp6+, is involved in both autophagy and sexual development in Schizosaccharomyces pombe. Curr Genet 49: 403–413. [DOI] [PubMed] [Google Scholar]
- 42. Davey J, Nielsen O (1994) Mutations in cyr1 and pat1 reveal pheromone-induced G1 arrest in the fission yeast Schizosaccharomyces pombe. Curr Genet 26: 105–112. [DOI] [PubMed] [Google Scholar]
- 43. Imai Y, Yamamoto M (1994) The fission yeast mating pheromone P-factor: its molecular structure, gene structure, and ability to induce gene expression and G1 arrest in the mating partner. Genes Dev 8: 328–338. [DOI] [PubMed] [Google Scholar]
- 44. Cooper TG (2002) Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev 26: 223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matsuo T, Otsubo Y, Urano J, Tamanoi F, Yamamoto M (2007) Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol Cell Biol 27: 3154–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mochizuki N, Yamamoto M (1992) Reduction in the intracellular cAMP level triggers initiation of sexual development in fission yeast. Mol Gen Genet 233: 17–24. [DOI] [PubMed] [Google Scholar]
- 47. Wilkinson MG, Samuels M, Takeda T, Toone WM, Shieh JC, et al. (1996) The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev 10: 2289–2301. [DOI] [PubMed] [Google Scholar]
- 48. Shiozaki K, Russell P (1996) Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev 10: 2276–2288. [DOI] [PubMed] [Google Scholar]
- 49. Lawrence CL, Maekawa H, Worthington JL, Reiter W, Wilkinson CR, et al. (2007) Regulation of Schizosaccharomyces pombe Atf1 protein levels by Sty1-mediated phosphorylation and heterodimerization with Pcr1. J Biol Chem 282: 5160–5170. [DOI] [PubMed] [Google Scholar]
- 50. Wang L, Griffiths K Jr, Zhang YH, Ivey FD, Hoffman CS (2005) Schizosaccharomyces pombe adenylate cyclase suppressor mutations suggest a role for cAMP phosphodiesterase regulation in feedback control of glucose/cAMP signaling. Genetics 171: 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of the genes up-regulated in unstarved (+N) gaf1 Δ cells (Group −G).
(PDF)
List of the genes up-regulated in nitrogen-starved wild-type ( gaf1 +) cells (Group −N).
(PDF)
List of the genes up-regulated in nitrogen-starved (−N) gaf1 Δ cells (Group −N/−G).
(PDF)
List of the genes in Subgroup A.
(PDF)
List of the genes in Subgroup B.
(PDF)
List of the genes in Subgroup C.
(PDF)
List of the genes in Subgroup D.
(PDF)
List of the genes in Subgroup E.
(PDF)
List of the genes in Subgroup F.
(PDF)
List of the genes in Subgroup G.
(PDF)