Abstract
There is a lack of fast and high resolution methods to measure metabolic activity of single cells in their native environment. Here we develop a straightforward, non-invasive and sensitive method to measure metabolic phenotype of single cells in a live tissue. By using NADH as optical biomarker and the phasor approach to Fluorescence Lifetime microscopy (FLIM) we identify cellular metabolic fingerprints related to different rates of oxidative phosphorylation and glycolysis. For the first time we measure a three dimensional metabolic gradient in the small intestine (SI) epithelia that appears tightly associated with epithelial cell proliferation, differentiation and the Wnt gradient. The highest free/bound NADH ratios are measured at the base of the crypt within the highly proliferative stem cells, indicating high levels of glycolysis. For the first time mouse small intestinal stem cells in intact live crypts are identified within the tissue by their metabolic fingerprint.
Although several methodologies to measure physiological processes in tissues and gene expression in single cells have been developed in the last decades, the major limitation remains the absence of robust and non-noninvasive methods that provide a fast quantitative readout of single cells metabolism within the native microenvironment of the living tissue.
Metabolic activity in proliferating cells, such as cancer cells and stem cells, is fundamentally different from non-proliferating cells. Warburg first observed that most cancer cells ferment glucose into lactate regardless of the presence of oxygen1. This effect, known as aerobic glycolysis, supports the efficient synthesis of macromolecular components necessary for rapidly dividing cells. Most proliferative cells rely on aerobic glycolysis in contrast to normal differentiated cells which rely primarily on oxidative phosphorylation2. During proliferation, the large increase in glycolytic flux rapidly generates cytosolic ATP resulting in high ATP/ADP and NADH/NAD+ ratios2,3,4.
The metabolic coenzyme nicotinamide adenine dinucleotide (NADH) is the principal electron acceptor in glycolysis and electron donor in oxidative phosphorylation. NADH ubiquity renders this coenzyme one of the most useful and informative intrinsic biomarkers for metabolism, mitochondrial function, oxidative stress, aging and apoptosis in live cells and tissues5. Since the pioneering work of Britton Chance6 metabolic imaging of NADH levels and of the relative amounts of reduced and oxidized NADH is extensively used to monitor changes in metabolism. Chemical methods that infer NADH:NAD+ ratios indirectly from the concentrations of redox couples such as lactate and pyruvate7 require the use of cell extracts and are incompatible with studying dynamics in intact living cells and tissues. Instead, optical readouts of NADH autofluorescence allows real time and non-invasive monitoring of the metabolic state of a cell during (patho)physiological changes and reports on levels of oxidative phosphorylation and glycolysis. Monitoring the NADH fluorescence intensity provides useful information on NADH/NAD+ ratios8, since NADH loses fluorescence upon oxidation to NAD+. However intensity-based measurements of NADH/NAD+ contain artifacts due to the heterogeneity of fluorophore concentration and to differing quantum yields of NADH in the free and bound form. This problem can be tackled by using fluorescence lifetime imaging (FLIM), since lifetime is a concentration-independent optical response and is minimally affected by tissue absorption and scattering and fluctuation in excitation intensity. FLIM reports on a fluorophore's micro-environment and can discriminate free or protein-bound NADH. The combination of FLIM and multi-photon excitation provides 3-D images of NADH lifetimes with cellular and subcellular resolution in living tissues with minimal photo damage and phototoxicity9,10, and is becoming a valuable technique to assess metabolic states of cells associated with carcinogenesis and cell differentiation in vitro11,12,13,14 and in vivo15,16. NADH has either a short or long lifetime component depending whether it is in a free or protein-bound state. Protein-bound NADH is characterized by a complex multi-exponential lifetime decay that has been related to its binding to different enzymes, such as malate dehydrogenase (MDH) and lactate dehydrogenase (LDH)17. Metabolic pathways related to carcinogenesis and differentiation are known to change NADH binding sites and enzymatic binding is directly related to NADH cycling through the energy production pathway18. For example, Bird et al. 2005 demonstrated that changes in the ratio of free to protein-bound NADH are associated with the NADH/NAD+ redox ratio in breast cancer cells11. Although technological advancements in time resolved microscopy19,20 are allowing for more sensitive measurements, one of the major limitations of NADH metabolic imaging remains the interpretation of complex multi-exponential lifetime decays for NADH. The traditional way of fitting intensity decays in every pixel with two exponentials assumes that the ratio of the “short” and “long” components is equivalent to the ratio of free/bound NADH. This approximation is misleading because free and protein-bound NADH have common exponential components and the NADH binding sites with different enzymes cannot be taken in account. We recently introduced the phasor approach to fluorescence lifetime imaging analysis that provides a fit-free, unbiased representation of the raw autofluorescence FLIM data16,21. We demonstrated that phasor FLIM is a very sensitive tool to quantify the relative concentrations of intrinsic fluorophores, regardless of the complexity of the lifetime decay and allows discrimination of small cellular redox ratio differences16.
In this work we develop a label-free method to measure non-invasively metabolic phenotypes of single cells in a live tissue. We quantify the rate of oxidative phosphorylation and glycolysis in single cells and we associate cellular metabolic fingerprint to their proliferation and differentiation state. We characterize the metabolic phenotype of native, living small intestinal crypt epithelium using the phasor approach to FLIM and the ratio of free to protein-bound NADH biomarker. The small intestine is an important model for stem cell biology and cancer research due the high proliferative rate of epithelial cells. The position and number of stem cells are strictly regulated in order to maintain tissue homeostasis and to prevent tissue degradation or tumor growth. One major regulator of cell proliferation, differentiation and stem cell self-renewal in the intestinal crypt is the Wnt signaling pathway22,23,24,25,26,27,28. Wnt signals are strongest at the base of each crypt where the Wnt target gene Lgr5 marks multipotent stem cells29. The tight regulation of self-renewal of these stem cells and proliferation of the committed progenitor cells they produce is subverted in cancer cells by aberrantly high levels of Wnt signaling leading to malignant transformation. Constitutive activation of the Wnt signaling pathway occurs most often via loss-of-function mutations in the adenomatous polyposis coli gene (APC) and when this critical event occurs in Lgr5+ stem cells, the resultant patterns of aberrant proliferation and differentiation lead to malignant transformation and colorectal cancer30,24.
Although recent work has characterized spectrally the intrinsic contrast in healthy and diseased gastric tissue31, to our knowledge no previous study has investigated the metabolism of intestinal tissue via the intrinsic biomarker NADH. Phasor approach to FLIM identifies different tissue components of the small intestine live tissue and differentiates the metabolic activity of crypt base epithelial cells (including Lgr5+ cells and Paneth cells) from terminally differentiated villus epithelial cells. We show that Lgr5+ stem cells have a unique metabolic fingerprint; their characteristic free/bound NADH ratio allows their label-free identification and separation both from the neighboring Paneth cells and from their differentiated progeny. By mapping the free/bound NADH concentration in the intestinal epithelium we measure the 3D metabolic gradients from the base of the crypt to the tip of the villi. For the first time we identify a unique metabolic fingerprint of proliferative small intestine stem cells and a metabolic trajectory (M-Trajectory) along the crypt-villus axis, which is strongly correlated with Wnt gradient and the level of cell proliferation and differentiation.
Results
Origin of the intrinsic contrast in the small intestine tissue
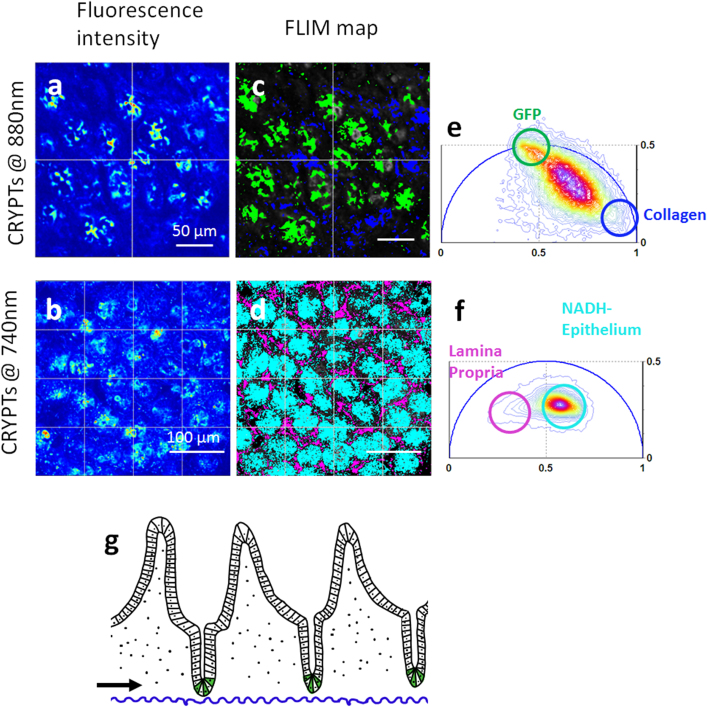
Two-photon microscopy and Phasor FLIM cluster analysis (See methods and ref.16) are used to visualize and identify the intrinsic contrast of the small intestine. Figure 1g shows a schematic diagram of small intestinal structure. The small intestine is covered by a single layer of epithelial cells organized into crypts. Each crypt is populated with 8–14 stem cells at the crypt base, whose progeny perpetually move upward and are lost from villus tips. All progeny are regenerated every 4–6 days. Crypt epithelia sit on a complex scaffold of myofibroblasts, vascular endothelia, and immune cells that comprise the lamina propria of the villus core. Figure 1 shows that different compartments of the small intestinal tissue are characterized by unique Phasor FLIM signatures that correspond to specific intrinsic sources. The two-photon intensity images and FLIM maps excited at 740 nm and 880 nm (Fig 1a–d) acquired at the base of the crypt (black arrow in Figure 1g) show round crypts that contain epithelial cells. Here we use transgenic mice expressing eGFP in Lgr5+ stem cells in intestinal crypt bases29. Individual GFP-Lgr5+ stem cells intercalate between adjacent Paneth cells (Fig 1b) at crypt bases, consistent with their previously described pattern29. Two-photon fluorescence intensity excited at 740 nm highlights NADH contrast of single epithelial cells within the round crypt base, showing subcellular details, such as the dim nuclei and bright mitochondria. (Fig 1d) The Phasor FLIM signature of the epithelial cells excited at 740 nm (Fig 1f) corresponds to the typical complex multi-exponential lifetime distribution of NADH located in the center of the phasor plot16. The blue-peaked emission spectrum of small intestine colon crypts (Fig SM1b) and the free NADH accumulation upon blocking cellular respiration (Fig SM1a) confirms that the primary contributor to the epithelial fluorescence signature is NADH, in agreement with previous work31. The regular spacing between round crypts is constituted by the lamina propria. When excited at 880 nm, lamina propria is characterized by a short lifetime (Fig 1a–c) and Second Harmonic Generation (SHG) signal (SM2), that corresponds to collagen fibers16. The Phasor FLIM signature of the lamina propria excited at 740 nm is instead characterized by a longer lifetime (purple in Fig 1d–f) that indicates a mixture of erythrocyte porphyrins16 within capillaries and NADH within fibroblasts and macrophages.
Figure 1. Intrinsic contrast in the small intestine.
(a-b) Fluorescence Intensity images at the base of small intestine crypts excited at 880 nm (a) and 740 nm (b). Images are acquired at the level of the black arrow in fig g. (c–d) Phasor FLIM maps of the tissue excited at 880 nm (c) and 740 nm (d) highlight Lgr5+GFP stem cells (green), collagen (blue), porphyrins within the lamina propria (purple) and NADH within the epithelial cells (cyan). Phasor plot histogram of the FLIM image at 880 nm (e) and 740 nm (f). The color scale (from blue to purple  ) corresponds to the 64 levels of the contours that indicate the percent occurrence in the phasor histogram of the pixels of the image. Four clusters corresponding to different tissue components are identified in the phasor distribution with different colors. (g) Epithelium is organized with differentiated villi and crypts that contain the stem cell (in green) while the lamina propria (dotted area) forms the core of each villus and is composed of loose connective tissue containing lymphocytes, myofibroblasts and a network of capillaries and lymphatics.
) corresponds to the 64 levels of the contours that indicate the percent occurrence in the phasor histogram of the pixels of the image. Four clusters corresponding to different tissue components are identified in the phasor distribution with different colors. (g) Epithelium is organized with differentiated villi and crypts that contain the stem cell (in green) while the lamina propria (dotted area) forms the core of each villus and is composed of loose connective tissue containing lymphocytes, myofibroblasts and a network of capillaries and lymphatics.
Proliferative stem cells in the small intestine have a unique NADH metabolic fingerprint
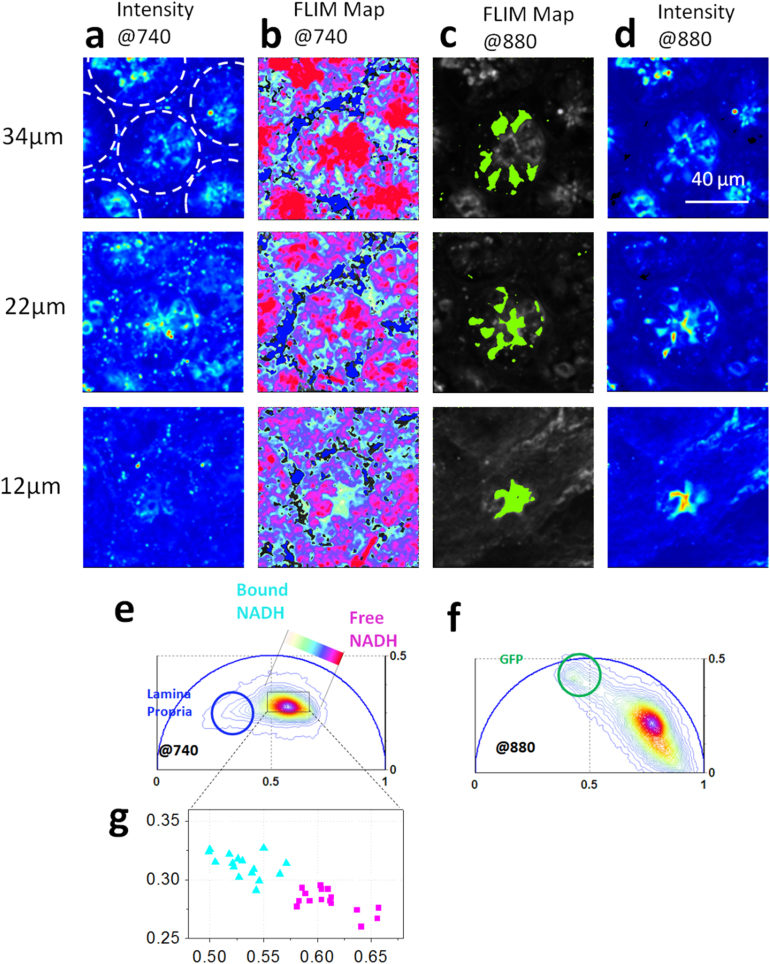
Here we show that the epithelial stem cells located at the base of the crypt have a unique NADH Phasor FLIM signature that allows their identification in living tissue without any extrinsic labeling. Stem cells at the base of the crypt alternate with Paneth cells, differentiated epithelial cells that function in innate immunity32 and as ‘nurse’ cells to the stem cells33. To distinguish between Paneth cells and stem cells, we use Lgr5-GFP mice to mark the Lgr5+ stem cell population at the base of small intestine (SI). (See Material and Methods). The excitation wavelength of the Ti:Sapphire laser is tuned between 880 nm and 740 nm to excite alternatively GFP and NADH9. Fig. 2 shows the 2-photon excited fluorescence images and FLIM maps acquired in ex-vivo small intestine from an Lgr5-GFP mouse excited at 740 nm (Fig. 2a–b) and at 880 nm (Fig. 2c–d). The broad lifetime distribution measured in crypt epithelial cells excited at 740 nm (Fig 1.f and Fig 2.b) has a characteristic linear-elongated pattern that reflects a mixture of free and bound NADH, yielding information on different distributions of metabolic states and redox ratios of the cells within the same crypt. In Fig 2 we map the relative concentration of free and bound NADH within the epithelial cells at the base of SI crypts, according to the FLIM phasor location of the free NADH measured in solution16 and the bound NADH measured in epithelial cells (Fig.SM1.a). Different ratios of free and protein-bound NADH reflect different rates of glycolysis and oxidative phosphorylation (SM4 and SM5) in agreement with ref.11.
Figure 2. Label-free identification of stem cells at the base of the small intestine.
Two-photon fluorescence intensity images excited at 740 nm (a) and 880 nm (d) of the small intestine from Lgr5-GFP mice that express eGFP in the crypt base stem cells. Dotted lines indicate the border of the round crypts within the SI tissue. (b) Phasor color maps at 740 nm of porphyrin (blue) and of the relative concentrations of free NADH and bound NADH in the lamina propria. Red-purple color indicates a high free/bound NADH ratio, while violet, cyan and white indicate linearly and progressively decreasing ratios free/bound NADH ratio, as shown in f. (c) Phasor color map at 880 nm highlights GFP (green) stem cells. (e) Phasor histogram of the FLIM image excited at 740 nm. Linear cluster represents all possible relative concentrations of free NADH (purple) and bound NADH (white). (f) Phasor histogram of the FLIM image excited at 880 nm, with a circular cluster that highlights the GFP (green). (g) Scatter plot of the cell phasor of all stem cells (cyan triangles) and Paneth cells (purple squares) excited at 740 nm.
The metabolic phasor FLIM signature at the base of the crypts excited at 740 nm (Fig 2.b) strikingly follows the map of stem cells intercalated between adjacent Paneth cells. Stem cells (cyan) are characterized by a lower free/bound NADH ratio with respect to the Paneth cells (purple) (Fig 2.b, Fig 2e,g). The unique NADH FLIM signature of stem cells and the alternated pattern of high and low free/bound NADH cells within the crypts is present in all crypts of the small intestine tissue, regardless of GFP expression (Fig 2.b and Fig 2.c). We calculate the average phasor values of stem cells and Paneth cells by performing manual image segmentation in the Lgr5-GFP positive crypts of Fig 2.b and Fig 2.c (See Material and Methods). We select the region of interest's stem cells using a cursor of arbitrary shape that marks the outlines of the single Lgr5+GFP stem cells in Fig 2.c. Paneth cells are identified at the base of the round crypts by the absence of Lgr5+GFP expression. The average phasor value of cells is calculated within the cursor and plotted in the scatter diagram of Fig 2g. The average FLIM phasor values of stem cells are significantly different from the values of Paneth cells, indicating a different metabolism (t-test, p<0.05, Fig 2g).
NADH metabolic trajectory from glycolysis to oxidative phosphorylation is associated with cellular proliferation and differentiation along the small intestine crypt-villus axis
Here we show that the epithelial cells in the small intestine follow a precise metabolic trajectory associated with free and protein-bound NADH. Proliferative cells at the base of the crypt are characterized by a glycolytic metabolic phenotype, while differentiated cells have an oxidative phosphorylation phenotype (Fig 3, Fig 4, SM4 and SM5).
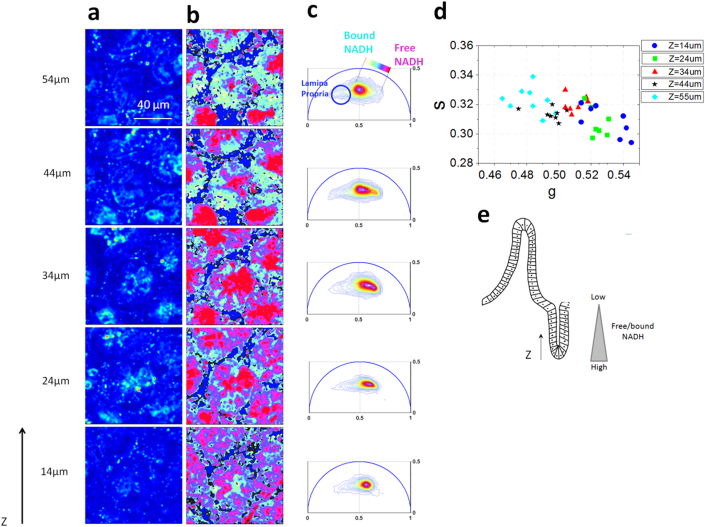
Figure 3. Metabolic gradient in the epithelial cells in the small intestine crypt.
Two-photon fluorescence intensity images (a) and free/bound NADH FLIM maps (b) of the ex-vivo small intestine crypts excited at 740 nm at different depths (z) of the tissue starting from the base of the crypt. (c) Phasor FLIM distribution at different depths (z). The circular blue cluster is used to highlight the porphyrin in the lamina propria. A linear cluster represents relative concentrations of free NADH (purple) and bound NADH (white). Red-purple color indicates a high free/bound NADH ratio, while violet, cyan and white indicate linearly and progressively decreasing ratios of free/bound NADH. (d) Scatter plot of the cell phasor of stem cells and differentiated epithelial cells at different depths from the collagen fibers of the basal membrane (Figure SM3) excited at 740 nm. Cyan diamond for Z = 55 µm, black stars for Z = 44 µm, red triangles for Z = 34 µm, green squares for Z = 24 µm and blue circles for Z = 14 µm). Along the Z axis the cell phasor shifts toward a longer lifetime indicating an increase of bound NADH with respect to free NADH. i.e. a decrease in NADH/NAD+ ratio. (e) Schematic diagram of the small intestine epithelia shows a free/bound NADH gradient along the epithelial cells of the SI crypt.
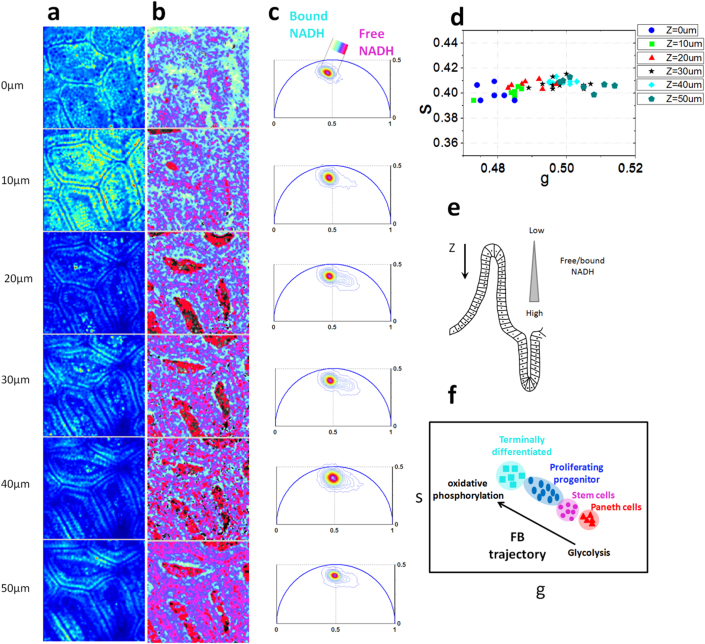
Figure 4. Metabolic gradient in the epithelial cells in the small intestine villi.
Two-photon fluorescence intensity images (a) and free/bound NADH FLIM maps (b) of ex-vivo small intestine mucosa excited at 740 nm at different depths (z) of the tissue starting from the tip of the villi. (c) Phasor FLIM distribution at different depths z. Circular blue cluster is used to highlight the porphyrin in the lamina propria vessels. A linear cluster represents the relative concentrations of free NADH (purple) and bound NADH (white). Purple color indicates a high free/bound NADH ratio, while violet, cyan and white indicate linearly and progressively decreasing ratios free/bound NADH ratio. (d) Scatter plot of the cell phasor of stem cells and differentiated epithelial cells at different depths from the tip of the villi. Along the Z the cell phasor shifts toward the shorter lifetime indicating an increase of free/bound NADH ratio. (e) Schematic diagram of the small intestine epithelia shows a free/bound NADH gradient along the epithelial cells of the SI villi. (f) Metabolic trajectory (M-trajectory) from free to protein-bound NADH indicated a shift from a glycolytic phenotype to an oxidative phosphorylation phenotype.
In Figure 3 and 4 we measure the three-dimensional NADH FLIM signature of small intestine and we map the metabolic gradients of epithelial cells in the small intestine from the base of the crypt to the villus tips. The stem cell pool is located at the base of the intestinal crypt where they self-renew and generate differentiated cell types (Fig 1g). When stem cells divide they frequently undergo asymmetric cell division, forming a new stem cell and a committed daughter cell. Stem cell cycling produces rapidly proliferating “committed progenitor” (CP) cells capable of differentiation towards all cell lineages. CP cells then undergo a limited number of cell divisions before they terminally differentiate on the villus surface.
In Figure 3 we imaged ex-vivo small intestine tissue from the ‘outside in’ using an intact section of small intestine. (Fig 3e and Material and Methods). Starting from the external serosal surface of intact small intestine we first imaged submucosal collagen fibers, then the crypt bases and interspersed lamina propria. (Fig SM1). In Figure 4 we open the SI tube to image the mucosal surface from the villus tips toward the SI crypt bases (Fig 4e and Material and Methods).
The NADH lifetime distribution measured in the epithelial cells excited at 740 nm is different at various depths (z) along the crypt/villus axis (Fig 3c–d and Fig 4c–d). Their three dimensional FLIM phasor is distributed along a linear trajectory in the phasor plot that corresponds to the mixture of free and bound NADH (Fig SM1.a and ref16). We define “M-Trajectory” (metabolic trajectory) the trajectory from a glycolytic phenotype with high free/bound NADH ratios to an oxidative phosphorylation phenotype with high free/bound NADH ratios (Fig 4g and SM4 and SM5). Mapping the relative concentration of free and bound NADH within the epithelial cells of the SI crypts and villi shows a shift toward lower ratios of free/bound NADH with differentiation from the base of the crypt to the villus tip (In Fig 3b and 4b). We calculate the average phasor values of stem cells and the differentiating progenies by performing manual image segmentation. (See Material and Methods). The average phasor value of epithelial cells at different depths is calculated within the cursor and plotted in the scatter diagram of Fig 3g and 4g. The FLIM phasor values of stem cells are statistically different from differentiated epithelial cells at different depths, (t-test, p<0.05 Fig 3d) showing a gradual decrease of the free/bound NADH ratio along the crypt. Stem cells (z = 14 um and z = 24 um) (Fig 3d) and Paneth cells (Fig 2b), located within the Wnt-rich environment at the crypt base, are characterized by a glycolytic metabolic phenotype within the M-Trajectory (Fig 4g, SM4 and SM5) with the shortest lifetime and the highest free/bound NADH (i.e NADH/ NAD+) ratio. Movement up the crypt to the committed progenitor compartment (Fig 3b–d) and on to areas of fully differentiated cells on the mucosal surface (Fig 4b–d) corresponds to a movement along the M-Trajectory, with decreasing free/bound NADH (i.e NADH/ NAD+) ratios (white Fig 3b–d), thus indicating a metabolic shift from glycolysis to oxidative phosphorylation (SM4 and SM5). The same trend is observed in five different animals (SM6 and SM7).
Discussion
Here we developed a non-invasive, label-free optical method to measure and identify the NADH metabolic signature associated with glycolysis and oxidative phosphorylation within a living tissue. Our method provides the identification of metabolic state associated with cell proliferation and differentiation within the live murine small intestine and identifies the proliferative intestinal stem cells based on their unique metabolic fingerprint.
By performing ex-vivo two-photon excited Fluorescent lifetime microscopy and Phasor analysis we show that we can identify different SI cell populations and structures based on their unique FLIM signatures (Fig 1). Collagen fibers (Fig 1, SM2 and SM3) are imaged at the base of the crypts supporting the stem cell niche, and the lamina propria is identified by its peculiar long lifetime that indicates the presence of porphyin within the vascular network of the capillaries (Fig 1d, f). We show that the major source of intrinsic contrast of epithelium is the metabolic coenzyme NADH (Fig 1d–f and Fig. SM1). These findings are consistent with the measurements performed on intrinsic contrast by hyper-spectral microscopy in gastric tissue31. Our method provides a straightforward way to identify and target the intrinsic contrast of intestinal tissue by assigning FLIM contrast to intracellular and extracellular physiological fluorophores (Fig 1). More importantly our method provides physiological information related to the metabolism of single cells within the tissue. We show that the Phasor FLIM signature of epithelial cells yields information on the content of free and protein-bound forms of NADH associated with the glycolysis and oxidative phosphorylation (SM4 and SM5) and with cellular proliferation and differentiation (Fig 3 and Fig 4).
Our results show that Phasor FLIM identifies the highly proliferative intestinal stem cells based on their metabolic signature without the need of any extrinsic label or genetic marker. We showed that the Lgr5 positive stem cells have a unique NADH FLIM signature that distinguishes them both from the neighboring Paneth cells (Fig 2) and from differentiated epithelial cells along the crypt (Fig 3) and the villi (Fig 4). FLIM maps of free and bound NADH (Fig 2b and Fig 3b) and Phasor FLIM metabolic fingerprints (Fig 2g and Fig 3d) sort intestinal stem cells and differentiated progenies by their metabolic state. The metabolic state of intestinal stem cells (Fig 3) is characterized by a high ratio of free/bound NADH, indicating that they mainly rely on glycolysis, as expected in highly proliferative cells2,3. We demonstrate that the three-dimensional Phasor FLIM signature of small intestine allows straightforward mapping of the relative concentration of free and bound NADH and provides information on the metabolic gradients associated with proliferation and differentiation (Fig 3c and Fig 4c). The evolution of the cell phasor fingerprint from the base of the crypt to the top of the villi (Fig 3d and Fig 4d) reflects a decrease of free NADH with respect to protein-bound NADH along the M-Trajectory (Fig 4g). Our findings therefore indicate a decrease in the relative contribution of the glycolytic metabolic pathway with respect to oxidative phosphorylation during epithelial stem cell differentiation in the living small intestine (Fig 3, Fig 4, SM4 and SM5). The observed shift from glycolysis to oxidative phosphorylation is consistent with the increased NADH lifetime measured during adult stem cell differentiation in vitro12,13 and in living tissue16. The change in the NADH FLIM fingerprint during differentiation (Fig 3 and Fig 4) may also suggest a change in the binding sites of NADH with different enzymes such as malate dehydrogenase (MDH) and lactate dehydrogenase (LDH)17. The metabolic gradient at the tip of the villi (Fig 4d) could indicate a contribution of FAD, while cell-to-cell metabolic heterogeneity at the tip of the villi (Fig 4b) might indicate cell types with different function, such as goblet cells, enterocytes or enteroendocrine cells. Alternatively, it could indicate epithelial cells under oxidative stress and undergoing apoptosis.
Interestingly the free/bound NADH FLIM gradient that we measure (Fig 3 and Fig 4) is tightly associated with the Wnt gradient along the crypt-villus axis, which shows the highest signaling activity at the crypt bottom where it controls cell proliferation, stem cell self-renewal and cell fate decisions24,34. Epithelial cells at the crypt base (Lgr5+ stem cells and Paneth cells) are both characterized by high Wnt signaling26,35, and we observe them to have the highest free/bound NADH ratio, (Fig 2 and Fig 3) i.e the highest glycolysis/oxidative phosphorylation ratio (SM4 and SM5). Interestingly, Paneth cells are characterized by a higher free/bound NADH ratio than Lgr5+ stem cells, indicating that they utilize a strong, active glycolytic metabolism for their functions. Although Paneth cells are differentiated cells, they express several Wnt target genes such as defensins and cryptdins26,36 and they provide essential niche signals for stem cell support and self renewal such as EGF, Wnt3, R-Spondin and Notch33,37.
To our knowledge this is the first time that intestinal stem cells have been identified label-free within a living tissue based on their metabolic fingerprint. Metabolic states of single cells are measured and the rate of glycolysis and oxidative phosphorylation are mapped in the living tissue. We report for the first time that a metabolic gradientassociated with free and protein-bound NADH correlates with cellular proliferation, differentiation and Wnt signaling in the small intestine. The ability to identify intestinal stem cells and to discriminate the metabolic state of highly proliferative cells associated with high Wnt signaling without any extrinsic marker might greatly impact the field of stem cell biology, cancer research and clinical diagnosis. Our methods would allow non-invasive imaging of metabolic shifts in any given cell compartment, associated with epithelial growth dysregulation in any model of colorectal cancer. Mutated cells with constitutively active oncogenic pathways that allow a metabolic phenotype of biosynthesis independent of normal physiological conditions could be isolated and detected at early states of the disease. In future work we plan to characterize the metabolic signatures of colon and small intestine tissues in transgenic animal models of inflammatory bowel disease and colorectal cancer. Since our approach is label-free and non-invasive, our method could have tremendous utility in the diagnosis and management of cancer enabling the development of “metabolic histology” signatures and eventually in-vivo and in-situ endoscopic microscopy38. Miniaturized fiber optic approaches have been adapted for intravital microscopy and endoscopic clinical imaging39 with recent advancement in fluorescence lifetime imaging endoscopy40. FLIM endoscopes would allow in vivo diagnostic imaging with molecular contrast- and label-free identification of cancer cells and possibly stem cells based on their metabolic states.
Our method provides a straightforward and quantitative measurement of oxidative phosphorylation and glycolysis metabolic rates (SM4 and SM5) and associates metabolic phenotypes with proliferation and differentiation within the M-Trajectory (Fig 3 and Fig 4). By providing high sensitivity to the free/bound NADH molecular gradient our method represents a promising tool to monitor (patho)physiological processes correlated to metabolic changes. Our methods could be used to identify single highly proliferative stem cells and cancer cells and to measure metabolic gradients and dynamic metabolic adaptation to the level of oxygen and nutrients uptake during development and cancer progression in a variety of living tissues.
Methods
Animal model and procedures
We used Lgr5+GFP mice that express GFP within the stem cells of the small intestine and colon epithelium (Jackson Labs strain 008875). Ex-vivo tissue imaging is performed immediately after mice euthanasia. Selected freshly excised small intestine is cleaned by flushing with Hank's Balanced Salt Solution (HBSS) (Thermo Scientific Part NumberSH30031.02). To image the SI crypt, images were acquired from the outside (serosal) surface. To acquire images of the SI villi, a length of SI tissue was opened flat, gently rinsed and imaged, villi up, in a Mat-Tek dish with a #1 coverslip. Tissue was bathed in HBSS while imaging and all imaging occurred immediately after isolation. All procedures followed were reviewed and approved by the Irvine University Institutional Animal Care and Use Committee (IACUC animal protocol number 2002-2357-3).
We block the respiratory chain of the DLD-1 colon cancer cells by means of potassium cyanide (KCN) to inhibit the Oxidative phosphorylation and increase the mitochondrial concentration of NADH. KCN in PBS was added to the culture medium with a final concentration of 4 mM. Cells were imaged immediately after the addition of KCN.
DLD-1 colon cancer cells are treated for 24 hours with 50 mM Sodium dichloroacetate (Sigma #34779) to inhibit glycolysis.
NIH3T3 fibroblasts are treated for one hour with Low Glucose (4.4 mM glucose) and High Glucose (22 mM glucose) media. Low Glucose medium (Invitrogen Dulbecco's Modified Eagle Medium (D-MEM), # 11885-092, no Fetal Bovine Serum). High Glucose medium (Invitrogen Dulbecco's Modified Eagle Medium (D-MEM) # 11965-092, no Fetal Bovine Serum).
Imaging
Fluorescence lifetime images are acquired with a Zeiss 710 microscope coupled to a Ti:Sapphire laser system (Spectra-Physics Mai Tai) and a ISS A320 FastFLIM system20. A 40×0.8 NA water immersion objective (LUMPlanFl Olympus.) is used. For image acquisition the following settings are used: image size of 256×256 pixels or 1024v1024 pixels and scan speed of 25 μm/pixel. A dichroic filter (690 nm) is used to separate the fluorescence signal from the laser light and the fluorescence. For the acquisition of FLIM images, fluorescence is detected by a photomultiplier (H7422P-40 of Hamamatsu) and a 610 nm short pass filter is placed in front of the detector. FLIM data are acquired and processed by the SimFCS software developed at the Laboratory of Fluorescence Dynamics. The excitation wavelengths used were 880 nm and 740 nm. An average power of about 5 mW was used to excite the live tissue. FLIM calibration of the system is performed by measuring the known lifetime of the fluorescein with a single exponential of 4.04 ns. FLIM data are collected until 100 counts in the brightest pixel of the image are acquired. Typically the acquisition time was of the order of few seconds.
FLIM Phasor data analysis
Every pixel of the FLIM image is transformed in one pixel in the phasor plot as previously described in ref16,21. The coordinates g and s in the phasor plot are calculated from the fluorescence intensity decay of each pixel of the image by using the transformations defined in ref16,21. The analysis of the phasor distribution is performed by cluster identification. Clusters of pixel values are detected in specific regions of the phasor plot. The cluster assignment is performed by taking in account not only the similar fluorescence properties in the phasor plot but also exploiting the spatial distribution and localization in cellular substructures or tissues, as described in ref16. Fractional intensities of chemical species in every pixel of the image are evaluated with a graphical analysis in the phasor plot as described in ref21. We perform image segmentation on the FLIM data by selecting the region of interest of cells within the tissue. The region of interest of cells is selected by using a cursor with arbitrary shape. We calculate the phasor average value within these regions of interest. When measuring the cell phasor, all pixels of the cell (about 1000) are taken in account and the signal to noise ratio of the FLIM signature of cells is higher than in single pixels. All phasor transformation and the data analysis of FLIM data are performed using SimFCS software developed at the LFD.
All procedures followed were reviewed and approved by the Irvine University Institutional Animal Care and Use Committee (IACUC animal protocol number 2002-2357-3).
Author Contributions
CS performed experiments, wrote the manuscript. RE, KP, MW, PD prepared samples and participated to the experiments. EG wrote the software for phasor analysis. All authors discussed the experiment and reviewed the paper.
Supplementary Material
Supplemental Information
Acknowledgments
Work supported in part by NIH-P41 P41-RRO3155, P41 GM103540, P50-GM076516, RO1 HD49488, PO1 HD47675, CIRM RC1-00110 , RO1 CA096878, RO1 CA108697, P30 CA62203.
References
- Warburg O. On the origin of cancer cells. Science 123, 309–314 (1956). [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C. & Thompson C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324(5930), 1029–33 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis R. J., Lum J. J., Hatzivassiliou G. & Thompson C. B. The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell Metab. 7(1), 11–20 (2008). [DOI] [PubMed] [Google Scholar]
- Christofk H. R. et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452(7184), 230–3 (2008). [DOI] [PubMed] [Google Scholar]
- Heikal A. A. Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomark Med 4(2), 241–63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B., Cohen, P., Jobsis F. & Schoener B. Intracellular oxidation-reduction states in vivo. Science. 137, 499–508 (1962). [DOI] [PubMed] [Google Scholar]
- Williamson D. H. L., Patricia. & Krebs H. A. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J. 103(2), 514–27 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasischke K. A., Vishwasrao H. D., Fisher P. J., Zipfel W. R. & Webb W. W. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science 305(5680), 99–103 (2004). [DOI] [PubMed] [Google Scholar]
- Zipfel W. R. et al. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci U S A 100(12), 7075–80 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squirrell J. M., Wokosin D. L., White J. G. & Bavister B. D. Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability. Nat Biotechnol. 17(8), 763–7 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird D. K. et al, Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 65(19), 8766–73 (2005). [DOI] [PubMed] [Google Scholar]
- Guo H. W. et al. Reduced nicotinamide adenine dinucleotide fluorescence lifetime separates human mesenchymal stem cells from differentiated progenies. J Biomed Opt. 13(3), 050505 (2008). [DOI] [PubMed] [Google Scholar]
- König K., Uchugonova A. & Gorjup E. Multiphoton fluorescence lifetime imaging of 3D-stem cell spheroids during differentiation. Microscopy Research and Technique 74(1), 9–17, 2011. [DOI] [PubMed] [Google Scholar]
- Stringari C., Sierra R., Stringari C., Sierra R., Donovan P. J. & Gratton E. Label-free separation of human embryonic stem cells and their differentiating progenies by Phasor Fluorescence Lifetime Microscopy. J Biomed Opt. 17(4), 046012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skala M. C. et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A 104(49), 19494–9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringari C. et al. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc Natl Acad Sci U S A. 108(33), 13582–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q. & Heikal A. A. Two-photon autofluorescence dynamics imaging reveals sensitivity of intracellular NADH concentration and conformation to cell physiology at the single-cell level. J Photochem Photobiol B. 95(1), 46–57 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S. & Bhatt D. K. Histochemical studies on the distribution of certain dehydrogenases in squamous cell carcinoma of cheek. Indian J Cancer 26(1), 21–30 (1989). [PubMed] [Google Scholar]
- Becker W. et al. Fluorescence lifetime imaging by time-correlated single-photon counting. Microsc Res Tech. 63(1), 58–66 (2004). [DOI] [PubMed] [Google Scholar]
- Colyer R., Lee C. & Gratton E. A novel fluorescence lifetime imaging system that optimizes photon efficiency. Microsc Res Tech. 71(3), 201–13 (2008). [DOI] [PubMed] [Google Scholar]
- Digman M. A., Caiolfa V. R., Zamai M. & Gratton E. The phasor approach to fluorescence lifetime imaging analysis. Biophys J. 94(2), L14–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald B. T., Tamai K. & He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 17, 9–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M. et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111, 241–50 (2002). [DOI] [PubMed] [Google Scholar]
- Reya T. & Clevers H. Wnt signalling in stem cells and cancer. Nature 434(7035), 843–50 (2005). [DOI] [PubMed] [Google Scholar]
- Klaus A. & Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 8, 387–98 (2008). [DOI] [PubMed] [Google Scholar]
- van der Flier L. G. & Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 71, 241–60 (2009). [DOI] [PubMed] [Google Scholar]
- Korinek V. et al, Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 19(4), 379–83 (1998). [DOI] [PubMed] [Google Scholar]
- Najdi R., Holcombe R. F. & Waterman M. L. Wnt signaling and colon carcinogenesis: beyond APC. J Carcinog. 10(5) (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449(7165), 1003–7 (2007). [DOI] [PubMed] [Google Scholar]
- Barker N. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457(7229), 608–11 (2009). [DOI] [PubMed] [Google Scholar]
- Grosberg L. E., Radosevich A. J., Asfaha S., Wang T. C. & Hillman E. M. Spectral characterization and unmixing of intrinsic contrast in intact normal and diseased gastric tissues using hyperspectral two-photon microscopy. PLoS One 6(5), e19925 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette A. J. Paneth cell α-defensins in enteric innate immunity. Cell Mol Life Sci. 68(13), 2215–29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469(7330), 415–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E. et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 111(2), 251–63 (2002). [DOI] [PubMed] [Google Scholar]
- Gregorieff A. et al. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology 129(2), 626–38 (2005). [DOI] [PubMed] [Google Scholar]
- vanEs J. H. et al.Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol 7, 381–6 (2005). [DOI] [PubMed] [Google Scholar]
- de Lau W B. N. et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476(7360), 293–7 (2011). [DOI] [PubMed] [Google Scholar]
- Dela Cruz J. M., McMullen J. D., Williams R. M. & Zipfel W. R. Feasibility of using multiphoton excited tissue autofluorescence for in vivo human histopathology. Biomed Opt Express 1, 1320–1330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. T. et al. Point-of-care pathology with miniature microscopes. Anal Cell Pathol (Amst) 34(3), 81–98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy G. T. et al. A fluorescence lifetime imaging scanning confocal endomicroscope. J Biophotonics. 3(1–2), 103–7 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information