Abstract
Pancreatic ductal adenocarcinoma (PDAC) is characterized by overexpression of Enhancer-of Zeste-Homolog-2 (EZH2), which plays a pivotal role in cancer-stem-cell (CSC) self-renewal through methylation of histone-H3-lysine-27 (H3K27m3). Against this background, EZH2 was identified as an attractive target and we investigated the interaction of the EZH2-inhibitor DZNeP with gemcitabine.
EZH2 expression was detected by quantitative-RT-PCR in 15 PDAC cells, including 7 primary cell cultures, showing expression values correlated with their originator tumors (Spearman-R2=0.89, P=0.01). EZH2 expression in cancer cells was significantly higher than in normal ductal pancreatic cells and fibroblasts.
DZNeP (5 μM, 72-hour-exposure) modulated EZH2 and H3K27m3 protein expression, and synergistically enhanced the antiproliferative activity of gemcitabine, with combination index values of 0.2 (PANC-1), 0.3 (MIA-PaCa-2) and 0.7 (LPC006). The drug combination reduced the percentages of cells in G2/M phase (e.g., from 27 to 19% in PANC-1, P<0.05), and significantly increased apoptosis compared to gemcitabine-alone. Moreover, DZNeP enhanced the mRNA and protein expression of the nucleoside transporters hENT1/hCNT1, possibly because of the significant reduction of deoxynucleotides content (e.g., 25% reduction of deoxycytidine-nucleotides in PANC-1), as detected by LC-MS/MS.
DZNeP decreased cell migration, which was additionally reduced by DZNeP/gemcitabine combination (-20% in LPc006, after 8-hour exposure, P<0.05), and associated with increased E-cadherin mRNA and protein expression. Furthermore, DZNeP and DZNeP/gemcitabine combination significantly reduced the volume of PDAC spheroids growing in CSC-selective-medium, and decreased the proportion of CD133+ cells.
All these molecular mechanisms underlying the synergism of DZNeP/gemcitabine combination support further studies on this novel therapeutic approach for treatment of PDAC.
Keywords: Pancreatic ductal adenocarcinoma, EZH2, DZNeP, gemcitabine
Introduction
With a 5-year survival rate of less than 5 percent, pancreatic ductal adenocarcinoma (PDAC) is the most lethal among the major solid tumors (1). Despite extensive clinical and scientific efforts, the grim prognosis of this disease has not improved over the past decade (2). The main reasons for the lack of efficient therapeutic strategies include invasive behavior and intrinsic resistance to most chemo-/radio- and immuno-therapy regimens (3).
Recently PDAC emerged as a Cancer Stem Cell (CSC) – driven disease. Pancreatic CSCs are highly tumorigenic and have the abilities to self-renew and produce differentiated progeny. Pancreatic CSCs possess the ability to undergo epithelial-mesenchymal transition (EMT) (4) and form spheroids in serum-free-medium containing well-defined growth factors. The PDAC spheroids showed increased proliferation, invasiveness, and metastasis (5). CSCs have also been associated with chemoresistance to gemcitabine (6-8). Against this background, studies on key determinants in CSCs can provide both biomarkers of PDAC aggressiveness and optimal novel targets to overcome chemoresistance.
Enhancer of Zeste Homolog 2 (EZH2) is a histone methyltransferase essential for self-renewal of CSCs (9). This protein is the catalytic subunit of Polycomb Repressive Complex 2 (PRC2), one of the two multimeric repressive complexes in the organization of the Polycomb group (PcG). PcG proteins act as important epigenetic mediators that can repress gene expression by forming multiple complexes leading to histones methylation, resulting in epigenetic control of gene expression (10-11). In particular, EZH2 can silence several tumor suppressor genes by trimethylation at lysine 27 of histone H3 (H3-K27) (10), playing an important role in tumor development (12).
EZH2 is overexpressed in many cancer types, including PDAC. Recently, EZH2 expression has been associated with decreased E-cadherin expression and poor prognosis in PDAC patients (13). Furthermore, Ougolkov and colleagues showed that EZH2 is an important factor in PDAC cell chemoresistance. In particular, EZH2 depletion by RNA-interference sensitized PDAC cells to gemcitabine (14), which is used in the first-line treatment for PDAC. Since gemcitabine has very limited efficacy (5, 15), novel therapeutic strategies combining gemcitabine with targeted agents against EZH2 are warranted.
Recently the cyclopentenyl analog of 3-deaza-neplanocin A (DZNeP) was identified as a compound capable of reducing the levels of EZH2. This compound is a global histone methylation inhibitor by suppressing the activity of S-adenosyl-L-homocysteine (AdoHcy) hydrolase, the enzyme responsible for the reversible hydrolysis of AdoHcy to adenosine and homocysteine (16). This results in intracellular accumulation of AdoHcy, which leads to inhibition of the S-adenosyl-L-methionine–dependent lysine methyltransferase (KMTase) activity. S-adenosyl-methionine (AdoMet) metabolism represents a key cellular mechanism for methyl-group donation for a variety of methylation dependent metabolic processes that are disrupted by DZNeP, including production of methyltransferases such as EZH2 (17).
In the present study, we evaluated the EZH2 expression in PDAC tissues and cells, and the growth inhibition by DZNeP in combination with gemcitabine in monolayer cell cultures and cells growing as spheroids in serum-free-CSC-medium. Furthermore, we characterized several factors, including cell cycle perturbation, apoptosis induction and inhibition of cell migration as well as modulation of the expression of several genes involved in the DZNeP/gemcitabine interaction.
Material and methods
Drugs and chemicals
DZNeP was provided by Dr. Victor E. Marquez (NCI, NIH, Frederick, MD), while gemcitabine was a gift from Eli Lilly Corporation (Indianapolis, IN). The drugs were dissolved in sterile water, and diluted in culture medium before use. RPMI medium, foetal bovine serum (FBS), penicillin (50 IU/ml) and streptomycin (50 μg/ml) were from Gibco (Gaithersburg, MD). All other chemicals were purchased from Sigma-Aldrich (Zwijndrecht, The Netherlands).
Cell culture
Eight PDAC cell lines (PANC-1, MIA-PaCa-2, BxPc3, Capan-1, PL45, HPAC, HPAF-II, and CFPAC-1), the human pancreatic duct epithelial-like cell line hTERT-HPNE and skin fibroblasts Hs27 were obtained from the American Type Culture Collection (ATCC, Manassas, VA), while seven primary PDAC cultures (LPc006, LPc028, LPc033, LPc067, LPc111, LPc167 and PP437) were isolated from patients at the University Hospital of Pisa (Pisa, Italy), as described previously (18). Cells were cultured in RPMI-1640, supplemented with 10% heat-inactivated-FBS and 1% streptomycin/penicillin at 37°C, and harvested with trypsin-EDTA in their exponentially growing phase. The cell lines were tested for their authentication by PCR profiling.
Quantitative Real-Time PCR
Total RNA was extracted using the TRIREAGENT-LS (Invitrogen, Carlsbad, CA, US) and its yields and purity were checked at 260-280 nm with NanoDrop®-1000-Detector (NanoDrop-Technologies, Wilmington, NC). In order to prevent RNA degradation, the cells were harvested quickly on ice. One μg of RNA was reverse transcribed using the DyNAmo-cDNA-Synthesis Kit (ThermoScientific, Vantaa, Finland), according to the manufacturers’ instruction. In order to evaluate whether the expression of EZH2 was similar in the primary cells and their originator tumors, we also extracted RNA from these 7 tumors, after laser-microdissection with a Leica-LMD6000 instrument (Leica, Wetzlar, Germany), using the QiaAmp-RNA-micro-Kit (Qiagen, San Diego, CA), as described previously (18).
Primers and probes to specifically amplify EZH2, hENT1, CD133, and E-cadherin (Hs01016789_m1, Hs00191940_m1, Hs01009250_m1, and Hs01023894_m1, respectively). were obtained from Applied Biosystems (Forster City, CA).
The real-time quantitative PCR reactions were performed in the ABIPRISM-7500 sequence detection system instrument (Applied Biosystems).
We performed a preliminary analysis of 3 housekeeping genes (β-actin, GAPDH and Beta-2-microglobulin) in all our PDAC cells. Since the values of β-actin were the closest to the geometric mean values of these housekeeping genes, we used this housekeeping for the normalization of all the following analysis. Preliminary experiments were carried to demonstrate that the efficiencies of amplification of target and reference genes are approximately equal (18).
Western blot
In order to evaluate modulation of EZH2 and H3K27me3 protein expression, the PANC-1, MIAPaCa-2, and LPC006 cells were treated with 5 μM DZNeP for 72 hours, as reported (19). Blotting procedures were performed as described previously (18). Membranes were incubated overnight at 4°C with purified mouse anti-EZH2 mAb (BD Biosciences, Breda, The Netherlands) at 1:1000 dilution in blocking solution (Rockland in PBS-T), rabbit anti-H3K27me3 (1:1000; Upstate Biotechnology, Waltham, MA), rabbit anti-hENT1 and rabbit anti-hCNT1 (1:1000, kindly provided by Prof. M. Pastor-Anglada), and mouse anti-β-actin (1:50.000; Sigma–Aldrich Chemicals). The membrane was then probed for 1-hour with the goat-anti-mouse-InfraRedDye (1:10.000, Westburg, The Netherlands) or goat-anti-rabbit-InfraRedDye (1:10.000, Westburg) secondary antibodies. Fluorescent proteins were detected by an Odyssey Infrared Imager (LI-COR Biosciences, Lincoln, NE), at 84-μm resolution, 0-mm offset, using high quality settings. Then, the intensities of protein bands were quantified using the Odyssey v.3.0 software (LI-COR, Bioscience).
Immunocytochemistry
The LPc006 cells were grown in Chamber Slides System (Lab-Tek, Chicago, IL) in a humidified incubator. After 48 hours the cells were fixed with 70% ethanol for 10 minutes. Immunocytochemistry was performed using a monoclonal mouse-anti-human E-cadherin antibody (Cell Signaling, Euroclone, Milan, Italy; 4°C overnight incubation and 1:30 dilution in PBS). Cells were then stained with avidin-biotin-peroxidase complex (UltramarqueTM-HRP-Detection, Greenwood, AR). Negative controls were obtained replacing the primary antibody with PBS. The sections were reviewed and scored blindly by comparing the staining of treated cells versus untreated cells (positive control, basal expression), using a system based on staining intensity and on the number of positively stained cells, as described (18).
Growth inhibition studies
Cell growth inhibitory effects of the DZNeP, gemcitabine and their combination were studied using the sulforhodamine-B (SRB) assay. Cells were seeded in triplicate at 5×103 cells/well and kept at 37°C for 24 hours. Then the cells were treated for 72 hours with DZNeP (0.001-20 μM), gemcitabine (0.001-500 nM) and DZNeP at fixed concentration of 5 μM simultaneously with gemcitabine (0.001-500 nM). After 72 hours, plates were processed for the SRB assay as described earlier (20). In order to determine whether the drug could kill cells, we also measured Optical Density (OD) of the day of drug addition, since cell kill could lead to a decrease in the OD of day 0. Therefore, for the cell growth inhibition curves, the OD after 72 hours was corrected for the mean OD observed for the control wells at the day of drug addition (day 0 value). The 50% growth inhibitory concentration (IC50) was calculated by non-linear least squares curve fitting (GraphPad PRISM, Intuitive Software for Science, San Diego, CA).
Evaluation of synergistic/antagonistic interaction of DZNeP with gemcitabine
The drug interaction of DZNeP and gemcitabine was evaluated by the median-drug effect analysis method originally described by Chou (21). Cell growth inhibition of the combination was compared with the cell growth inhibition of each drug alone using the combination index (CI), where CI<0.9, CI=0.9-1.1, and CI>1.1 indicated synergistic, additive and antagonistic effects, respectively. Data analysis was carried out using CalcuSyn software (Biosoft, Oxford, UK). Since we considered growth inhibition lower than 50% as not relevant, CI values at fraction affected (FA) of 0.5, 0.75 and 0.9 were averaged for each experiment, and this value was used to calculate the mean between experiments.
Cell-cycle analysis and measurement of cell death
Cell-cycle modulation and cell death induced by treatments with 5 μM DZNeP, gemcitabine at IC50 values and their combination was investigated using 105 cells, after 24 and 72 hours, by Propidium Iodide (PI) staining using a FACScaliber flow-cytometer (Becton-Dickinson, San Jose‘, CA). Data analysis was carried out with CELLQuest (Becton-Dickinson), while cell cycle distribution was determined using Modfit (Verity-Software, Topsham, ME), as described (22). DZNeP, gemcitabine and their combination were also characterized for their ability to induce cell death, by evaluating the sub-G1 region of the FACS analysis (23), and by fluorescence microscopy analysis with bisbenzimide staining (22).
In vitro migration assay (Wound-healing assay)
Migration was evaluated using the LeicaDMI300B (Leica) migration station integrated with the Scratch-Assay 6.1 software (Digital-Cell Imaging Labs, Keerbergen, Belgium). Briefly, cells were plated at a density of 3×104 cells/well onto 96-well plates, and, after 24 hours, artificial wound tracks were created by scraping with a specific scratcher within the confluent monolayers. After removal of the detached cells by gently washing with PBS, the cells were fed with fresh medium and exposed to 5 μM DZNeP, gemcitabine at IC50, or their combination. The ability of the cells to migrate into the wound area was assessed by comparing the pixels in the images taken at the beginning of the exposure (time 0), with those taken after 4, 6, 8, 24 and 48 hours.
DZNeP/Gemcitabine activity in multicellular spheroids
PANC-1, MIA-PaCa-2 and LPc006 spheroids were established seeding 10000 cells/ml in DMEM/F12+GlutaMAX-I (1:1) with insulin-transferrin-selenium (1:1000, Gibco, Invitrogen), in 24-well ultra-low attachment plates (Corning, NY, US), according to manufacturers’ protocol, as described earlier (24). Spheroids were generated for 10-14 days, and then harvested for growth inhibition studies in 96-well plates, as well as for RNA isolation.
After checking their growth rate and stability, the spheroids were treated with 5 μM of DZNeP, IC50s of gemcitabine and their combination for 72 hours. The cytotoxic effects were evaluated by measuring the size and number of spheroids with the microscope LeicaDMI300B (Leica), taking 9 pictures for each well. Spheroid volume (V) was calculated from the geometric mean of the perpendicular diameters D=(Dmax+Dmin)/2, (V=4/3π(D/2)3).
Liquid chromatography–mass spectrometry (LC-MS/MS) measurement of adenosine and phosphorylated deoxynucleosides
Analysis by LC-MS/MS was used to determine total cytosolic adenosine as well as total phosphorylated deoxynucleosides. The latter were calculated from the difference before and after alkaline phosphatase treatment, as described previously (25). Approximately 2×106 cells were seeded into 6-well plates, and exposed to either 5 μM DZNeP, gemcitabine at IC50 values, or DZNeP/gemcitabine combination for 24 hours, prior to being snap frozen as a pellet. Cell pellets were re-suspended in a known aliquot of phosphate buffer and precipitated with excess isopropyl alcohol. The supernatant was removed and evaporated to dryness via freeze-drying. The dry samples were reconstituted in 200 μl of water and 20 μl aliquots were used for analysis. The remaining samples were treated quantitatively with alkaline phosphatase (4 units) at 37°C overnight. Chromatography was conducted using a Dionex Ultimate 3000 micro HPLC coupled via a Turbo spray ionization source to a SCIEX API 3000 mass spectrometer (Applied). Data analysis was performed with v.1.52 Analyst software (AB Sciex, Nieuwerkerk aan den Ijssel, Netherlands) controlled by Dionex Mass Spectrometry Link combined with Chromeleon management software modules (Thermo Scientific).
Statistical analysis
All experiments were performed in triplicate and repeated at least twice. Data were expressed as mean values±S.E. and analysed by Student’s t-test or ANOVA followed by the Tukey’s multiple comparison test. The level of significance was P<0.05.
Results
EZH2 overexpression and modulation by DZNeP
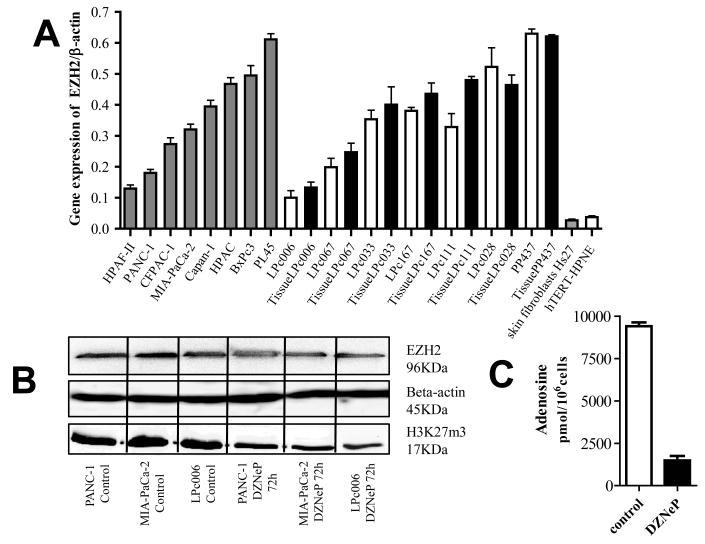
The mRNA expression of EZH2 was detectable in all PDAC cells, as well as in the originator tissues of the primary tumor cell cultures. This expression differed among cells, ranging from 0.100 a.u. in LPc006 cells to 0.644 a.u. in PL45 cells (Fig. 1A). The mean expression in the tumor cells (0.372±0.174 a.u.) was similar to the median (0.360 a.u.), and significantly higher (P<0.01) than the expression detected in hTERT-HPNE cells (0.037 a.u.), and in fibroblasts (0.027 a.u.).
Figure 1. EZH2 expression and DZNeP activity in PDAC cells.
(A) EZH2 mRNA expression in cell lines (grey-bars), primary tumor cultures (white-bars), and their originator tissues (black-bars); (B) Effect of 5 μM DZNeP on the expression of EZH2 and H3K27me3 after 72 hours; (C) Modulation of adenosine by 5 μM DZNeP after 72 hours, as detected by LC-MS/MS. Columns, mean values obtained from three independent experiments.
EZH2 gene expression in the 7 primary tumor cells and their originator tumors showed a similar pattern and were highly correlated with Spearman analysis (R2=0.89, P=0.01).
PANC-1, MIA-PaCa-2 and LPc006 cells were selected for further studies, because of previous studies on expression of the CD133 CSC marker (24, 26), and their differential levels of EZH2. The expression of EZH2 was also studied at protein level, both in untreated cells and in cells treated with 5 μM DZNeP for 24 or 72 hours. As shown in Fig. 1B, DZNeP reduced the expression of EZH2, especially after 72 hours (e.g. 48%, 32% and 36% reduction of EZH2 in PANC-1, MIA-PaCa-2 and LPc006 cells, respectively). In addition, we investigated the expression of the H3K27me3 protein, which was also reduced after 72 hours exposure.
Finally, since previous studies reported that DZNeP is a S-adenosylhomocysteine hydrolase inhibitor, we verified this inhibition and measured the intracellular concentration of its product adenosine in PANC-1 cells by a specific LC-MS/MS method (16). Adenosine was significantly reduced after 72-hour exposure to DZNeP (Fig. 1C), indicating that S-adenosylhomocysteine hydrolase was significantly inhibited.
Synergistic interaction of DZNeP with gemcitabine
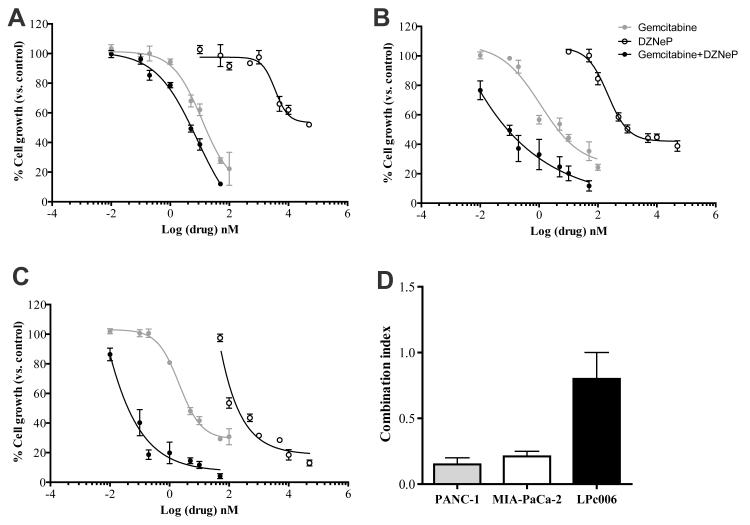
Treatment with DZNeP showed minimal growth inhibition in PANC-1 cells. More than 50% of these cells were still growing after exposure at the highest concentration (20 μM). MIA-PaCa-2 and LPc006 cells were much more sensitive, with IC50 values of 1.0±0.3 and 0.10±0.03 μM, respectively (Fig. 2A-C). To the contrary, gemcitabine was highly cytotoxic, with IC50s of 17.9±1.3 nM (PANC-1), 5.9±0.8 nM (MIA-PaCa-2), and 7.2±1.3 nM (LPc006).
Figure 2. Inhibition of cell proliferation and pharmacological interaction of DZNeP and gemcitabine.
Representative curves of growth inhibitory effects after 72 hours exposure to 5 μM DZNeP, gemcitabine at IC50 or their combination (drug concentrations on the X-axis are referred to gemcitabine) in PANC-1 (A), MIA-PaCa-2 (B) and LPc006 (C); (D) Mean CI of the DZNeP/gemcitabine combination. CI values at FA of 0.5, 0.75 and 0.9 were averaged for each experiment, and this value was used to calculate the mean between experiments, as described in the Materials and Methods section. Points and columns, mean values obtained from three independent experiments; bars, SEM.
Based on these results, as well as the modulation of EZH2 protein by 5 μM DZNeP, combination studies were performed using a fixed concentration of DZNeP at 5 μM. DZNeP enhanced the antiproliferative activity of gemcitabine, reducing the IC50s of gemcitabine to 5.02±1.31, 0.12±0.04 and 0.03±0.01 nM in PANC-1, MIA-PaCa-2, and LPc006. The mean CI showed slight-to-moderate synergism in LPc006 cells, and strong synergism in the PANC-1 and MIAPaCa-2 cells (Fig. 2D).
In order to evaluate the mechanisms underlying this synergistic interaction, several biochemical analyses were performed with the simultaneous combination, as detailed below.
DZNeP/gemcitabine combination enhanced apoptosis
DZNeP, gemcitabine and their combination affected the cell cycle of PDAC cells (Table 1). In particular, DZNeP significantly reduced the percentage of PANC-1 cells in the G2/M phase from 27 to 19%, after 72 hours, while gemcitabine increased cells in G2/M phase to 36% (P<0.05). The drug combination significantly reduced the percentage of cells in the G2/M phase. Hence, DZNeP blocked PANC-1 cells in the G1-S boundary. Conversely gemcitabine reduced the cells in this phase and no modulation was detected after drug combination in the PANC-1 cells. Similar perturbations of the cell cycle were observed in MIA-PaCa-2 cells. However, in LPc006 the percentage of cells in the G2/M phase was significantly reduced both after DZNeP-alone and DZNeP/gemcitabine combination. Moreover, the DZNeP/gemcitabine combination significantly increased cells in the S-phase, while reducing cells in the G0/G1 phase.
Table 1.
Effects of gemcitabine and DZNeP and their combination on cell cycle distribution and apoptotic index
| Cells | Treatment | G0/G1 Phase (%) |
S Phase (%) |
G2/M Phase (%) |
Apoptotic Index |
|---|---|---|---|---|---|
| PANC-1 | Control Gemcitabine DZNeP DZNeP+Gemcitabine |
55.2±1 46.4±2.8 60.4±0.2 54.4±1.4 |
17.7±1.3 17.8±2.8 20.8±0.2 24.5±2.7 |
27.2±2.2 35.9±5.6 18.9±0.1 21.2±1.3 |
3.2±0.4 8.7±1.2* 13.0±0.3* 29.6±2.8** |
| MIA-PaCa-2 | Control Gemcitabine DZNeP DZNeP+Gemcitabine |
60.5±2.6 44.9±0.9 73.2±2 62.8±0.1 |
15.9±0.9 19.7±2.1 14.6±1.4 25.3±1.0 |
23.5±1.8 35.3±1.2 12.2±0.6 11.9±0.9 |
3.9±0.9 9.4±1.3* 16.5±0.9* 34.1±0.8** |
| LPc006 | Control Gemcitabine DZNeP DZNeP+Gemcitabine |
65.5±1.3 60.9±7.5 72.2±1 52.5±1.7 |
12.5±1.8 11.8±1.1 12.4±0.9 30.8±1.1 |
22.1±0.4 27.2±8.5 15.4±0.1 16.8±0.7 |
2.1±0.6 11.8±0.8* 17.5±1.2* 29.2±0.4** |
P<0.05 with respect to control cells
P<0.05 with respect to gemcitabine Cells were exposured to IC50s values of gemcitabine, 5μM DZNeP and their combination The apoptotic index was calculated as the percentage from the ratio between the number of cells displaying apoptotic features and the number of counted cells.
Analysis of the sub-G1 region demonstrated that drug treatments significantly enhanced cell death compared to control (Table 1). In particular, MIA-PaCa-2 cells treated with the combination exhibited the largest sub-G1 signal (e.g., 34%).
Further analysis with fluorescence microscopy showed that cells exposed to DZNeP, gemcitabine and their combination presented a typical apoptotic morphology with cell shrinkage, nuclear condensation and fragmentation, and rupture of cells into debris, after 72-hour exposure. In all cell lines, 5-9% of apoptotic cells were observed after gemcitabine treatment, whereas DZNeP exposure was associated with a higher percentage (6-15%) of apoptotic cells; drug combination significantly increased the apoptotic index with respect to both control cells and gemcitabine-treated cells.
DZNeP/gemcitabine combination inhibited cell migration and up-regulated E-cadherin
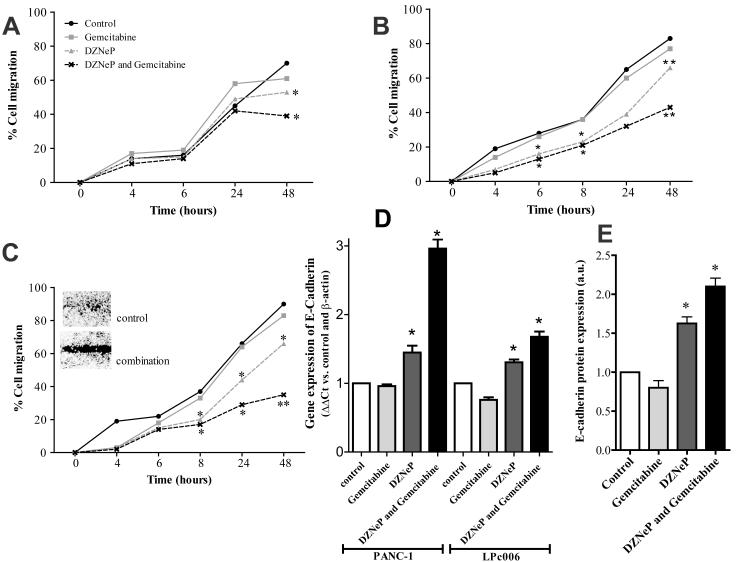
To investigate the effects of DZNeP, gemcitabine and their combination on migratory behavior, a scratch assay was performed in PANC-1, MIA-PaCa-2 and LPc006 cells. After exposure of PANC-1 cells to gemcitabine at IC50, 5 μM of DZNeP and their combination, a significant reduction of migration was observed after 48 hours (Fig. 3A and Supplementary Fig. S1). In particular, the percentages of cellular migration in PANC-1 were approximately 70%, 60%, 53% and 38%, in untreated, gemcitabine, DZNeP and their combination treated cells, respectively. Inhibition of migration of MIA-PaCa-2 and LPc006 with DZNeP or DZNeP/gemcitabine combination was much more effective than in PANC-1 cells (Fig. 3B-C and Supplementary Fig. S1). DZNeP significantly reduced cells migration with respect to controls, after 8 hours, with inhibition of about 20% in the reduction of scratch-area in LPc006 cells. In addition, the combination was also significantly more effective than DZNeP-alone after 48 hours in both MIA-PaCa-2 and LPc006 cells.
Figure 3. Effects of DZNeP, gemcitabine and their combination on PDAC cells migration.
Results of wound-healing assay in PANC-1 (A), MIA-PaCa-2 (B) and LPc006 (representative picture at 48 hours) (C) cells. Cells were exposed to 5 μM DZNeP, gemcitabine at IC50, and to their combination. Modulation of E-cadherin after 24 hours as determined by real-time RT-PCR (D) and immunocytochemistry (E) Columns, mean values obtained from three independent experiments; bars, SEM. *Significantly different from controls.
Since previous studies suggested that EZH2 repressed E-cadherin expression (27), we investigated whether DZNeP could affect the levels of this target at both mRNA and protein level. DZNeP and its combination with gemcitabine significantly enhanced E-cadherin mRNA expression (Fig. 3D). Similarly, immunocytochemistry analysis in LPc006 cells revealed a significant increase of E-cadherin protein staining after exposure to both DZNeP and DZNeP/gemcitabine combination (Fig. 3E and Supplementary Fig. S2).
DZNeP/gemcitabine combination reduced PDAC spheroids and CD133+ cells
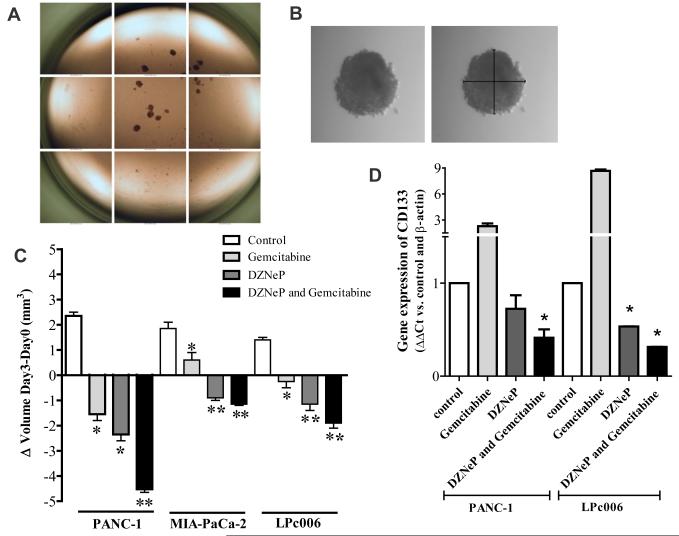
Earlier studies illustrated that results of sensitivity to anticancer drugs, including gemcitabine, in two-dimensional monolayer cell culture models were different from three-dimensional (3-D) culture models (28). Moreover, the use of serum-free-cancer-stem cell medium should select a population harboring CSCs characteristics, which should be selectively targeted by inhibitors of DZNeP. Thus, in order to determine whether DZNeP would enhance the efficacy of gemcitabine in 3-D systems, we tested these drugs in spheroids of PANC-1, MIA-PaCa-2 and LPc006 cells. After 10 days of culture, we transferred in each well of 96-well plates about 10 spheroids that were approximately 500 μm in diameter (Fig. 4A-B). These growing spheroids were exposed to DZNeP, gemcitabine and their combination for 72 hours. The growth of these spheroids was only slightly inhibited by gemcitabine (Fig. 4C), while DZNeP significantly reduced their volume. However, the DZNeP/gemcitabine combination remarkably increased the disintegration of these spheroids, which were significantly reduced in size compared to the spheroids exposed to gemcitabine-alone in all our three PDAC models. Spheroids from each treatment group were collected and used for PCR evaluation of CD133, which was significantly higher than in adherent cells (data not shown). This CSC-marker was significantly increased after gemcitabine exposure, whereas DZNeP reduced its mRNA levels. Moreover the DZNeP/gemcitabine combination significantly reduced CD133 expression in both PANC-1 and LPc006 cells, as shown in Fig. 4D.
Figure 4. Effects of DZNeP, gemcitabine and their combinations on PDAC spheroids.
Representative pictures of PANC-1 spheroids in a 96-well plate (A), and example (original magnification 40X) of the measurement of the diameters of one spheroid (B); Effect of 5 μM DZNeP, gemcitabine at IC50 values, and their combination on the volumes of PDAC spheroids (C) and CD133 mRNA expression (D), after 72 hours exposure. Columns, mean values obtained from three independent experiments; bars, SEM. *Significantly different from controls.
DZNeP and DZNeP/gemcitabine combination reduced deoxynucleotides and increased hENT1 and hCNT1 expression
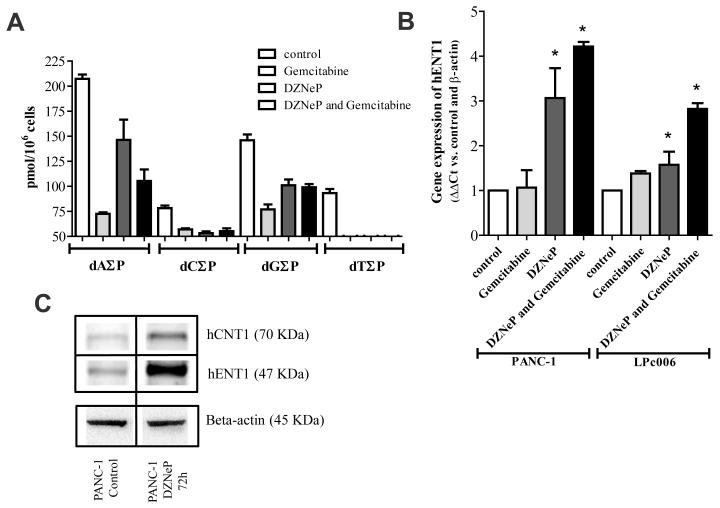
Treatment of PANC-1 cells with gemcitabine, DZNeP and their combination decreased the cellular concentrations of all deoxynucleotides from -25% (dA∑P after DZNeP) to -65% (dA∑P after gemcitabine). Of note, the levels of dT∑P were depleted by all treatments to not detectable levels (Fig. 5A). These changes in the pools of deoxynucleotides were associated with a significant increase in the mRNA expression of hENT1, after exposure to DZNeP and its combination with gemcitabine (Fig. 5B). An increased expression of both hENT1 and hCNT1 transporters was also observed at the protein level (Fig. 5C).
Figure 5. Effects of DZNeP, gemcitabine and their combination on phosphorylated deoxynucleosides and nucleoside transporters.
(A) Modulation of the intracellular deoxynucleotides, dA∑P (dAMP, dADP and dATP), dC∑P (dCMP, dCDP and dCTP), dG∑P (dGMP, dGDP and dGTP) and dT∑P (dTMP, dTDP and dTTP), as detected by the LC-MS/MS. Modulation of hENT1 mRNA expression (B) and representative blot of the modulation of hENT1 and hCNT1 protein levels after 72 hours exposure to 5 μM DZNeP (C). Columns, mean values obtained from three independent experiments; bars, SEM. *Significantly different from controls.
Discussion
The present study demonstrates that the combination of the EZH2 inhibitor DZNeP, and the cytotoxic compound gemcitabine, was strongly synergistic in a panel of PDAC cells characterized by different molecular properties.
The highly lethal nature of PDAC makes multiple areas of research a priority, including assessment of novel targets that might prevent or suppress the proliferative, invasive and chemoresistant behavior of PDAC cells.
EZH2 has a master regulatory role in the fate of native embriogenic cells (29), as well as in cancer development via methylation-mediated repression of transcription of several genes (9, 30). Overexpression of EZH2 is a marker of advanced and metastatic disease in many solid tumors, including PDAC (13, 31-32), and EZH2 nuclear accumulation is strongly associated with poor differentiation and prognosis of PDAC (13-14).
Previous studies on PANC-1 and SW1990 showed that suppression of EZH2 expression by RNA interference with Lentiviral-shEZH2 markedly inhibited cellular proliferation in vitro, and drastically diminished both tumorigenecity and liver metastasis in vivo (33). Furthermore, the transfection of shEZH2 construct cells sensitized MIA-PaCa-2 and Pac04.02 to doxorubicin and gemcitabine (14), suggesting that combination of EZH2 inhibitors with gemcitabine might overcome the intrinsic chemoresistance of PDAC.
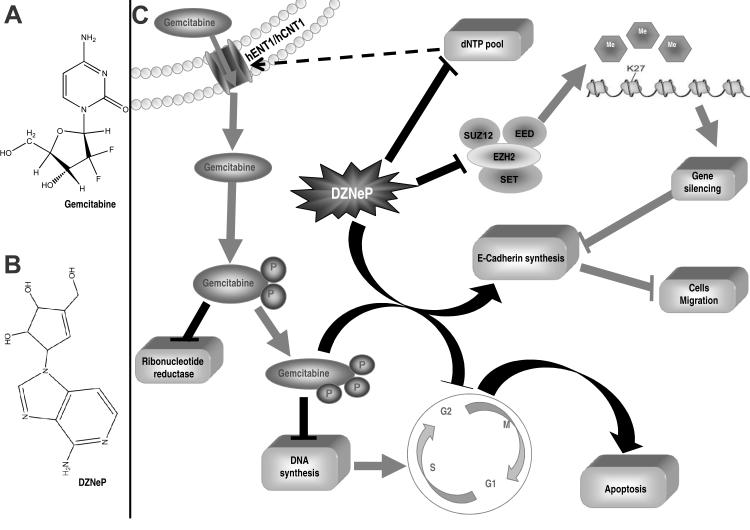
To our knowledge, this is the first study evaluating the pharmacological interaction of the small molecule EZH2 inhibitor DZNeP with gemcitabine in PDAC cells (Fig. 6).
Figure 6. Molecular mechanisms involved in the synergistic interaction of DZNeP with gemcitabine.
(A) Structures of gemcitabine and (B) DZNeP (C); DZNeP enhanced the growth inhibitory effects of gemcitabine through its pronounced pro-apoptotic, anti-invasive effects, as well as by inhibiting spheroids growth. Furthermore, modulation of phosphorylated deoxynuclosides and nucleoside transporters promotes gemcitabine uptake.
The expression of EZH2 was detectable in all our PDAC cells, including 7 primary tumor cell cultures, in their first passages, where the levels of EZH2 mRNA were comparable to their originator tumors, suggesting that these cells represent optimal preclinical models for our pharmacological studies. Conversely, EZH2 levels were significantly lower in both fibroblasts and in the normal pancreatic ductal cells HPNE, in agreement with earlier data on normal pancreatic tissue and specimens from patients affected by pancreatitis (13).
Since DZNeP inhibits S-adenosylhomocysteine hydrolase, a component of the methionine cycle, resulting in accumulation of the inhibitory S-adenosylhomocysteine, its effects on histone methylation is global rather than EZH2 specific (31, 34), and we evaluated both the modulation of H3K27me3 expression and the perturbation of intracellular adenosine.
In our PDAC cells, using concentrations and exposure time (5 μM, 72 hours) similar to those used in other tumor cells (35), we observed a significant reduction of both EZH2 and H3-K27 expression, as well as a dramatic decrease of intracellular adenosine content. Although DZNeP alone did not significantly affect proliferation of PANC-1 and MIA-PaCa-2 cells, these data suggested that DZNeP effectively reached its targets.
A recent phase-III trial showed that the oxaliplatin/irinotecan/fluorouracil/leucovorin (FOLFIRINOX) regimen is an option for the treatment of metastatic patients with good performance status, but was associated with increased toxicity (36). Thus, gemcitabine is still the standard first-line agent (37), and several studies are evaluating novel strategies to improve its activity against PDAC.
In the present study we demonstrated that DZNeP/gemcitabine combination was synergistic in two representative PDAC cell lines, PANC-1 and MIA-PaCa-2, and in the primary cell culture LPc006.
This synergistic interaction against cell proliferation was associated with a significant increase in apoptosis induction. This effect may be related to cell cycle modulation, which was also important for the efficacy of the combination of the histone-deacetylase inhibitor trichostatin-A with gemcitabine (38). Cellular damage induced by chemotherapeutic drugs such as gemcitabine can convert some targets of EZH2 into critical survival factors. In this context, the blockade of EZH2 after the exposure to cytotoxic drugs could prevent cell-damage repair, leading to apoptosis. In particular, previous studies in breast cancer cells resistant or sensitive to DZNeP led to the identification of a set of PRC2 target genes including TGFBI, IGFBP3, and PPPIR15A, which are involved in apoptosis (16), while TGFB signaling pathway is frequently deregulated in PDAC (39).
However, our findings show that the synergistic interaction of DZNeP with gemcitabine is also mediated by other mechanisms, which reduced PDAC aggressiveness and enhanced sensitivity to gemcitabine.
Since one of the major hallmarks and problems in the therapy of PDAC is its early local and systemic dissemination, we evaluated whether DZNeP might affect cell migration. In agreement with previous studies, showing that inhibition of EZH2 by DZNeP, attenuated glioblastoma and mesothelioma cell migration/invasion (36, 40), we observed that inhibition of EZH2 by DZNeP and its combination with gemcitabine significantly reduced cell migration, as detected with wound-healing assay.
Several classes of proteins are participating to invasive PDAC phenotype, including cell-cell adhesion molecules like members of immunoglobulin and calcium-dependent cadherin families and integrins. One widely observed alteration in cell-to-environment interaction in PDAC involves E-cadherin, which couples adjacent cells by E-cadherin bridges, and a recent study showed that recruitment of histone deacetylases HDAC1/HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in PDAC (41).
Keeping with previous evidence on inverse relationship between EZH2 and E-cadherin expression (13), also our data show that DZNeP-induced EZH2 inhibition resulted in an increase in both mRNA and protein expression of E-cadherin.
Recently PDAC also emerged as a CSC-driven disease (42). This might at least partially explain its chemoresistant nature (8), and compounds targeting critical developmental genes keeping self-renewal in CSCs, including Sonic hedgehog, BMI-1 and EZH2, seem promising anticancer agents. For example the Curcumin-analog CDF inhibited formation of pancreatospheres as well as PDAC growth by switching on several suppressor microRNAs and attenuating EZH2 expression (43).
DZNeP significantly reduced the volume of PDAC spheroids growing in serum-free-stem-cell medium. Gemcitabine only slightly reduced the volume of these spheroids, possibly by affecting some remaining bulk tumor cells, but it increased the expression of the CSC-marker CD133, as observed previously (44), suggesting that exposure to gemcitabine might select a population of more aggressive cells. Conversely, DZNeP was able to effectively deplete the most aggressive subpopulation of PDAC cells, as suggested by the significant reduction of both spheroids and CD133 expression.
In addition to the effects of DZNeP on migration and spheroids, the present study also showed that it interfered with pivotal determinants for the activity of gemcitabine. In particular, different thymidylate synthase inhibitors upregulated hENT1 and increased gemcitabine sensitivity by depleting intracellular nucleotide pools (45-46). Therefore, we analyzed the cellular deoxynucleotides pools and modulation of the expression of key nucleoside transporters (47). Gemcitabine, DZNeP and their combination significantly depleted all the endogenous deoxynucleotides. The results achieved after exposure to gemcitabine might be explained by gemcitabine-induced inhibition of ribonucleotide reductase, as reported previously (48). DZNeP is not phosphorylated and does not get incorporated into DNA (49), but it markedly reduced endogenous deoxynucleotides. This might at least in part explain the significant up-regulation of both hENT1 and hCNT1, potentially facilitating gemcitabine cytotoxicity.
In conclusion, inhibitors of EZH2, such as DZNeP, seem very promising anticancer agents, by attacking key mechanisms involved in the proliferation, cell cycle control, apoptosis and of migration properties of PDAC cells. Moreover, the favorable modulation of hENT1/hCNT1 transporter makes DZNeP an optimal candidate for combination with gemcitabine. The synergistic results observed in the present study may have critical implications for the rational development of innovative regimes including DZNeP and gemcitabine to improve the efficiency of the actual treatment of PDAC.
Supplementary Material
Acknowledgements
Authors would like to thank M. Smits and Dr. T. Wurdinger (Dept. Neuro-Oncology, VUmc, Amsterdam) for their advice on EZH2, Dr. C. Fedrigo (Dept. Radiobiology, VUmc, Amsterdam) for his help on experiments with spheroids, and Prof. A. Griffioen (Angiogenesis-group, VUmc, Amsterdam) for the migration-station to perform wound-healing assays.
Grant Support: This work was supported by grants from the Netherlands-Organization for Scientific Research (Veni grant#91611046, E. Giovannetti), AIRC-Marie Curie (International Fellowship, E. Giovannetti), Iran’s National-Elites-Foundation (Award, A. Avan), and CCA-VICI foundation (grant#2012-5-07, A. Avan, G.J. Peters, E. Giovannetti); and by the Intramural Research Program of the NIH, NCI, Center for Cancer Research (V.E. Marquez).
Footnotes
Conflicts of interest. The authors have no conflict of interest to disclose.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ottenhof NA, de Wilde RF, Maitra A, Hruban RH, Offerhaus GJ. Molecular Characteristics of Pancreatic Ductal Adenocarcinoma. Patholog Res Int. 2011;2011:620601. doi: 10.4061/2011/620601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamacher R, Schmid RM, Saur D, Schneider G. Apoptotic pathways in pancreatic ductal adenocarcinoma. Mol Cancer. 2008;7:64. doi: 10.1186/1476-4598-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duhagon MA, Hurt EM, Sotelo-Silveira JR, Zhang X, Farrar WL. Genomic profiling of tumor initiating prostatospheres. BMC Genomics. 2010;11:324. doi: 10.1186/1471-2164-11-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaviraghi M, Tunici P, Valensin S, Rossi M, Giordano C, Magnoni L, et al. Pancreatic cancer spheres are more than just aggregates of stem marker-positive cells. Biosci Rep. 2011;31:45–55. doi: 10.1042/BSR20100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong SP, Wen J, Bang S, Park S, Song SY. CD44-positive cells are responsible for gemcitabine resistance in pancreatic cancer cells. Int J Cancer. 2009;125:2323–31. doi: 10.1002/ijc.24573. [DOI] [PubMed] [Google Scholar]
- 7.Ottinger S, Klöppel A, Rausch V, Liu L, Kallifatidis G, Gross W, et al. Targeting of pancreatic and prostate cancer stem cell characteristics by Crambe crambe marine sponge extract. Int J Cancer. 2012;130:1671–81. doi: 10.1002/ijc.26168. [DOI] [PubMed] [Google Scholar]
- 8.Rajeshkumar NV, Rasheed ZA, García-García E, López-Ríos F, Fujiwara K, Matsui WH, et al. A combination of DR5 agonistic monoclonal antibody with gemcitabine targets pancreatic cancer stem cells and results in long-term disease control in human pancreatic cancer model. Mol Cancer Ther. 2010;9:2582–92. doi: 10.1158/1535-7163.MCT-10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 11.Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;16:871–74. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 12.Santos-Rosa H, Caldas C. Chromatin modifier enzymes, the histone code and cancer. Eur J Cancer. 2005;41:2381–402. doi: 10.1016/j.ejca.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Toll AD, Dasgupta A, Potoczek M, Yeo CJ, Kleer CG, Brody JR, et al. Implications of enhancer of zeste homologue 2 expression in pancreatic ductal adenocarcinoma. Hum Pathol. 2010;41:1205–9. doi: 10.1016/j.humpath.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Ougolkov AV, Bilim VN, Billadeau DD. Regulation of Pancreatic Tumor Cell Proliferation and Chemoresistance by the Histone Methyltransferase EZH2. Clin Cancer Res. 2008;14:6790–6796. doi: 10.1158/1078-0432.CCR-08-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong HQ, Carr K, Abbruzzese JL. Cytotoxic chemotherapy for pancreatic cancer: advances to date and future directions. Drugs. 2006;66:1059–1072. doi: 10.2165/00003495-200666080-00003. [DOI] [PubMed] [Google Scholar]
- 16.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi, et al. Pharmacologic disruption of polycomb-repressive complex 2- mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayden A, Johnson P, Packham G, Crabb SJ. S-adenosylhomocysteine hydrolase inhibition by 3-deazaneplanocin A analogues induces anti-cancer effects in breast cancer cell lines and synergy with both histone deacetylase and HER2 inhibition. Breast Cancer Res Treat. 2011;127:109–119. doi: 10.1007/s10549-010-0982-0. [DOI] [PubMed] [Google Scholar]
- 18.Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–38. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 19.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–88. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giovannetti E, Del Tacca M, Mey V, Funel N, Nannizzi S, Ricci S, et al. Transcription analysis of human equilibrative nucleotide transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res. 2006;66:3928–35. doi: 10.1158/0008-5472.CAN-05-4203. [DOI] [PubMed] [Google Scholar]
- 21.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 22.Giovannetti E, Mey V, Danesi R, Basolo F, Barachini S, Deri M, et al. Interaction between gemcitabine and topotecan in human non-small-cell lung cancer cells: effects on cell survival, cell cycle and pharmacogenetic profile. Br J Cancer. 2005;92:681–9. doi: 10.1038/sj.bjc.6602382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mousavi SH, Moallem SA, Mehri S, Shahsavand S, Nassirli H, Malaekeh-Nikouei B. Improvement of cytotoxic and apoptogenic properties of crocin in cancer cell lines by its nanoliposomal form. Pharm Biol. 2011;49:1039–45. doi: 10.3109/13880209.2011.563315. [DOI] [PubMed] [Google Scholar]
- 24.Mathews LA, Cabarcas SM, Hurt EM, Zhang X, Jaffee EM, Farrar WL. Increased expression of DNA repair genes in invasive human pancreatic cancer cells. Pancreas. 2011;40:730–9. doi: 10.1097/MPA.0b013e31821ae25b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honeywell RJ, Giovannetti E, Peters GJ. Determination of the Phosphorylated Metabolites of Gemcitabine and of Difluorodeoxyuridine by LCMSMS. Nucleosides Nucleotides Nucleic Acids. 2011;30:1203–13. doi: 10.1080/15257770.2011.632389. [DOI] [PubMed] [Google Scholar]
- 26.Yao J, Cai HH, Wei JS, An Y, Ji ZL, Lu ZP, et al. Side population in the pancreatic cancer cell lines SW1990 and CFPAC-1 is enriched with cancer stem-like cells. Oncol Rep. 2010;23:1375–82. doi: 10.3892/or_00000774. [DOI] [PubMed] [Google Scholar]
- 27.Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–84. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padrón JM, van der Wilt CL, Smid K, Smitskamp-Wilms E, Backus HH, Pizao PE, et al. The multilayered postconfluent cell culture as a model for drug screening. Crit Rev Oncol Hematol. 2000;36:141–57. doi: 10.1016/s1040-8428(00)00083-4. [DOI] [PubMed] [Google Scholar]
- 29.Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332:963–6. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–8. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 31.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 32.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Xie D, Yin Li W, Man Cheung C, Yao H, Chan CY, et al. RNAi targeting EZH2 inhibits tumor growth and liver metastasis of pancreatic cancer in vivo. Cancer Lett. 2010;297:109–16. doi: 10.1016/j.canlet.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Zoabi M, Sadeh R, de Bie P, Marquez VE, Ciechanover A. PRAJA1 is a ubiquitin ligase for the polycomb repressive complex 2 proteins. Biochem Biophys Res Commun. 2011;408:393–8. doi: 10.1016/j.bbrc.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 35.Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, Niers JM, et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget. 2010;1:710–20. doi: 10.18632/oncotarget.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–25. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 37.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–57. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 38.Gahr S, Ocker M, Ganslmayer M, Zopf S, Okamoto K, Hartl A, et al. The combination of the histone-deacetylase inhibitor trichostatin A and gemcitabine induces inhibition of proliferation and increased apoptosis in pancreatic carcinoma cells. Int J Oncol. 2007;31:567–76. [PubMed] [Google Scholar]
- 39.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemp CD, Rao M, Xi S, Inchauste S, Mani H, Fetsch P, et al. Polycomb Repressor Complex-2 is a Novel Target for Mesothelioma Therapy. Clin Cancer Res. 2012;18:77–90. doi: 10.1158/1078-0432.CCR-11-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aghdassi A, Sendler M, Guenther A, Mayerle J, Behn CO, Heidecke CD, et al. Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut. 2012;61:439–48. doi: 10.1136/gutjnl-2011-300060. [DOI] [PubMed] [Google Scholar]
- 42.Simeone DM. Pancreatic cancer stem cells: implications for the treatment of pancreatic cancer. Clin Cancer Res. 2008;14:5646–8. doi: 10.1158/1078-0432.CCR-08-0584. [DOI] [PubMed] [Google Scholar]
- 43.Bao B, Ali S, Banerjee S, Wang Z, Logna F, Azmi AS, et al. Curcumin analog CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 2012;72:335–45. doi: 10.1158/0008-5472.CAN-11-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crea F, Hurt EM, Mathews LA, Cabarcas SM, Sun L, Marquez VE, et al. Pharmacologic disruption of Polycomb Repressive Complex 2 inhibits tumorigenicity and tumor progression in prostate cancer. Mol Cancer. 2011;10:40. doi: 10.1186/1476-4598-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rauchwerger DR, Firby PS, Hedley DW, Moore MJ. Equilibrative-sensitive nucleoside transporter and its role in gemcitabine sensitivity. Cancer Res. 2000;60:6075–9. [PubMed] [Google Scholar]
- 46.Giovannetti E, Mey V, Nannizzi S, Pasqualetti G, Marini L, Del Tacca M, et al. Cellular and pharmacogenetics foundation of synergistic interaction of pemetrexed and gemcitabine in human non-small-cell lung cancer cells. Mol Pharmacol. 2005;68:110–8. doi: 10.1124/mol.104.009373. [DOI] [PubMed] [Google Scholar]
- 47.Danesi R, Altavilla G, Giovannetti E, Rosell R. Pharmacogenomics of gemcitabine in non-small-cell lung cancer and other solid tumors. Pharmacogenomics. 2009;10:69–80. doi: 10.2217/14622416.10.1.69. [DOI] [PubMed] [Google Scholar]
- 48.Van Moorsel CJ, Smid K, Voorn DA, Bergman AM, Pinedo HM, Peters GJ. Effect of gemcitabine and cis-platinum combinations on ribonucleotide and deoxyribonucleotide pools in ovarian cancer cell lines. Int J Oncol. 2003;22:201–7. doi: 10.3892/ijo.22.1.201. [DOI] [PubMed] [Google Scholar]
- 49.Glazer RI, Knode MC, Tseng CK, Haines DR, Marquez VE. 3-Deazaneplanocin A: a new inhibitor of S-adenosylhomocysteine synthesis and its effects in human colon carcinoma cells. Biochem Pharmacol. 1986;35:4523–7. doi: 10.1016/0006-2952(86)90774-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.