Abstract
Mosquito saliva carries a large number of factors with anti-hemostatic, anti-inflammatory and immuno-modulatory activities. The cE5 protein was initially identified during an Anopheles gambiae salivary gland transcriptome study and later shown to share sequence similarity with anophelin, a thrombin inhibitor from the saliva of the New World mosquito Anopheles albimanus. The cE5 gene was found to encode different mRNA isoforms coexisting in several tissues of both male and female mosquitoes, a highly unusual profile for a gene potentially encoding an anti-thrombin and involved in blood feeding. Expression of the cE5 protein and assessment of its activity and inhibitory properties showed that it is a highly specific and tight-binding thrombin inhibitor, which differs from the An. albimanus orthologue for the fast-binding kinetics. Despite the widespread occurrence of cE5 transcripts in different mosquito tissues the corresponding protein was only found in female salivary glands, where it undergoes post-translational modification. Therefore, tissue-specific restriction of the An. gambiae cE5 is not achieved by transcriptional control, as common for mosquito salivary genes involved in hematophagy, but by post-trascriptional gene regulatory mechanisms. Our observations provide a paradigm of post-transcriptional regulation as key determinant of tissue specificity for a protein from an important disease vector and point out that transcriptomic data should be interpreted with caution in the absence of concomitant proteomic support.
Keywords: Anopheles, salivary protein, anti-thrombin, anophelin, hematophagy, post-transcriptional regulation
1. INTRODUCTION
The ability to exploit blood as a food source provides hematophagous insects with the advantage of using a highly nutritious resource for egg development, which translates into a high reproductive capacity and evolutionary success. However, the blood feeding style of life requires a complex armamentarium of behavioral, structural and physiological adaptations which enable these insects to locate suitable hosts, overcome the epidermal barrier, reach blood vessels and efficiently suck/digest the blood meal (Ribeiro, 1996). In this context hematophagous insect’s saliva, discharged into the host skin both in the exploratory phase (probing) and during blood ingestion, plays a crucial role by counterbalancing the hemostatic, inflammatory and immune responses of vertebrate hosts to tissue injury. Salivary transcriptome analyses performed in the last ten years disclosed the complexity of the salivary repertoires (sialomes) of blood feeding insects, allowing for the identification and classification of over 150 salivary protein families (Ribeiro et al., 2010; Ribeiro and Arcà, 2009). It is noteworthy that for a large number of these proteins a possible function cannot even be hypothesized to date.
We have previously explored the salivary repertoire of the African malaria vector Anopheles gambiae and, among other components, identified a cDNA encoding the putative salivary protein cE5 (Arcà et al., 1999). Its possible function stayed unknown until a thrombin inhibitor, named anophelin, was identified from the salivary extracts of the New World mosquito Anopheles albimanus and shown to possess significant sequence similarity to the An. gambiae cE5 (Valenzuela et al., 1999). It is known that anopheline mosquitoes use as a salivary anti-clotting strategy a thrombin inhibitor (Stark and James, 1996), and it is likely that the An. gambiae cE5 may represent the orthologue of the An. albimanus anophelin. However, the following observations encouraged further investigation of expression profile and biological properties of the cE5 protein. First, putative anophelin homologs from members of the subgenus Cellia (An. gambiae, Anopheles funestus and Anopheles stephensi) are 18–22 amino acids longer in their C-terminal region in comparison to the An. albimanus protein (Calvo et al., 2007); this C-terminal tail is enriched in polar aminoacids, especially Ser (~30–33%) and Asn/Gln (~17–35%). Second, although expression of mosquito salivary genes involved in hematophagy is mostly restricted to female salivary glands, cE5 transcripts were also detected by Reverse Transcription-Polymerase Chain Reaction (RT-PCR) in adult males and other female tissues (Arcà et al., 1999; Arcà et al., 2005). Third, database searches retrieved multiple cDNAs, all potentially encoding the An. gambiae cE5 protein but carrying 3′-untranslated regions (UTRs) of different length, most likely as a result of a different polyadenylation site usage.
The absence of selective expression in mosquito female salivary glands, the presence of an additional C-terminal region and the expression of alternative transcripts raises the possibility that the An. gambiae cE5 may act on a broader range of serine proteases as compared to anophelin and/or that some sort of transcriptional/post-transcriptional gene regulation may take place. In order to address some of the above questions we performed a detailed analysis of the cE5 tissue-specific expression profile, expressed the An. gambiae cE5 protein in recombinant form and assayed its potential serine protease inhibitory properties.
2. MATERIAL AND METHODS
2.1 Mosquito colonies and tissue dissection
Anopheles gambiae (M-form, population GACAM: Xag, 2R+, 2L+, 3R+, 3L+) and Ae. albopictus (colonized in Rome in 2000, provided by R. Romi, Istituto Superiore di Sanità) were reared under standard insectary conditions (25°–28°C, 60%–70% humidity). Adults were fed either on 5% glucose or on guinea pigs, and larvae on dry cat food. Tissues were dissected from 1–6 days-old mosquitoes in Phosphate Buffered Saline (PBS), frozen in liquid nitrogen and stored at −80°C until needed. Salivary glands were typically frozen in batches of twenty salivary gland pairs in 20 μl of Hepes Buffered Saline (HBS, Hepes 20 mM pH 7.5, NaCl 150 mM).
2.2 Protein extracts
Salivary gland extracts (SGE) for biochemical assays were prepared by freezing and thawing (3X) followed by addition of 80 μl of HBS, brief vortexing and centrifugation (15000g, 4°C, 10 min). Supernatants were transferred to clean tubes and kept in ice for max 4–5 hours before use. SGE for SDS-PAGE analysis were obtained by freezing and thawing as above, followed by addition of 1 volume (V) of 2x SDS sample buffer, boiling 5–10 min and centrifugation (15000g, 10 min). Protein extracts from the other tissues were obtained by homogenization in PBS containing protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany). After centrifugation (15000g, 10 min) supernatants were recovered and boiled 5–10 min after addition of 1V of 2x SDS sample buffer. Hemolymph and hemocytes were obtained from adult females by proboscis-clipping followed by thorax squeezing: hemolymph (containing circulating hemocytes) was collected in PBS and, after gentle centrifugation (5 min, 1000g), separated from hemocytes. Protease inhibitor and 2x SDS sample buffer were added to hemolymph and hemocytes as above and, after boiling 5–10 min, supernatants were used for SDS-PAGE.
2.3 RNA extraction and expression analysis
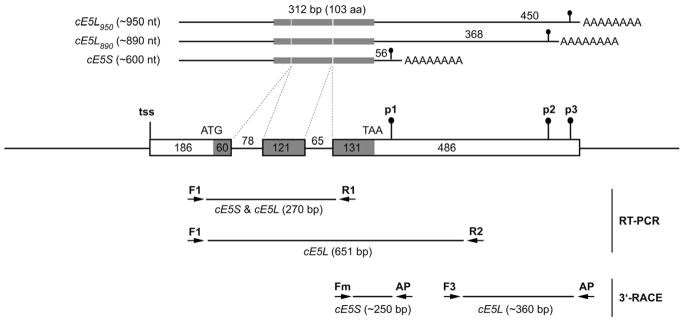
If not otherwise specified, experimental procedures were performed following standard protocols (Ausubel et al., 1991; Sambrook et al., 1989) and/or according to manufacturer’s instruction. Total RNAs were extracted using the Trizol reagent and treated with RNAse-free DNAse I. Typically 80 ng or 1 μg of DNAseI-treated total RNA were used as template for the One-step RT-PCR or the first strand cDNA synthesis, respectively. For the rapid amplification of cDNA 3′-end (3′-RACE), first strand cDNA synthesis was primed by an Adapter Primer (AP). After heat inactivation of the reverse transcriptase (94°C, 2 min) PCR amplifcations were performed by 25 (rpS7 mRNA) or 35 (cE5 mRNA) PCR cycles: 45 sec at 94°C, 45 sec at 55°C, 45 sec at 72°C. PCR amplifications without reverse transcription were included as control and all reactions were performed at least twice on different RNA batches. Representative PCR amplification products were gel purified (QIAGEN Gel Purification Kit), cloned into the pCR2.1 vector and sequenced. Trizol reagent, RNAse-free DNAse I, SuperScript One-step RT-PCR system, Superscript II Reverse Transcriptase and the pCR2.1 vector were purchased from Invitrogen (CA, USA). Oligonucleotide primers used for the expression analysis as indicated in Figure 1 are reported in the Table S1 (Online Resource 1).
Figure 1. Schematic representation of the genomic region encoding the An. gambiae cE5.
The putative transcription start site (tss), start (ATG) and stop (TAA) codons, alternative polyadenylation signals (p1–p3) are shown. Exons are boxed and coding region is shaded. Introns and genomic flanking regions are represented as lines. Numbers represents length in base pairs. On the top the three cE5 transcripts with different 3′UTRs are represented. The distance between stop codon and polyadenylation signals is indicated (56, 368, and 450 nucleotides, respectively). At the bottom the different primer pairs used for the RT-PCR and 3′-RACE are indicated along with the size of the expected PCR fragments (see also text for additional details).
2.4 Protein Expression and Purification
The region encoding the An. gambiae cE5 was amplified using Pfx DNA polymerase (Invitrogen, CA, USA) from an available cDNA clone (Arcà et al., 2005) with the oligonucleotides TEV_cE5-F (5′-GAGAATCTTTATTTTCAGGGCGCACCGCAGTATGCACGCGG -3′), which contains the recognition sequence for the TEV protease (underscored) fused in frame to the first 23 nt of the mature cE5 coding sequence (i.e. without signal peptide), and cE5_Hind-R (5′-GTCTAAGCTTTTATTCGTCCGACTCCGAAG-3′), carrying the 3′-end of the cE5 coding region followed by the HindIII restriction site (italic bold). The amplified fragment was directionally cloned into the XmnI/HindIII-digested pMAL-c4X vector (New England Biolabs, UK) yielding the pMALT-cE5 expression vector, which can drive the expression of a recombinant An. gambiae cE5 fused to the C-terminus of the E. coli Maltose Binding Protein. After sequence verification pMALT-cE5 was transformed into TB1 E. coli cells and a single colony was grown over night at 37°C in 20 ml of LB medium. Five ml of the saturated culture were transferred in 400 ml of LB medium supplemented with 0.2% glucose and grown at 37°C to 0.5 OD600 before induction with IPTG (0.3 mM). After 3 hours the cells were harvested by centrifugation, resuspended in 20 ml of buffer S (50 mM Tris-HCl pH 8.0, 1 mM PMSF, 50 μg/ml lysozyme) and sonicated. After centrifugation (25000 g, 30 min, 4°C) the soluble fraction was subjected to anion exchange chromatography onto an HiTrapQ HP column (5 ml, GE Healthcare) previously equilibrated in buffer L (20 mM Tris-HCl pH 8.0). Elution was carried out with a linear gradient 0–500 mM NaCl in Buffer L (15 column volumes). Fractions containing the fusion protein (MBP-FT-cE5) were identified by SDS-PAGE, pooled, dialyzed against MBP binding buffer (20 mM Tris-HCl pH 7.4, 200 mM NaCl, 1 mM EDTA) and further purified by affinity chromatography on a MBP-Trap column (5 ml, GE Healthcare). After SDS-PAGE analysis the fractions containing the recombinant protein, which was essentially homogeneous, were pooled and the protein concentration estimated by the Bio Rad Protein Assay (Bio Rad Laboratories, MI, Italy). The MBP was removed by cleavage for 16h at 25°C with the TEV protease in C buffer (50 mM Tris-HCl pH 8.0, 0.5 mM EDTA, 1 mM DTT; MBP-cE5 : TEV ratio 100: 1 w/w). Recombinant cE5 was purified from MBP and TEV protease by RP-HPLC on a Juppiter C5 column (Phenomenex Inc., USA) with a linear gradient 5%–95% acetonitrile, 0.1% TFA. The cE5 containing fractions were pooled, lyophilized and protein concentration was estimated by the extinction coefficient calculated at the ExPASy Proteomic Server (Gasteiger E., 2005). The recombinant purified cE5 carries, in comparison to the mature native protein, an additional N-terminal Glycin and its homogeneity was assessed by SDS-PAGE and further confirmed by mass spectrometry (CEINGE Biotecnologie Avanzate, Naples).
2.5 Antibody Production
Purified recombinant cE5 protein was used to immunize BALB/c mice. Briefly, 50 μg of protein were mixed to complete Freund’s adjuvant and injected intraperitoneally. Immunization was repeated after 28, 42, 56 days using 25 μg of antigen in incomplete Freund’s adjuvant. At day 70 the immunized mice were bled to obtain immune serum. Pre-immune serum was obtained before mice immunization by blood collection (~100 μl) from the submandibular vein. The immune serum was purified by the Melon Gel IgG Purification System (Thermo Scientific, Rockford, IL) to remove highly abundant non-relevant proteins.
2.6 Western blot
Protein extracts were fractionated by SDS-PAGE on 15% polyacrylamide gels and electro-blotted on to nitrocellulose membranes. Immuno-staining was performed according to standard procedures using the mouse anti-cE5 polyclonal serum (1:500), a rabbit anti-mouse horseradish peroxidase-conjugated secondary antibody (1:20.000, Sigma-Aldrich) and the chemiluminescent peroxidase substrate-1 (Sigma-Aldrich). For the western blot on tissue extracts (Figure 4B) the mouse anti-cE5 polyclonal serum was purified by the Melon Gel IgG Purification System (see above) and used at a 1:30 dilution. Typically SDS gels mirroring the ones used for western blots but containing approximately 1/5th to 1/10th the protein amounts were silver stained as sample loading and integrity control.
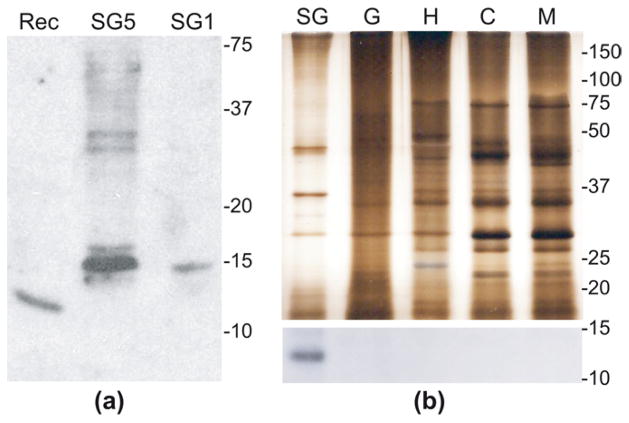
Figure 4. Tissue-specific expression of the cE5 protein in adult female salivary glands.
(a) Western blot analysis on female salivary gland extracts and recombinant cE5 protein. Rec, ~100 ng recombinant cE5; SG5, salivary extracts from 5 pairs of glands; SG1, salivary extracts from 1 pair of glands. (b) Silver staining (top) and western blot (bottom) on protein extracts from different tissues. The silver stained gel contains approximately 1/10th the protein amount as compared to western blot. The exact amount of protein extracts loaded on the gels for silver staining (s) and western analysis (w) were as follows. SG, female salivary glands: 8 pairs (w), 1 pair (s). G, midguts: 35 (w), 3.5 (s). H, heads: 35 (w), 3.5 (s). C, carcass (leftover after salivary glands, heads and midgut dissection): 11 (w), 1 (s). M, adult males: 11 (w), 1.5 (s). Numbers on the right of each panel refer to molecular weights in kDa.
2.7 Chromogenic assay for Anti-Thrombin activity
Thrombin inhibition by recombinant cE5 or by An. gambiae and Ae. albopictus SGE was evaluated following the hydrolysis of the chromogenic substrate Benzoyl FVR-pNA (Calbiochem, 02-32-0078). Reactions were carried out in Falcon 3912 96-well plates in 100 μl total volume in HBS. Briefly, 10 nM thrombin (Calbiochem, 605195) was pre-incubated for 3 min at 37°C with increasing amounts of recombinant cE5 (1 to 20 nM) or salivary gland extracts (0.1 to 2.0 salivary gland pairs). The reaction was initiated by addition of substrate (150 μM), whose hydrolysis was followed for 5 min (405 nm, 37°C) on a microplate reader (BioTek, Synergy HT). Data represent three independent experiments, each one in triplicate. Statistical analysis and graphics were performed using GraphPad Prism 5.0® (GraphPad Software, Inc. La Jolla, CA).
2.8 Serine protease inhibition assays and IC50 determination
All enzymes used were of human origin, purified or recombinant. Source and concentration of enzymes, composition of assay buffers and substrates used are given in detail as supplementary materials and methods (Online Resource 1). Unless otherwise stated, all reactions were performed at 37°C and the standard substrate for thrombin was the fluorogenic substrate Boc-Asp-Pro-Arg-AMC (Sigma-Aldrich, St. Louis, MO). The amount of enzyme used in each assay is shown in Table 1. All substrates were used at a final concentration of 250 μM. For the initial screen, the cE5 protein (600 nM) was pre-incubated 10 min at room temperature with each enzyme in Microfluor™ 96-Well Fluorescence Plates (Thermo Fisher Scientific, Pardubice, Czech republic) before addition of the corresponding substrate. The amount of enzyme used in the assays was the lower possible to obtain a linear substrate hydrolysis rate (r2 > 0.95). After addition of the substrate, the plate was incubated for 5 min at 30°C and substrate hydrolysis was followed for 20 min at 30°C in a plate reader (Tecan Infinite M200, Tecan group Ltd, Switzerland) using 365 nm excitation and 450 nm emission wavelengths, with a cutoff at 435 nm. Wells containing only inhibitor and substrate were used as negative control. Student’s t-test was used to evaluate statistical significance (p < 0.05) of the observed inhibition.
Table 1.
Specificity of cE5 to thrombin
| Enzyme (nM) | % activity |
|---|---|
| Thrombin (0,03) | 13.3 ± 0.6 |
| Factor Xa (0,1) | 103.8 ± 4.0 |
| Kallikrein (0,04) | 105.2 ± 2.7 |
| Chymase (0,9) | 102.7 ± 7.9 |
| Trypsin (0,1) | 101.7 ± 4.9 |
| a-Chymotrypsin (0,075) | 102.5 ± 1.4 |
| b-Tryptase (0,01) | 104.7 ± 4.3 |
| Elastase (0,18) | 90.5 ± 5.2 |
| Cathepsin G (6,7) | 95.6 ± 2.0 |
| u-PA (0,5) | 98.9 ± 5.1 |
| Plasmin (0,4) | 102.3 ± 2.7 |
| Matriptase (0,1) | 102.7 ± 8.3 |
| Factor XIa (0,06) | 100.6 ± 1.6 |
| Factor XIIa (0,1) | 106.3 ± 3.7 |
| t-PA (0,06) | 106.9 ± 2.1 |
cE5 (600 nM) was incubated with the amount of protease indicated in parentheses in the presence of the specific fluorogenic substrate as described in the supplementary materials and methods (Online Resource 1). Enzyme activities in the absence of inhibitor were set as 100%. Data represent the mean ± SEM (Standard Error of the Mean) of triplicate experiments. Bold denotes statistically significant difference (t-test, P<0.05).
The IC50 of cE5 and anophelin for thrombin was estimated as previously described (Chmelar et al., 2011). Briefly, thrombin and decreasing concentrations of inhibitor were pre-incubated at room temperature for 10 min and the reaction was started by adding the corresponding substrates. All experiments were performed in triplicate. The mean residual thrombin activity (assuming as 100% the activity in the absence of inhibitor) was plotted against the concentration of inhibitor (in logarithmic scale) and the sigmoidal fit of the data gave the estimate of the IC50 of the inhibitors for thrombin. To examine whether cE5 is a tight-binding inhibitor, thrombin was used at nanomolar concentration (2 nM, 5 nM and 15 nM) and the assay was performed using the fluorogenic substrate Boc-Ile-Glu-Gly-Arg-7-amido-4-methylcoumarin hydrochloride (Sigma, St. Louis, MO). To examine whether cE5 and anophelin bind thrombin with fast or slow kinetic, the inhibitors were pre-incubated 10 min at room temperature either with the standard fluorogenic substrate or with thrombin and the enzymatic reactions were started with the addition of thrombin or the fluorogenic substrate, respectively. Plate reading was performed as described above. All experiments comparing the activity of anophelin with cE5 were performed simultaneously on the same plate. The An. albimanus anophelin was synthesized as previously described (Valenzuela et al., 1999). Statistical analysis and graphics were performed using GraphPad Prism 5.0® (GraphPad Software, Inc. La Jolla, CA).
3. RESULTS
3.1 Transcriptional profile of the An. gambiae anophelin
Using the cE5 genomic region (VectorBase: AGAP008004) for BLASTN searches (Zhang et al., 2000) against the NCBI nucleotide collection and EST datasets, cDNAs of different length could be retrieved. These cDNAs, potentially encoding the same polypeptide, differed in their 3′-UTRs, presumably as a consequence of different polyadenylation site usage. The most represented transcript in the databases was a short form, originated by the usage of a polyadenylation signal located 56 nts downstream of the stop codon (polyadenylation signal 1, p1, Figure 1) and exemplified by GenBank entry BX023608. However, a few additional longer transcripts, deriving from the recognition of alternative polyadenylation signals located 368 nts and 450 nts downstream of the stop codon (as example see GenBank ID XM317463 and CX819455), were also detected (p2 and p3, Figure 1). Assuming that transcription starts in the region identified from the ESTs CD747374 and BM649960, these in silico observations led to the conclusion that transcription of the cE5 gene yields three different cDNA variants: a short form of approximately 600 nucleotides, which will be named as cE5S, and two longer forms of about 890 and 950 nucleotides, which will be indicated collectively as cE5L forms and, when needed, distinguished as cE5L890 and cE5L950.
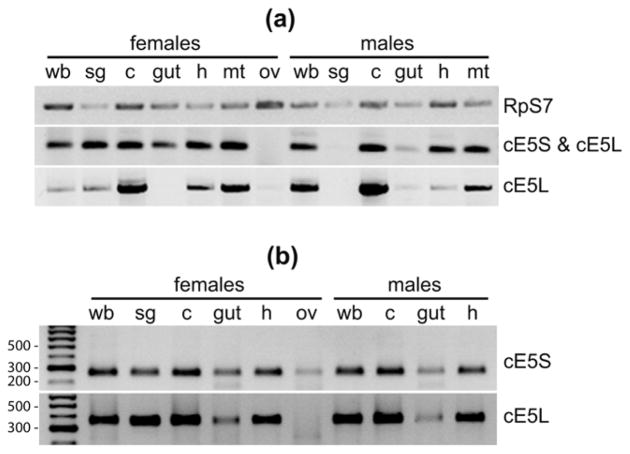
Previous analyses by RT-PCR revealed poor tissue-specificity of the cE5 gene, whose transcript was found not only in female salivary glands, but also in adult males and in female carcass, i.e. remaining female tissues after salivary glands dissection (Arcà et al., 1999; Arcà et al., 2005). To extend this initial analysis we performed some additional RT-PCR using as template first strand cDNA synthesized from RNA extracted from ovaries and a few other male and female tissues: salivary glands, guts, heads, malpighian tubules, carcasses (i.e. the leftover after dissection of the above mentioned tissues) and whole bodies. A common forward primer (F1) and two different reverse primers (R1 and R2) were designed to amplify both cE5S and cE5L (F1+R1) or cE5L only (F1+R2) (see Figure 1). Electrophoretic analysis of the RT-PCR products showed that cE5 transcripts were absent in ovaries and in male salivary glands (Figure 2A), which is fully in agreement with a previous male salivary gland transcriptome study where not a single cE5 transcript was found (Calvo et al., 2006). Little expression was found in the gut, with the exception of an enrichment of cE5S in female guts. In the remaining tissues cE5 transcripts were detected, although with different intensities, with both primer pairs (Figure 2A). A total of nine PCR products obtained with the two primer pairs from different tissues were randomly selected, cloned and sequenced; with just one exception (a small deletion of 5 codons falling in the signal peptide region) all other products corresponded to the expected, correctly spliced cDNA sequences. Within the limits of our analysis, which is not quantitative, the relative intensities of the PCR products obtained with the two different primer pairs suggested an enrichment of cE5S in male heads and in female salivary glands and guts, whereas the cE5L forms were apparently enriched in both male and female carcasses (Figure 2A). Very similar results were independently obtained on different RNA sets using one-step RT-PCR rather than amplification from first strand cDNAs (not shown). To confirm the coexistence of cE5S and cE5L we also performed a 3′-RACE using two different forward primers Fm and F3, which allow for the independent detection of the two forms, in combination with an oligodT-containing reverse primer (AP, see Figure 1). The results of this analysis are in substantial agreement with the RT-PCR data confirming the presence in several tissues of both sexes of short and long versions of cE5 (Figure 2B). In addition, a more accurate electrophoretic analysis revealed that the band labeled in Figure 2B as cE5L included two fragments, corresponding to cE5L890 and cE5L950 as verified by cloning and sequencing of the respective PCR products (data not shown). Overall, our results fit well with those reported in a recent microarray-based tissue- and sex-specific expression study where the cE5 gene was found to be expressed with a rather promiscuous pattern and up-regulated in female salivary glands and female carcasses (Baker et al., 2011, Figure S1, Online Resource 1). Moreover, the cE5 transcript was previously reported as differentially expressed and up-regulated in An. gambiae hemocytes versus heads and carcasses (Pinto et al., 2009).
Figure 2. Tissue- and sex-specific expression of cE5S and cE5L transcripts.
(a) Expression pattern as obtained by RT-PCR expression analysis. (b) 3′-RACE on selected tissues. wb, whole body; sg, salivary glands; c, carcass (leftover after removal of salivary glands, gut, heads, malpighian tubules and ovaries); gut, midgut; h, head; mt, malpighian tubules; ov, ovaries. RpS7, ribosomal protein S7.
The similarity of the An. gambiae cE5 to the An. albimanus anophelin (43% identity, 57% similarity, see Figure S4, Online Resource 1) and the expression in female salivary glands suggest a potential blood feeding role as thrombin inhibitor. However, the co-existence of different cE5 transcripts (cE5S, cE5L890 and cE5L950) in several tissues, as well as the longer carboxy-terminal region of cE5 as compared to the anophelin protein, is intriguing. These observations raise the possibility that the An. gambiae cE5 gene may have acquired some additional or modified function in this mosquito species and/or that post-transcriptional gene regulatory mechanisms may determine the role of this gene in mosquito biology.
3.2 Expression and purification of the A. gambiae cE5 protein
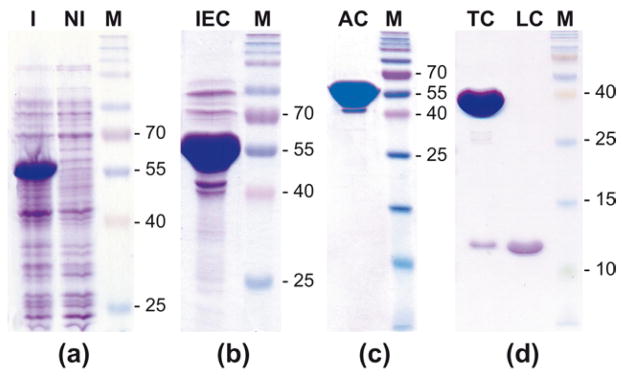
After several previous attempts in both procaryotic and yeast-based systems, the successful expression of the An. gambiae cE5 protein was achieved in E. coli only after fusion to the Maltose Binding Protein (MBP). Because of the presence within the cE5 sequence of secondary sites for Factor Xa (FXa), a cleavage site for the Tobacco Etch Virus (TEV) protease was engineered just downstream to the FXa recognition site in the E. coli expression vector pMAL-c4X. This MBP-FT-cE5 version could be expressed in soluble form at a good level (see band of ~55 kDa in Figure 3A) and was essentially pure after a step of ion-exchange-chromatography (IEC) followed by an affinity-chromatography (AC) on an amylose resin (Figure 3B–C). Cleavage by the TEV protease followed by Reverse Phase HPLC (RP-HPLC) purification allowed for the efficient removal of the fusion partner (Figure 3D). This way the cE5 protein was obtained in its mature form with just an additional Gly at the N-terminus. Therefore, the recombinant cE5 was 83 amino acids in length with an expected molecular mass of 8988 Daltons, which fits very well with the results obtained by mass spectrometry (not shown), although it should be noticed that the recombinant protein migrates slower than expected in SDS-PAGE, i.e. in the range of ~12 kDa (Figure 3D).
Figure 3. Expression and purification of the recombinant cE5 protein.
SDS-PAGE analysis of protein fractions containing the recombinant cE5 after the different purification steps. M, Prosieve QuadColor Protein Marker, 4.6–300 kDa (Lonza, Cologne, GmbH). (a) Expression levels of the MBP-cE5 fusion protein after induction. I, induced; NI, non induced. (b) Purification of MBP-cE5 by Ion-Exchange Chromatography (IEC). (c) Recombinant MBP-cE5 after Affinity Chromatography (AC). (d) Final purification by TEV cleavage (TC) and RP-HPLC (LC).
The purified cE5 protein was used to immunize BALB/c mice and obtain an anti-cE5 polyclonal serum. Although its sensitivity was rather poor, the immune serum allowed the detection of both the native protein in salivary gland extracts (SGE) and the recombinant polypeptide (Figure 4A). The different mobility after SDS-PAGE between the native and recombinant forms suggests that the cE5 protein may be post-translationally modified in the salivary glands, most likely undergoing a mucin-type O-glycosylation on C-terminal selected Ser or Thr residues, as also predicted by the NetOGlyc 3.1 Server (Julenius et al., 2005).
Next, protein extracts from a few different tissues were analyzed by western blot to verify whether the cE5 protein was also present elsewhere in the mosquito apart from the salivary glands. Even with some complication due to the relatively high background of the anti-cE5 polyclonal serum, the cE5 protein, as expected, was present in female SGE. On the contrary, despite several attempts, we never detected any band potentially corresponding to cE5 in extracts from guts, heads, carcasses, hemocytes or hemolymph, even when large amounts of protein extracts were used (not shown). The western blot reported in Figure 4b (bottom) was obtained after IgG purification of the mouse anti-cE5 immune serum and clearly shows the presence of cE5 only in female SGE. The accompanying silver stained gel (Figure 4b, top) testifies integrity and large excess of tissue extracts in comparison to SGE. Therefore, although cE5 transcripts were detected in several tissues both by our RT-PCR expression analysis and by a recent microarray study (Baker et al., 2011), surprisingly, the cE5 protein was only found in mosquito female SGE. Overall, these observations support the hypothesis that post-transcriptional regulation may prevent the translation of the cE5 protein or make it extremely unstable in tissues other than female salivary glands.
3.3 cE5 is a specific thrombin inhibitor
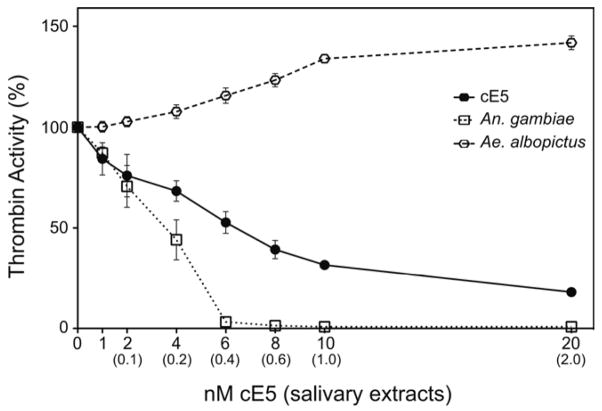
To test whether the An. gambiae cE5 acts as a specific thrombin inhibitor or not, we first measured the amidolytic activity of thrombin (10 nM) on the chromogenic substrate benzoyl-FVR-pNA. Progressive inhibition of thrombin activity was observed when increasing concentrations of cE5 protein (1 to 20 nM) were included in the assay (Figure 5). In these conditions the residual thrombin activity dropped to 53 % when 6 nM cE5 were added to the assay (molar ratio cE5 : thrombin ~0.6 : 1), reaching a minimum of ~18% at the maximum cE5 concentration tested (20 nM, cE5 : thrombin = 2 : 1). When SGE from An. gambiae were tested in the same assay, the residual thrombin activity was ~70% and ~44% after addition of 0.1 and 0.2 salivary gland pair equivalents (SGeq), respectively. Almost complete inhibition of thrombin was obtained when 0.4 SGeq were added to the assay (3% residual activity), and very little changes were observed after further addition of An. gambiae SGE. Also salivary extracts from the mosquito Aedes albopictus, known to be devoid of anti-thrombin activities (Stark and James, 1996), were analyzed as a negative control. As expected, no thrombin inhibition was detected in our assay. On the contrary, increased cleavage of the chromogenic substrate was observed, possibly due to the action of some other protease present in the saliva of the tiger mosquito (Figure 5).
Figure 5. Effects of cE5 and salivary extracts from An. gambiae and Ae. albopictus on thrombin activity.
The activity of thrombin (10 nM) in the presence of recombinant cE5 (1 to 20 nM) or salivary extracts from An. gambiae and Ae. albopictus (0.1 to 2.0 salivary gland pairs) was measured following the hydrolysis of the chromogenic substrate Benzoyl FVR-pNA. The activity of thrombin in the absence of inhibitor or SGE was assumed as 100%. The nanomolar amounts of cE5 protein used in the assay are shown on the X axis; numbers in parentheses refer to salivary gland pair equivalents. The mean of three independent experiments (each in triplicate) are reported. Error bars represent 95% CI.
To evaluate the specificity of cE5 for thrombin, a screen for potential inhibitor activity against a panel of fifteen different serine proteases was performed; in this assay fluorogenic substrates specific for each protease were used to determine the enzyme activity in the absence and in the presence of cE5. The residual activity of the fifteen proteases in the presence of a large excess of the cE5 protein (at least 100-fold) showed that cE5 was able to inhibit thrombin but did not affect any of the other enzymes tested (Table 1). From this screen we conclude that cE5 is a specific inhibitor of thrombin. However, it is worth noting that even with the large molar excess of cE5 used in this assay (600 nM versus 30 pM) the activity of thrombin did not drop below 12%.
3.4 cE5 is a tight- and fast-binding inhibitor of thrombin
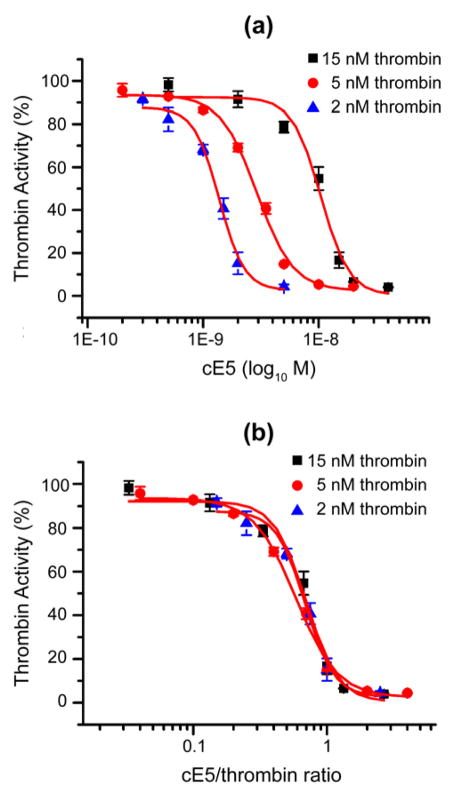
To further investigate the thrombin inhibitory properties of cE5 we titrated the loss of activity of three different concentrations of thrombin in the nanomolar range (2 nM, 5 nM and 15 nM) in the presence of different concentrations of cE5. The IC50 of cE5 for thrombin varied according to the amount of enzyme used in the assays (Figure 6A). More specifically, the IC50 was 10.43 ± 1 nM with 15 nM of thrombin and decreased to 2.9 ± 0.16 nM and 1.38 ± 0.09 nM when the concentration of thrombin was reduced to 5 nM and 2 nM, respectively. When the same data are replotted against the cE5/thrombin molar ratio the three curves shown in figure 6A widely overlap and the IC50 is achieved at 0.7 ± 0.07, 0.59 ± 0.03 and 0.69 ± 0.05 cE5/thrombin ratio for 15 nM, 5 nM and 2 nM of thrombin, respectively (Figure 6B). Therefore, in this thrombin nanomolar range the half maximal cE5 inhibitory activity is achieved around the 0.66 cE5/thrombin molar ratio. These observations clearly indicate that the An. gambiae cE5, similar to the An. albimanus anophelin (Valenzuela et al., 1999), is a tight binding inhibitor of thrombin.
Figure 6. IC50 of cE5 for thrombin.
(a) Residual thrombin activity as a function of different cE5 concentrations. Three different concentrations of thrombin (2 nM, 5 nM and 15 nM) were incubated with increasing amounts of cE5 in the presence of the fluorogenic substrate Boc-Ile-Glu-Gly-Arg-7-amido-4-methylcoumarin hydrochloride. IC50 were, respectively: 10,43 ± 1 nM for 15 nM of thrombin, 2,9 ± 0,16 nM for 5 nM of thrombin and 1,38 ± 0,09 nM for 2 nM of thrombin. (b) Residual thrombin activity as a function of the cE5/thrombin molar ratio. The IC50 was achieved at a cE5/thrombin ratio of 0.7 ± 0.07 for 15 nM thrombin, 0.59 ± 0.03 for 5 nM thrombin and 0.69 ± 0.05 for 2 nM thrombin, respectively. Data represent the mean of triplicates and error bars refer to Standard Error of the Mean (SEM). The activity of thrombin in the absence of inhibitor was assumed as 100%.
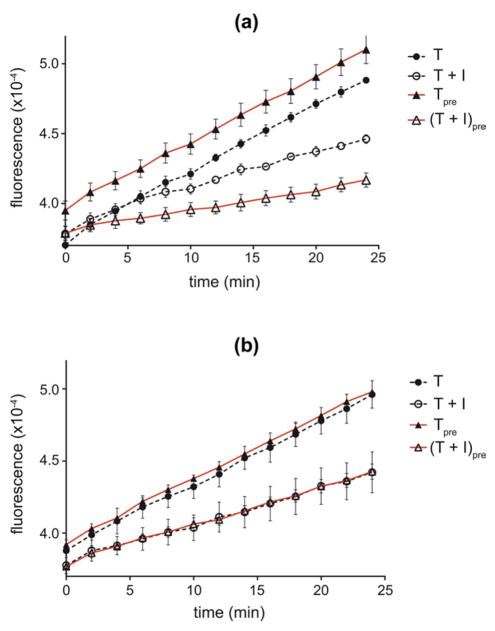
Moreover, the An. albimanus anophelin was previously shown to be a slow-binding inhibitor of thrombin (Francischetti et al., 1999). To verify if this was also true for the An. gambiae cE5 we compared the effects of a 10 minutes pre-incubation on thrombin inhibition. In this assay thrombin (10 pM) was incubated at 37°C with 1 nM of either cE5 or anophelin in the presence of 100 mM NaCl in the assay buffer. According to its slow-binding kinetics, more efficient inhibition was obtained when the An. albimanus anophelin was pre-incubated with thrombin before addition of the fluorogenic substrate (Figure 7A) . This was not the case for the An. gambiae cE5, for which the same effect on thrombin activity was obtained no matter whether the enzyme was pre-incubated or not with the inhibitor (Figure 7B). Similar results were obtained with both inhibitors at a picomolar range (20 pM anophelin, 5 pM cE5) in the absence of salt (not shown). These observations indicate different kinetics of interaction of the two proteins with their target, suggesting that cE5 is a fast-binding thrombin inhibitor.
Figure 7. Effect of pre-incubation on thrombin inhibition by the An. gambiae cE5 and the An. albimanus anophelin.
The activity of thrombin (10 pM) was measured following the hydrolysis of the fluorogenic substrate Boc-Ile-Glu-Gly-Arg-7-amido-4-methylcoumarin hydrochloride, with or without 10 minutes pre-incubation, in the presence of 100 nM NaCl and either 1 nM anophelin (a) or 1 nM cE5 (b) in the assay buffer. T, thrombin no preincubation; Tpre, thrombin with preincubation; T + I, thrombin + inhibitor no preincubation; (T + I)pre, thrombin + inhibitor with preincubation. Data represent the mean of triplicates. Error bars represent SEM. Activity of thrombin is reported as arbitrary fluorescence unit.
3.5 Salt effect on thrombin inhibition by cE5 and anophelin
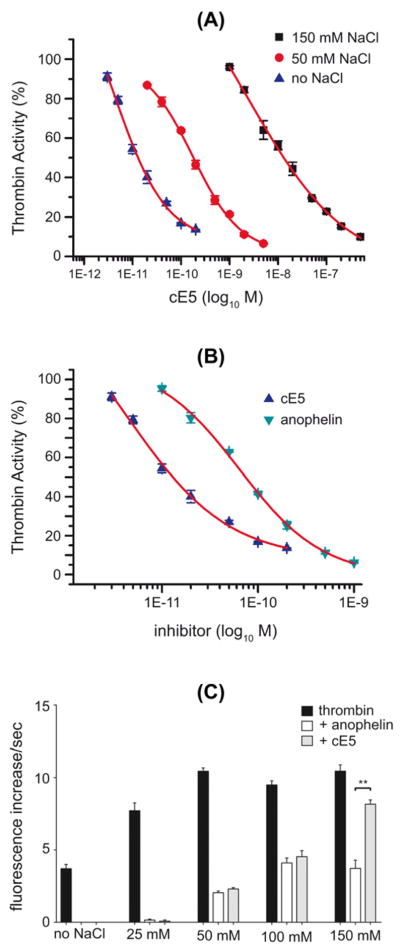
It has been previously shown that the An. albimanus anophelin is a dual inhibitor, binding both the catalytic site and the thrombin anion binding exosite 1 (TABE1), a region that is required for thrombin interaction with fibrinogen and with other proteins involved in hemostasis regulation (Francischetti et al., 1999). TABE1, which is rich in positively charged amino acids, is likely to interact with the highly negatively charged N-terminal half of anophelin. Accordingly, it was shown that increasing concentration of salt decreased the affinity of anophelin for thrombin, presumably by decreasing ionic interactions between the enzyme and the inhibitor (Francischetti et al., 1999). A similar effect was observed for the An. gambiae cE5. When thrombin (10 pM) was incubated with different amounts of cE5 in the absence or in the presence of NaCl in the assay buffer the inhibitory effect decreased at increasing salt concentrations. The cE5 IC50 was 6,25 ± 1,22 pM in the absence of salt, 173 ± 22 pM in the presence of 50 mM NaCl and 4,9 ± 1,22 nM in the presence of 150 mM NaCl (Figure 8A), suggesting that cE5 also interacts with TABE1. When the cE5 and anophelin IC50 were compared in the absence of salt and of pre-incubation with thrombin, cE5 showed ~10 times higher affinity for thrombin: the anophelin IC50 was 64 ± 9 pM as compared to 6,25 ± 1,22 pM of cE5 (Figure 8B). However, when thrombin (10 pM) and inhibitor (1 nM) were preincubated for 10 minutes before substrate addition, the two proteins showed very similar inhibitory properties up to 100 mM NaCl. A further increase of NaCl concentration (150 mM) had no effect on anophelin whereas the inhibitory power of cE5 decreased, indicating that at this salt concentration, after pre-incubation, the An. albimanus anophelin appears to be a better inhibitor than the An. gambiae cE5 (Figure 8C).
Figure 8. Effect of salt on thrombin inhibition by the An. gambiae cE5 and the An. albimanus anophelin.
The activity of thrombin (10 pM) was assayed in the presence of different NaCl concentrations measuring the hydrolysis of the fluorogenic substrate Boc-Ile-Glu-Gly-Arg-7-amido-4-methylcoumarin. (a) Thrombin inhibition as function of cE5 concentration in the presence and absence of NaCl as indicated. (b) Comparison of thrombin inhibition by the An. gambiae cE5 and the An. albimanus anophelin in the absence of salt in the assay buffer and without pre-incubation. (c) Comparison of thrombin inhibition by the An. gambiae cE5 and the An. albimanus anophelin in the presence of different NaCl concentrations (25 mM, 50 mM, 100 mM, 150 mM) in the assay buffer. Thrombin (10 pM) was pre-incubated (10 min) with 1 nM cE5 or anophelin before addition of the fluorogenic substrate. Data represent the mean of triplicates. Error bars represent SEM. In (A) and (B) the activity of thrombin in the absence of inhibitor was assumed as 100%. In (C) the activity of thrombin is expressed as increase per second of arbitrary fluorescence units. **, p<0.01 (unpaired two-tailed t-test).
4. DISCUSSION
With the aim of identifying genes potentially involved in hematophagy we have previously employed a simple RT-PCR assay to classify An. gambiae salivary gland genes in three main categories: (a) housekeeping; (b) involved in blood feeding; (c) playing non sex-specific roles in gland physiology (i.e. helping sugar digestion, preventing bacterial growth or lubricating mouthparts) (Arcà et al., 1999; Arcà et al., 2005; Lanfrancotti et al., 2002). This analysis, which worked successfully in most cases, failed to identify cE5 as involved in blood feeding because of its promiscuous expression pattern. Nevertheless, the identification of the An. albimanus anophelin as a thrombin inhibitor (Valenzuela et al., 1999) strongly suggested a similar function for cE5. However, a widespread expression of an anti-thrombin polypeptide did not make much sense, and for this reason we performed a more detailed analysis and showed that cE5 transcripts originated by alternative polyadenylation site usage and carrying 3′UTRs of different length coexist in various tissues (Figure 2). This expression profile is in substantial agreement with a recent microarray analysis reporting transcription of the cE5 gene in several tissues and upregulation in female salivary glands and carcasses (Figure S1, Online Resource 1) (Baker et al., 2011), a pattern highly unusual for a gene involved in hematophagy. In contrast other An. gambiae salivary genes encoding proteins with anti-hemostatic or anti-inflammatory activities, such as apyrase, salivary peroxidase or D7r1 exhibit expression profiles that both in our previous assays (Arcà et al., 2002; Arcà et al., 2005; Lombardo et al., 2000) and in the microarray study of Baker and colleagues (2011) were restricted or highly enriched in female salivary glands (Figure S1, Online Resource 1).
While the presence of cE5 transcripts in female salivary glands and in midguts was conceivable in terms of keeping the blood fluid during and after feeding, their presence in males and in other female tissues was of more problematic interpretation; moreover, an additional layer of intricacy derives from the presence of different cE5 mRNA isoforms. We reasoned that perhaps the cE5 protein could play some additional or slightly different function, for example acting on a broader range of serine proteases. To verify this hypothesis and characterize the biochemical activity of this anophelin family member from An. gambiae, we expressed the protein and obtained an anti-cE5 immune serum. Western blot analysis allowed the detection of the cE5 protein in female salivary glands but in no other tissue, even using large excess of protein extracts and repeating the experiment several times. Support to our observations also comes from a recent proteomic analysis reporting the presence of cE5-derived peptides only in female mosquito salivary glands and heads (Chaerkady et al., 2011). We believe that detection in head extracts is most likely due to the inclusion of salivary ducts in the dissected heads, and our interpretation is confirmed by the fact that many salivary genes involved in hematophagy and specifically expressed in female glands (i.e. apyrase, salivary peroxidase, D7-related, etc) were also present in head extracts in the same proteomic data set.
Overall, the above observations show that cE5 is transcribed in several tissues whereas the encoded polypeptide is only found in female salivary glands, implying that post-transcriptional gene regulatory mechanisms are preventing protein translation or accumulation in the other tissues. Eukaryotic messenger RNA 3′UTRs are known targets of regulatory mechanisms which may affect translation and stability (Fabian et al., 2010; Garneau et al., 2007; Licatalosi and Darnell, 2010), and the existence of cE5 mRNA isoforms carrying 3′UTRs of different length combined with our western blot results suggest a similar mechanism. Typical AU-rich elements that may induce accelerated mRNA degradation (Garneau et al., 2007) were not found in the cE5 3′UTR. On the contrary a search for potential microRNA binding sites by the microinspector program (Rusinov et al., 2005) (web server http://bioinfo.uni-plovdiv.bg/microinspector/) yielded two miRNAs: aga-miR-989 (miRBase: MI0006242), which pairs at short distance from the stop codon in a region common to all cE5 isoforms, and aga-miR-891 (miRBase: MI0008307) potentially targeting only cE5L isoforms (Figure S2, Online Resource 1). Inhibition of cE5 mRNA translation by aga-miR-989 in all tissues of both sexes with the exception of female salivary glands could well explain our observations. However, a female-restricted expression pattern has been reported for aga-miR-989 (Winter et al., 2007) and for the An. stephensi homologue ast-miR-989 (Mead and Tu, 2008), which excludes this possible explanation because it could not account for the presence of transcripts and absence of protein in males. The expression pattern of aga-miR-981 is not known, but the cE5L-specific targeting would not allow a proper justification of the absence of cE5S translation in males and in female tissues. An additional intriguing consideration is that the cE5L-specific 3′UTR matches a 99 nucleotide exon from a gene encoded on the other strand (VectorBase: AGAP008003, Figures S2 and S3, Online Resource 1). Although this gene of unknown function is up-regulated in female salivary glands (Baker et al., 2011) the significance of this observation in relation to potential regulatory mechanisms is completely unknown.
Under an evolutionary scenario cE5 is an anopheline-specific protein that evolved/was recruited to play blood feeding roles after the separation of anopheline from culicine mosquitoes, an event that is dated around 145–200 million years ago (Krzywinski et al., 2006). In the malaria mosquito An. gambiae most anopheline-specific proteins involved in hematophagy acquired tissue-specificity by mechanisms acting at transcriptional level. In this respect the cE5 protein appears unusual since salivary expression is not under tissue-specific transcriptional control, perhaps because of counteracting evolutionary constraints imposed from surrounding genomic regions, and instead was achieved by post-transcriptional regulatory mechanisms.
Expression of the cE5 recombinant protein was not straightforward since any attempt to express the protein in its native mature form or after the addition of short tags was unsuccessful, perhaps because high concentrations of the protein are toxic to E. coli. Similar problems were previously faced with the An. albimanus anophelin (Ribeiro JMC, personal communication), whose anti-thrombin properties were studied using a synthetic polypeptide (Francischetti et al., 1999; Valenzuela et al., 1999). The expression of the protein as a fusion to MBP allowed to circumvent this problem and to obtain, after cleavage by the TEV protease, a purified protein in its mature form carrying just an additional NH2-terminal Gly and with a yield of approximately 4–5 mg/liter of culture. This cE5 recombinant protein is active and exhibits highly specific thrombin inhibition, as shown by the absence of any activity against a panel of fourteen additional serine proteases. A similar result with a slightly different set of proteases was previously obtained with the anophelin from An. albimanus (Valenzuela et al., 1999), highlighting the stringent specificity of this family of thrombin inhibitors. Comparison of electrophoretic mobility of native and recombinant cE5 suggests that in the An. gambiae salivary glands the protein undergoes some sort of post-translational modification. The NetOGlyc 3.1 Server (Julenius et al., 2005) predicts ten potential mucin-type O-glycosylation sites, seven of which in the Serine-rich carboxy-terminal region. A similar prediction analysis on the An. albimanus anophelin, that lacks this C-terminal extension, yields three potential O-glycosylation sites (Figure S4, Online Resource 1). However, there are no evidence so far that anophelin may be glycosylated. When SGE and recombinant cE5 protein were compared for their thrombin inhibitory properties we observed total inhibition with An. gambiae SGE whereas, even using a very large excess of recombinant cE5, we never observed >90% inhibition of thrombin activity. An involvement of the N-terminal additional glycine seems unlikely, while the post-translational modification of the native protein may play some role in the inhibitory activity and thus explain this difference. However, the nature of the chromogenic or fluorogenic substrates used in our assays may offer an alternative, perhaps more likely explanation. These small molecules may gain some access to the catalytic site even in the presence of the inhibitor, instead the glycosylated protein may work better in preventing this access, yielding almost 100% inhibition. The situation may be significantly different with the large natural substrate, i.e fibrinogen; actually, this was the case for the synthetic anophelin that fully inhibits thrombin in a fibrinogen substrate assay whereas it reaches ~90% inhibition in a chromogenic substrate assay and only at higher inhibitor concentrations (Valenzuela et al., 1999).
Although An. gambiae and An. albimanus separated approximately 100 million years ago (Besansky N.J., 2008) and the two proteins, anophelin and cE5, are quite divergent (43% identity, 57% similarity), their anti-thrombin function is fully preserved, since they were both found to be highly specific and tight-binding inhibitors of thrombin. On the contrary, comparison of the inhibitory effect in the presence or absence of pre-incubation indicates that the two proteins differ for their binding kinetics. The An. albimanus anophelin is a slow-binding thrombin inhibitor, whereas the An. gambiae cE5 protein behaves like a fast-binding inhibitor, a property that may confer a significant advantage to a blood feeder like a mosquito. When, following a short preincubation, the activity of the two proteins was compared in the presence of salt it was evident that the addition of salt decreased their inhibitory effect. It has been previously shown that the An. albimanus anophelin binds both to thrombin catalytic site and TABE1, and the decreased affinity in the presence of salt was interpreted as the result of decreased ionic interactions between inhibitor and enzyme (Francischetti et al., 1999). The same seems to hold for the An. gambiae cE5, which can be predicted to similarly interact with both TABE1 (through its highly negatively charged N- terminal half) and the thrombin catalytic site. The two proteins still exhibited very similar inhibitory properties in the presence of 100 mM NaCl, but the An. albimanus anophelin was a better inhibitor when salt concentration was increased to 150 mM NaCl.
Mosquito salivary proteins are known to evolve at a very fast rate and the selective pressure of the vertebrate host immune system has been suggested to be the powerful driving force of this rapid evolution (Arcà et al., 2007; Ribeiro and Arcà, 2009; Valenzuela et al., 2003). Then, an attractive scenario is that after the ancestors of An. albimanus (subgenus Nyssorhynchus, including Center and South American Anopheles species) and An. gambiae (subgenus Cellia, including mainly Asian and African Anopheles species) separated, the proto-anophelin and proto-cE5 started to diverge by creating variants with different immunogenic properties in order to minimize inactivation by the immune response of their hosts. On the other side, the highly adaptive value of carrying an efficient anti-thrombin activity selected only those variants not affecting or improving the function. As a result of these two distinct and sometime opposing forces the An. albimanus anophelin evolved toward a variant able to function slightly better in salt concentrations close to physiological conditions (i.e. in 150 mM NaCl), whereas the An. gambiae cE5 acquired fast-binding properties, perhaps thanks to the longer carboxy-terminal region which is also found in the other Cellia subgenus members An. funestus and An. stephensi.
5. CONCLUSIONS
In conclusion we have expressed in recombinant form the An. gambiae salivary protein cE5 and obtained evidence that it is a highly specific, tight- and fast-binding dual inhibitor of thrombin. The setting up of conditions for large-scale expression and purification of the cE5 protein paves the way for structural studies on anophelin family members, which may shed further light on their mechanisms of thrombin inhibition. Moreover, we have shown that the tissue-specificity of the An. gambiae cE5 protein depends from some, so far undefined, post-transcriptional regulatory mechanism rather than being under transcriptional regulation, as is the case for many other An. gambiae salivary genes involved in hematophagy. These observations point out that transcriptomic data should be interpreted with care emphasizing the importance of associated proteomic support.
Supplementary Material
The Anopheles gambiae cE5 protein was expressed, purified and characterized.
cE5 is a highly specific tight- and fast-binding thrombin inhibitor.
Multiple cE5 transcript isoforms are found in several tissues of both males and females.
Post-transcriptional gene regulation restricts cE5 protein to mosquito female salivary glands.
Acknowledgments
This work was supported by the European Union BioMalPar (503578) and Infravec (228421) grants to BA. JMCR received support from the Intramural Research Program of the Division of Intramural Research, NIAID-NIH. MK received support by a Jan Evangelista Purkyne fellowship, by the National Academy of Sciences of the Czech Republic (Z60220518), by the Grant Agency of the Czech Republic (P502/12/2409), by a European Union MC Reintegration grant (PIRG07-GA-2010-268177) and from NIH (R01AI093653). RR was supported by the Infravec project. CR was partly supported by a short term fellowship of Istituto Pasteur-Fondazione Cenci-Bolognetti (Sapienza University, Rome).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arcà B, Lombardo F, de Lara Capurro M, della Torre A, Dimopoulos G, James AA, Coluzzi M. Trapping cDNAs encoding secreted proteins from the salivary glands of the malaria vector Anopheles gambiae. Proc Natl Acad Sci U S A. 1999;96:1516–1521. doi: 10.1073/pnas.96.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Francischetti IM, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JM. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem Mol Biol. 2007;37:107–127. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Lanfrancotti A, Spanos L, Veneri M, Louis C, Coluzzi M. A cluster of four D7-related genes is expressed in the salivary glands of the African malaria vector Anopheles gambiae. Insect Mol Biol. 2002;11:47–55. doi: 10.1046/j.0962-1075.2001.00309.x. [DOI] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, Ribeiro JMC. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J Exp Biol. 2005;208:3971–3986. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RF, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons Inc; New York: 1991. [Google Scholar]
- Baker DA, Nolan T, Fischer B, Pinder A, Crisanti A, Russell S. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics. 2011;12:296. doi: 10.1186/1471-2164-12-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ on behalf of the Anopheles Genomes Cluster Committee. Genome Analysis Of Vectorial Capacity in Major Anopheles Vectors of Malaria Parasites. 2008 http://www.vectorbase.org/Help/Anopheles_species_cluster_white_paper#tab=document.
- Calvo E, Dao A, Pham VM, Ribeiro JM. An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families. Insect Biochem Mol Biol. 2007;37:164–175. doi: 10.1016/j.ibmb.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Pham VM, Lombardo F, Arcà B, Ribeiro JMC. The sialotranscriptome of adult male Anopheles gambiae mosquitoes. Insect Biochem Mol Biol. 2006;36:570–575. doi: 10.1016/j.ibmb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Chaerkady R, Kelkar DS, Muthusamy B, Kandasamy K, Dwivedi SB, Sahasrabuddhe NA, Kim MS, Renuse S, Pinto SM, Sharma R, Pawar H, Sekhar NR, Mohanty AK, Getnet D, Yang Y, Zhong J, Dash AP, MacCallum RM, Delanghe B, Mlambo G, Kumar A, Keshava Prasad TS, Okulate M, Kumar N, Pandey A. A proteogenomic analysis of Anopheles gambiae using high-resolution Fourier transform mass spectrometry. Genome Res. 2011;21:1872–1881. doi: 10.1101/gr.127951.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmelar J, Oliveira CJ, Rezacova P, Francischetti IM, Kovarova Z, Pejler G, Kopacek P, Ribeiro JM, Mares M, Kopecky J, Kotsyfakis M. A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation. Blood. 2011;117:736–744. doi: 10.1182/blood-2010-06-293241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Ribeiro JMC. Anophelin: kinetics and mechanism of thrombin inhibition. Biochemistry. 1999;38:16678–16685. doi: 10.1021/bi991231p. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Gasteiger EHC, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Grushko OG, Besansky NJ. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol Phylogenet Evol. 2006;39:417–423. doi: 10.1016/j.ympev.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Lanfrancotti A, Lombardo F, Santolamazza F, Veneri M, Castrignano T, Coluzzi M, Arcà B. Novel cDNAs encoding salivary proteins from the malaria vector Anopheles gambiae. FEBS Lett. 2002;517:67–71. doi: 10.1016/s0014-5793(02)02578-4. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo F, Di Cristina M, Spanos L, Louis C, Coluzzi M, Arcà B. Promoter sequences of the putative Anopheles gambiae apyrase confer salivary gland expression in Drosophila melanogaster. J Biol Chem. 2000;275:23861–23868. doi: 10.1074/jbc.M909547199. [DOI] [PubMed] [Google Scholar]
- Mead EA, Tu Z. Cloning, characterization, and expression of microRNAs from the Asian malaria mosquito, Anopheles stephensi. BMC Genomics. 2008;9:244. doi: 10.1186/1471-2164-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto SB, Lombardo F, Koutsos AC, Waterhouse RM, McKay K, An C, Ramakrishnan C, Kafatos FC, Michel K. Discovery of Plasmodium modulators by genome-wide analysis of circulating hemocytes in Anopheles gambiae. Proc Natl Acad Sci U S A. 2009;106:21270–21275. doi: 10.1073/pnas.0909463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Mans BJ, Arcà B. An insight into the sialome of blood-feeding Nematocera. Insect Biochem Mol Biol. 2010;40:767–784. doi: 10.1016/j.ibmb.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC. Common problems to arthropod disease vectors. In: Beaty BJ, Marquardt WC, editors. The Biology of Disease Vectors. University Press of Colorado; Niwot, Colo: 1996. pp. 25–33. [Google Scholar]
- Ribeiro JMC, Arcà B. From Sialomes to the Sialoverse: An Insight into Salivary Potion of Blood-Feeding Insects. Adv Insect Physiol. 2009;37:59. [Google Scholar]
- Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:W696–700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsh EF, Maniatis Te. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Stark KR, James AA. Salivary gland anticoagulants in Culicine and Anopheline mosquitoes (Diptera: Culicidae) J Med Entomol. 1996;33:645–650. doi: 10.1093/jmedent/33.4.645. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JMC. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003;33:717–732. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Ribeiro JMC. Purification, cloning, and synthesis of a novel salivary anti-thrombin from the mosquito Anopheles albimanus. Biochemistry. 1999;38:11209–11215. doi: 10.1021/bi990761i. [DOI] [PubMed] [Google Scholar]
- Winter F, Edaye S, Huttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007;35:6953–6962. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.