Abstract
Mitochondrial dysfunction has been a hallmark of cancer. However, whether it plays a causive role awaits to be elucidated. Here, using an animal model derived from inactivation of SUV3, a mitochondrial helicase, we demonstrated that mSuv3+/− mice harbored increased mtDNA mutations and decreased mtDNA copy numbers, leading to tumor development in various sites and shortened lifespan. These phenotypes were transmitted maternally, indicating the etiological role of the mitochondria. Importantly, reduced SUV3 expression was observed in human breast tumor specimens compared to corresponding normal tissues in two independent cohorts. These results demonstrated for the first time that maintaining mtDNA integrity by SUV3 helicase is critical for cancer suppression.
Keywords: mitochondrial dysfunction, Suv3, animal model, tumorigenesis
Introduction
The mitochondria generate most of the cellular ATP and reactive oxygen species (ROS), regulate cellular calcium levels, and initiate cell death through the activation of the mitochondrial permeability transition pore (1, 2). Each mammalian cell contains hundreds of mitochondria and each mitochondrion contains multiple copies of circular mitochondrial DNA (mtDNA) molecules. The mtDNA encodes 13 polypeptides central to the mitochondrial energy production system, oxidative phosphorylation (OXPHOS), as well as the rRNAs and tRNAs necessary for mitochondrial protein synthesis. The mtDNA is replicated from two origins, one for the G-rich heavy (H) strand located in the control region and the other for the C-rich light stand located 2/3 of the way around the mtDNA (3). Cells have developed extensive systems for maintaining mitochondrial integrity and disturbance of mitochondrial homeostasis can lead to multiple diseases (2). In general, the mtDNA has a higher mutation rate than nuclear DNA (nDNA) genes. The accumulation of mtDNA mutations has not only been linked to ageing (2, 4, 5, 6), but also to cancer progression (7, 8). However, the causative roles of mitochondrial genome integrity in ageing and cancer remain to be substantiated.
SUV3 is a nuclear encoded ATP-dependent DNA/RNA helicase that primarily localizes to the mitochondrion. In Saccharomyces cerevisiae, Suv3 is a key component of the mitochondrial RNA degradosome besides the Dss1 exoribonuclease (9, 10, 11). Deficiency of SUV3 is associated with group I intron accumulation (10) and changes in the stability and processing of several mitochondrial RNAs (11). Yeast cells carrying null or mutant Suv3 genes gradually lose their capacity to grow on non-fermentable carbon sources and acquire a petite phenotype (12, 13). In human cells, depletion of SUV3 by siRNA leads to pleiotropic effects including decreased mtDNA copy number, a shift in mitochondrial morphology from tubular to granular, and eventual senescence or cell death (14). While the yeast Suv3 and mammalian SUV3 genes sharhe high sequence homology, salient differences between the two species such as the absence of Dss1 in human mitochondria and the lack of introns in mammalian mtDNA raises the question of whether the yeast and mammalian SUV3 serve analogous mitochondrial functions.
To clarify the function of mammalian SUV3, we generated a mouse Suv3 (mSuv3) knockout mouse. The homozygous mSuv3 knockout mice were embryonic lethal, suggesting an essential role of mSuv3 in embyronic development. Heterozygous (mSuv3+/−) animals, on the other side, had a shortened lifespan, reduced fecundity, and increased tumor incidence. More intriguingly, such effects were maternally transmitted, as only the mother but not the father, harboring the mutated mSuv3 gene, could transmit the defects to subsequent generations regardless the progenies’ nDNA genotype. The progenies of mSuv3+/− females showed elevated mtDNA mutation loads and reduced mtDNA copy numbers. These data suggest that SUV3 is a tumor suppressor gene that inactivation of one allele leads to destablization of mtDNA integrity, causing the observed detrimental effects in mice. Based on the fact that mSuv3+/− mice showed an increased tumor incidence, we quantitated the SUV3 mRNA level in two independent cohorts of human breast cancer specimens. We found that SUV3 mRNA expression was significantly reduced in tumor samples compared to adjacent normal tissue a panel of breast cancer specimens. Taken together, our data indicated that reduction of SUV3 expression level, either by genetic ablation of one allele of Suv3 in mice or other unknown mechanisms in human, could lead to destablization of mtDNA and promotes tumorigenesis, further supporting the notion that SUV3 is a potential human tumor suppressor that functions in a haploinsufficient manner.
Results
Targeted disruption of the mSuv3 locus leads to embryonic lethality
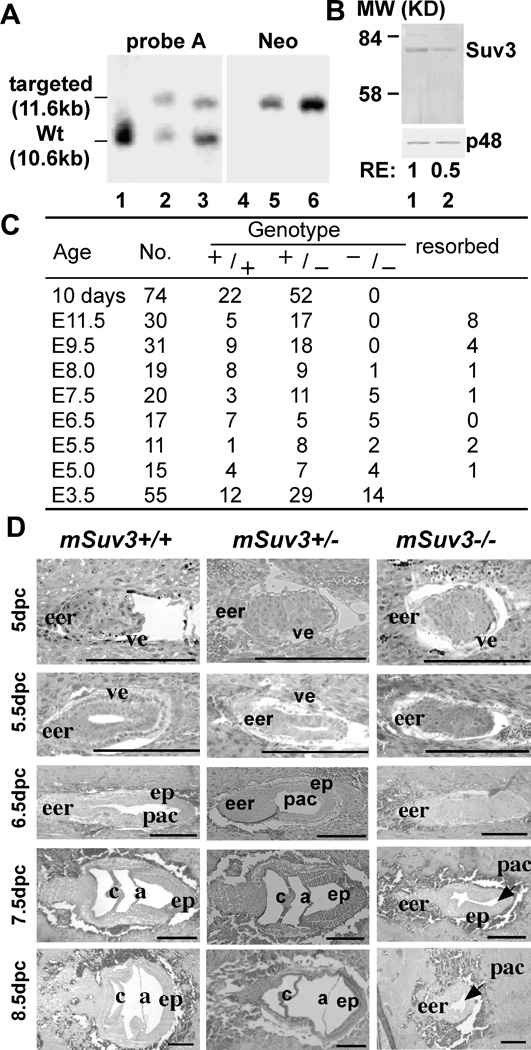
To test if deficiency in mammalian SUV3 would lead to detrimental effect in animal as a whole as it does in yeast, we inactivated the mSuv3 gene in embryonic stem (ES) cells to generate a knockout mouse model for SUV3. The mSuv3 gene was obtained by screening a 129/Sv-derived genomic library using the human SUV3 cDNA as a probe. The targeting vector, mSuv3-KO, was constructed by inserting an anti-parallel pgkneopA cassette into the mSuv3 location corresponding to codon 684 of the human SUV3, which rendered the SUV3 mRNA unstable and blocked SUV3 expression. The successfully targeted ES cells were used to generate chimeric mice, which ultimately resulted in germ line transmission of the knockout gene (Fig. 1A). An approximate 50% reduction in SUV3 protein level was observed in mSuv3 heterozygous (mSuv3+/−) mouse embryonic fibroblasts (MEF) when compared to the wild type MEF (Fig. 1B). There was no detectable truncated SUV3 protein in heterozygous MEF, further indicating that the knockout allele is a null depletion of SUV3.
Figure 1.
Targeted disruption of the mouse mSuv3 locus leads to embryonic lethality. (A) Confirmation of the targeted disruption of the mSuv3 allele. DNA samples from parental El4.1 cells (lanes 1 & 4) and two candidate recombinant clones [mSuv3-ko 5 (lanes 2 & 5) and mSuv3 ko 149 (lanes 3 & 6)] were digested with SalI-BamHI and probed with either probe A or the neo probe. An additional fragment of the expected size of 11.6 kb was found in each of the recombinant clones. (B) Western blot analysis of SUV3 protein level. Whole-cell lysate of MEFs of wild type (lane 1) and heterozygote (lane 2) were separated by SDS-PAGE and probed with an antibody against SUV3. p48 was used as a loading control. (C) Genotypes of offspring from heterozygous mSuv3+/− mice intercross. Pregnant females of mSuv3+/− intercrosses were sacrificed and the fetuses examined at different gestation times from E3.5 to E11.5 days. (D) Developmental abnormalities of mSuv3 mutant embryos. Histological analysis was performed on embryo sections of wild type and mSuv3−/− conceptuses. The uteri of mSuv3+/− females were dissected between 5.0 and 8.0 days after intercross, and 4 µm sections were prepared. All uterine decidua were sectioned transversely and labeled as described (48). The mesometrial to anti-mesometrial axis is from left to right. Abbreviations: (eer) extraembryonic region; (ve) viceral endoderm; (ep) epiblast; (pac) proamniotic cavity; (c) chorion; (a) amnion. Bar, 100 µm.
The mSuv3+/− mice were interbred and the genotypes of the offspring were determined at 10 days of age by PCR analysis of toe DNA. Out of the 74 animals screened, 52 were heterozygous (mSuv3+/−) and 22 were wild type (mSuv3+/+) with no homozygous mutant (mSuv3−/−), suggesting that the mSuv3−/− embryos are non-viable, which is consistent with previously described (15). To pinpoint the time of death, pregnant females of E5.0 to E11.5 from mSuv3+/− intercrosses were sacrificed and the fetuses were subjected to genotyping. At both E9.5 and E11.5, no mSuv3−/− embryos was found; instead, many resorbed embryos were present. Hence, the mSuv3 −/− embryos died by E9.5 (Fig. 1C).
To further characterize the developmental defects, decidual swellings from mSuv3+/− intercrossed females were obtained between embryonic E5.0 to E8.5, and examined histologically following staining with hematoxylin/eosin. As shown in Fig. 1D, both mSuv3+/+ and mSuv3+/− embryos exhibited normal growth, depicted by the elongation of the egg cylinder and formation of embryonic and extra-embryonic ectoderm, as well as distinct proamniotic cavities. By contrast, mSuv3−/− embryos presented abnormalities following implantation (E5.0 to E5.5), as they were smaller and failed to form the typical egg cylinders. By the time when mSuv3+/+ embryos underwent gastrulation (E7.5 to E8.0), the growth of mSuv3−/− embryos were remarkably delayed. By E8.5, the mSuv3−/− embryos started to degenerate and were resorbed, consistent with aforementioned genotyping results (Fig. 1C).
Shortened lifespan of mSuv3 heterozygotes at different intercross generations
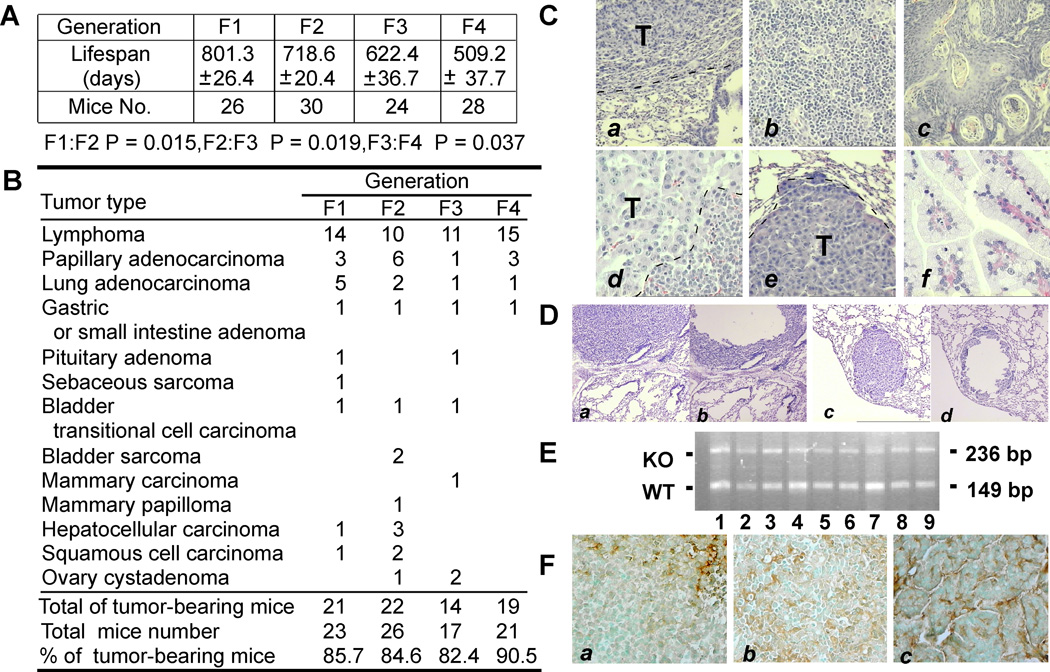
To monitor the effect of mSuv3 mutation over generations, we carried out a systematic studies on intercrossed mice. The first generation (F1) of heterozygous mSuv3+/− mice obtained by crossing the chimeric founder male with C57/BL6 female had normal lifespan comparable to the mSuv3+/+ wild type mice (Fig. 2A). However, the second generation (F2) of mSuv3+/− heterozygotes derived from intercrossing F1 heterozygotes had significantly shortened lifespan, which was aggravated in subsequent intercrosses of mSuv3+/− mice for the third and forth generations (F3 and F4, respectively). In addition, some of the intercross heterozygous (mSuv3+/−) mice showed signs of premature ageing including kyphosis (data not shown). These data suggested that inactivating one allele of mSuv3 leads to reduced longevity over generations.
Figure 2.
Lifespan and tumor spectra of the heterozygous offspring derived from mSuv3+/− intercross mice. (A) Shortened lifespan of mSuv3 heterozygotes at different intercross generations. (B) Tumor incidence and spectra of mSuv3+/− intercross mice. (C) H&E histology of mSuv3+/− mouse tumors: lung adenocarcinoma (a), high-grade lymphoma (b), squamous cell carcinoma (c), pituitary adenoma (d), metastasis – hepatocyte carcinoma (e) and papillary adenocarcinoma (f). (D) Microdissected tumors of mSuv3+/− mice. Micrographs showed paraffin-sections of lung adenocarcinoma (a & b) and lung metastatic HCC (c & d) before (a & c) and after (b & d) microdissection. (E) Genotyping of mSuv3 alleles in the tumor samples (wt: 149 bp: KO: 236 bp) (F) Immunostaining with anti-SUV3 antibodies demonstrated tumors derived from mSuv3+/− mice expressing SUV3.
SUV3 acts as a tumor suppressor with haploinsufficiency
Through pathological examinations, we found high incidence of solid tumors in mSuv3+/− mice. On autopsy, 90% of the mSuv3+/− intercross mice exhibited a broad spectrum of tumor types, including lymphoma and carcinoma (Fig. 2B & 2C). It was noted that about two thirds of the mSuv3+/− mice succumbed to lymphoma. The mSuv3+/− tumor incidence was substantially higher than the 30% incidence seen in the mSuv3+/+ mice from similar genetic crosses. To determine if the tumors in mSuv3+/− mice acquired a second mutation in the wild type mSuv3 allele, we isolated tumor cells by micro-dissections (Fig. 2D) and analyzed the wildtype mSuv3 locus by PCR. As shown in Fig. 2E, all of tumor samples retained the wild type mSuv3 allele. This was further confirmed by immunostaining with anti-SUV3 antibodies as the majority of tumor samples were SUV3 positive (Fig. 2F). These data combined indicated that loss of 50% of SUV3 protein, by genetic ablation of one mSuv3 allele, is sufficient to predispose the mice for tumorigenesis.
Mitochondrial function is a dominant determinant for mSuv3 inheritance
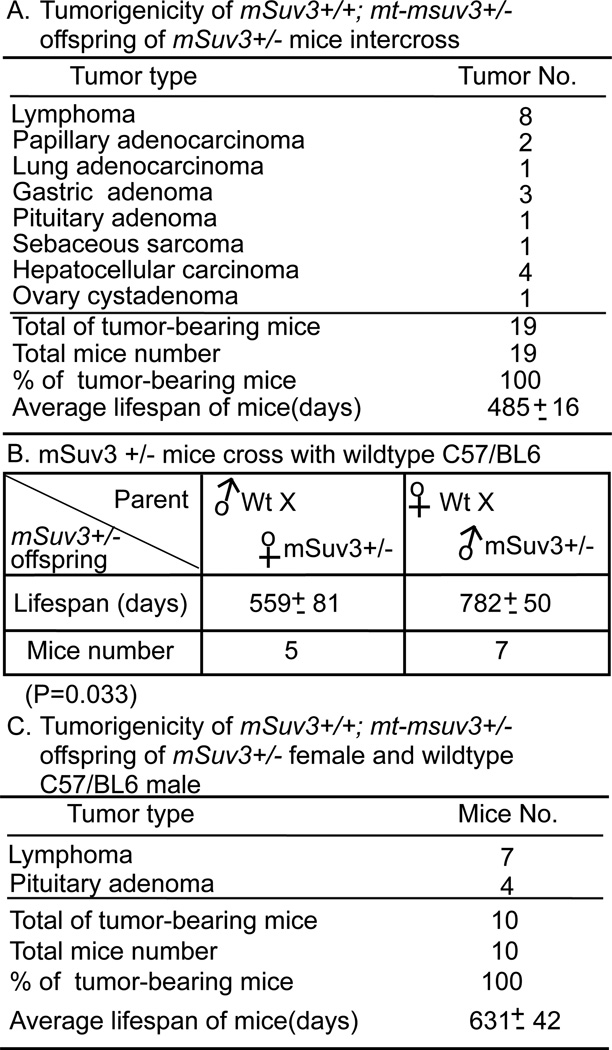
In the course of analyzing the third generation intercrossed mSuv3+/− mice, we found that even the mSuv3+/+, the mice carrying two wild type mSuv3 alleles, showed shortened lifespan and were prone to tumors at a comparable level to that of their mSuv3+/− littermates (Fig. 3A and Fig. 2B). This observation suggested that the reduced lifespan and increased tumor incidence were not simple consequences of the nuclear DNA genotype. Since SUV3 has been shown to be important in maintaining mitochondrial homeostasis (14), we investigated if a maternal factor might be contributing to the progressively declined lifespan and increased tumor incidence in mSuv3+/+ mice of F3. Accordingly, we crossed F3 mSuv3+/− males with C57/BL6 wildtype females. As shown in Fig. 3B, the average lifespan of the heterozygous offspring with wild type mitochondria inherited from their mothers was 782 days, which was comparable to that of the first generation cohort (801 days). In contrast, when F3 mSuv3+/− females were crossed with C57/BL6 males, the average lifespan of the their heterozygotes offsprings carrying mitochondria from their mSuv3+/− mothers was 559 days, simlar to the lifespan of the F3 mice (Fig. 3B). Moreover, the mSuv3+/+ wild type offsprings generated from crossing F3 mSuv3+/− females with C57/BL6 males also had reduced average lifespan of 631 days (Fig. 3C), which was significantly shorter than that of the heterozygous mice (mSuv3+/−) with mitochondria derived from wild type female (782 days), but not statistically different from the average lifespan (560 days) of offspring from mSuv3+/− females (P=0.37) (Fig. 3B). Autopsies performed on mSuv3+/+ offsprings derived from mSuv3+/− female by C57/BL6 male crosses revealed presence of tumor nodules, predominately lymphoma and pituitary adenoma, with nearly complete penetrance (Fig. 3C). Apparently, the status of mitochondria on the maternal side determines the outcome of the lifespan and tumor incidence in their offsprings, rather than their own nDNA genotype in terms of mSuv3.
Figure 3.
Mitochondrial function is a dominant determinant for mSuv3 inheritance. (A) Tumor incidence and spectra of mSuv3+/+; mt-msuv3+/− offspring of mSuv3 heterozygote intercross. (B) Lifespan of heterozygous offsprings derived from either crossing a mSuv+/− female to a wild type C57/BL6 male or crossing a wild-type female and a mSuv+/− male. (C) Tumor incidence of mSuv3+/+ offspring from the cross of mSuv3 heterozygous female and wild type C57/BL6 male (mSuv3+/+; mt-msuv3+/−).
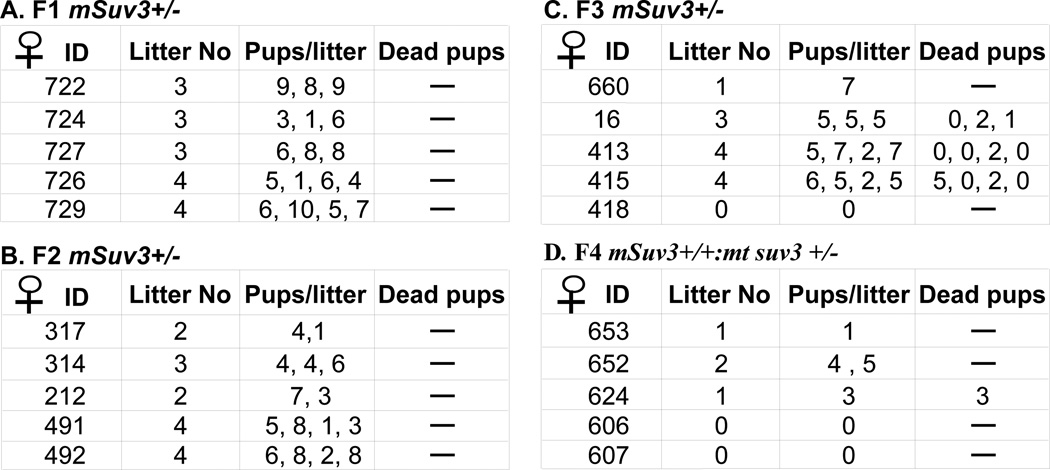
Similarly, the reproductive performance of the mSuv3+/− strain also depends on the status of mitochondria function. The litter size and number of viable pups of the F1 and F2 mSuv3+/− intercrossed mice were comparable to that of the wild type mice (6–8 pups) (Fig. 4A & B). However, the reproductive output of such female declined in F3 (Fig. 4C). Severe reduction in reproductive performance was also found in the mSuv3+/+ F4 female (Fig. 4D). Besides, these mSuv3+/+; mt-msuv3+/− pups that did survive were found to be sterile at 3–5 months of age, further suggesting the dominance of mitochondrial status over nuclear genotype. In the subsequent part of this communication, we designated the mitochondria derived from an mSuv3+/− female as (mt-msuv3+/−) and that from an mSuv3+/+ female as (mt-msuv3+/+).
Figure 4.
The fecundity is significantly reduced in the third generation of mSuv3 heterozygous mice. Designated female mice were housed together with C57/BL6 male since 8 weeks old; the number of pups of each delivery and litter were recorded to determine the fecundity of female mice.
Increased mutation load and reduced mtDNA copy number correlate with high tumor incidence and shortened lifespan
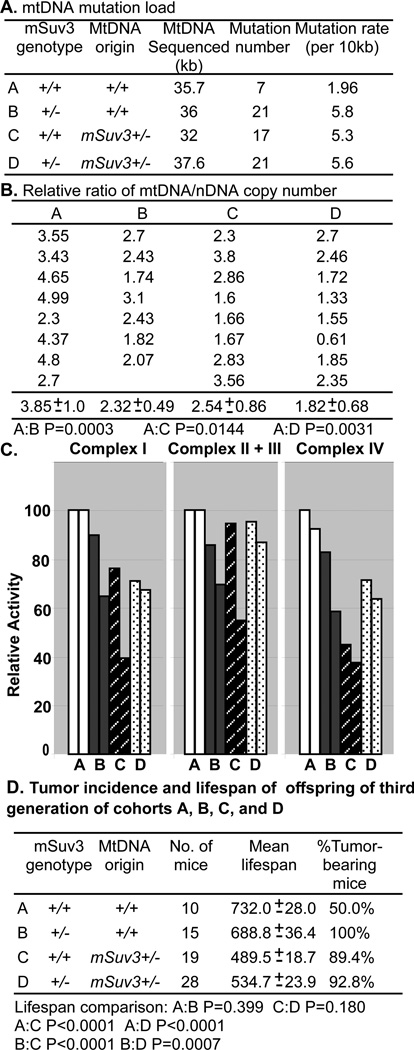
To test whether mtDNA serves as the key factor that determines the maternally transmitted defects, we generated four genotype cohorts of mice by crossing mSuv3+/− male or female mice with C57/BL6 mice: Cohort A: mSuv3+/+; mt-msuv3+/+; Cohort B: mSuv3+/−; mt-msuv3+/+; Cohort C: mSuv3+/+; mt-msuv3+/−; and Cohort D: mSuv3+/−; mt-msuv3+/− (Fig. 5A). Then, mtDNA mutation load and mtDNA copy number of these four cohorts of mice were analyzed. A DNA region surrounding the L-strand replication origin (nt 4418–6409) was sequenced to assess mtDNA mutation load. As shown in Fig 5A, the levels of new mtDNA mutations were increased about 3 folds in Cohorts B, C, and D compared to age-matched Cohort A or C57/BL6 mice. This data suggested that the deficiency of SUV3 increases mtDNA mutations load. Once the mtDNA mutations have been predisposed, they are stable and transmitted through the female germline, which may not be affected by nuclear genotype.
Figure 5.
Defective mitochondria inherited from mSuv3+/− female leads to increased tumorigenicity and reduced lifespan regardless of nuclear genotype. The offspring from the cross of mSuv3+/− female with C57/BL6 male and mSuv3+/− male with C57/BL6 female were designated as four cohorts: Cohort A (mSuv3+/+; mt-msuv3+/+); Cohort B, (mSuv3+/−; mt-msuv3+/+); Cohort C, (mSuv3+/+; mt-msuv3+/−); and Cohort D (mSuv3+/−; mt-msuv3+/−). (A) The region surrounding the mtDNA L-strand origin was sequenced and mtDNA mutation rates were calculated from brain samples collected from ten-month-old mice in each cohort. (B) The mtDNA to nDNA copy number ratio was determined from seven to eight spleens of 15-month-old mice of each cohort by quantitative real-time PCR using mitochondrial 12S rDNA and nuclear 18S rDNA gene primers (Student's t-test was used to assess P value). (C) Enzyme activities of OXPHOS complexes containing mtDNA encoded subunits, complexes I, II +III, and IV, assessed on brain mitochondria of 10-month-old animals from each cohort. (D) Lifespan and tumor incidence of the offspring (the third generation) derived from mSuv3+/− backcross with C57/BL6 mice. (Student's t-test was used to assess P value).
To further characterize the effects of the mSuv3+/− genotype on mtDNA metabolism, we analyzed the mtDNA to nDNA copy number ratio on the spleens of 15-month-old mice, quantified by real time PCR using mtDNA 12S rRNA and nDNA 18S rRNA primers (Fig. 5B). The mtDNA copy number was markedly reduced in Cohorts B, C, and D, relative to Cohort A. Hence, both the mSuv3+/− nuclear genotype and the mSuv3+/− female transmitted mtDNA contributed to the reduction in the mtDNA copy number. Hence, both the nuclear mSuv3 mutation and the maternally inherited defective mitochondria did contribute to the overall reduction in the mtDNA copy number in the mice.
Since these mitochondrial DNA aberrations may reduce mitochondrial activities, we then examined the biochemical activities of mitochondrial OXPHOS complexes I, II+III and IV, each containing one or more mtDNA encoded subunits. Mitochondria isolated from the brains of 10-month-old animals in Cohorts B, C, and D showed significantly reduced OXPHOS complex activities than those of Cohort A (Fig. 5C). Therefore, the increased mtDNA mutation rate and reduced mtDNA copy number indeed deteriorate mitochondrial energy production.
To further validate the contribution of mitochondrial aberration to the aforementioned phenotypes observed in mSuv3+/− mice, we crossed mSuv3+/− with C57/BL6 for two additional generations to obtain sufficient progenies for each cohort. Consistent with our earlier observation, the average lifespan of the third generation of Cohorts C (489 days) and D (534 days) were significantly shorter than that of Cohorts A (732 days) and B (688 days) (P<0.0001) (Fig. 5D). Similar to the observation shown in Fig. 3C, we found that Cohorts B, C, and D had tumor incidences approaching 100%, which were significantly higher than the 50% tumor incidence seen in Cohort A. These data further indicated that the reduced lifespan and increased tumor incidence correlate with the mtDNA aberration, rather than the mSuv3 nDNA genotype, supporting the hypothesis that the defective mtDNA is a key factor to the observed phenotypes.
Mouse embryonic fibroblasts (MEFs) derived from mice with aberrant mtDNA exibit impaired proliferation capability
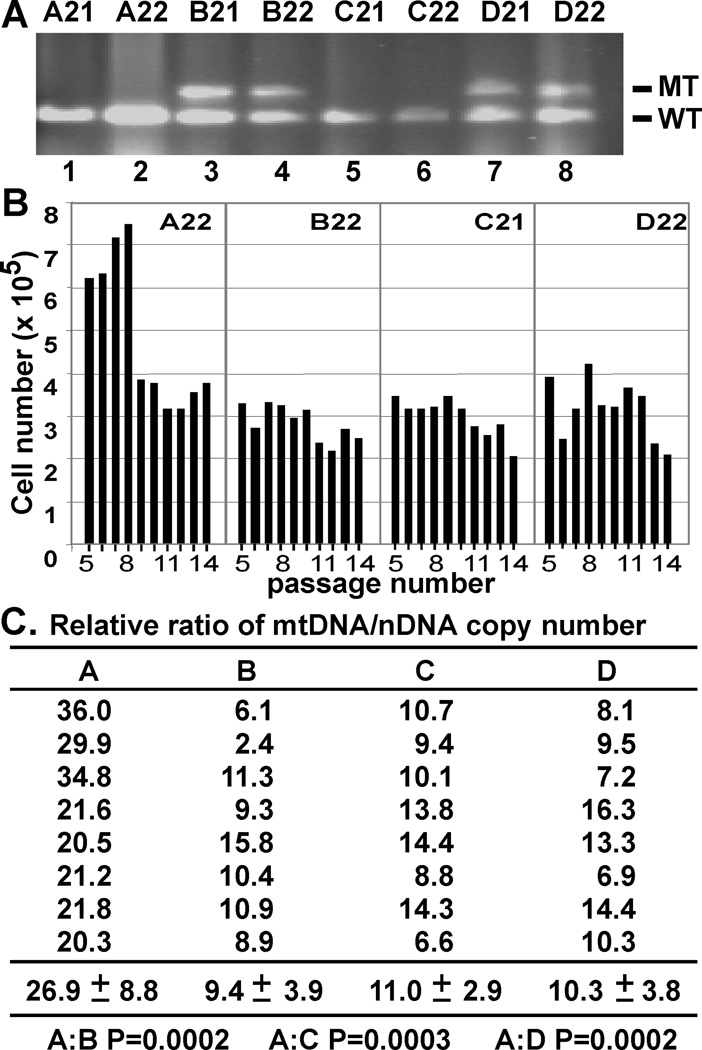
To determine if this mitochondrial dysfunction would impede cellular function, we generated MEFs from the four cohorts (genotyping shown in Fig. 6A) and measured their proliferation capacity following the 3T3 protocol (16). By this protocol, the Cohort A MEF cells doubled in number every three days for eight passages, after which the cells were arrested in crisis state and the three-day cell counts were comparable to the inoculation levels. In contrast, the cells from Cohorts B, C, and D did not have an initial proliferative phase, but immediately showed low proliferation potential (Fig. 6B), suggesting these fibroblasts might already enter senescence when initially explanted. To determine if the reduced proliferative capacity of the Cohorts B, C, and D could be correlated to the loss of mtDNA integrity, we assessed the mtDNA copy numbers of the four cell cohorts at passage four. The mtDNA copy numbers of the MEFs from Cohorts B, C, and D were apparently less than half of that of Cohort A (Fig. 6C). Collectively, our data suggested that the mSuv3+/− nDNA mutation and the maternally transmitted mtDNAs abnormalities leads to impaired MEF proliferation reminiscent of premature cellular senescence.
Figure 6.
Reduction of mtDNA copy number correlates with reduced MEF cell proliferation. (A) Genotyping of mSuv3 MEFs. (wt: 149bp, KO: 236bp): Cohort A (mSuv3+/+:mt-msuv3+/+) = A21 and A22; Cohort B (mSuv3+/−: mt-msuv3+/+) = B21 and B22; Cohort C (mSuv3+/+: mt-msuv3+/−) = C21 and C22; and Cohort D (mSuv3+/−: mt-msuv3+/−) = D21 and D22. (B) Proliferation capacity of MEFs. MEFs prepared from an embryo of each strain were serially passaged following 3T3 protocol. Each plot represents MEFs from a single embryo, with cell numbers on the Y-axis against passage numbers on the X-axis. Each data point was the mean of two calculations. Standard deviations were too small to be visible. (C) Quantification of relative mtDNA copy number of MEFs from each cohorts at forth passage (mean +/− s.e.m; n=8).
Breast tumor cells express low amounts of SUV3 compared to nonmalignant cells
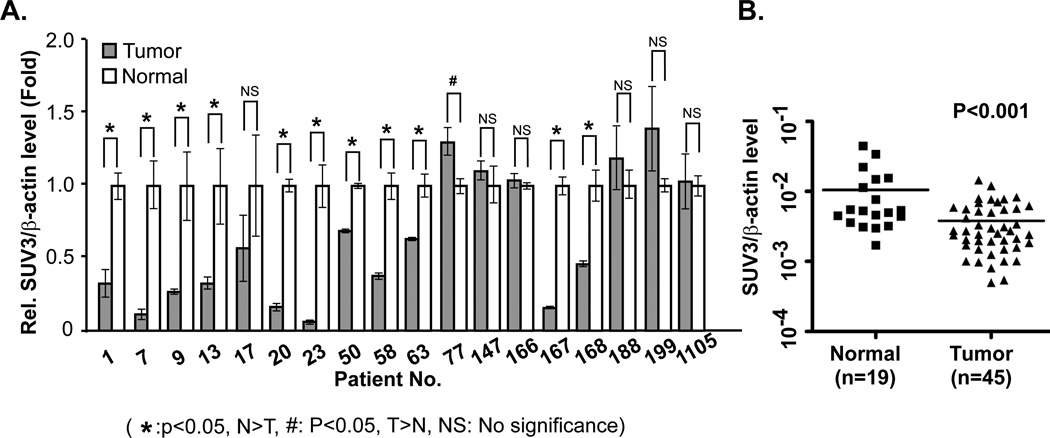
As mentioned above, SUV3 is a highly conserved protein during evolution. Given the substantial stimulation in tumor incidence after inactivation of only one allele of mSuv3 in mice, it is likely that deficiency of SUV3 may also play a similar role in human tumorigenesis. To validate this, we compared the expression of Suv3 mRNA within a cohort of eighteen pairs of breast tumor and adjacent normal tissues. As shown in Fig. 7A, eleven tumor specimens contained significantly reduced Suv3 mRNA. In another cohort with 19 normal tissues and 45 breast tumor specimens, the expression of Suv3 mRNA in tumors was also significantly lower than in normal tissues (Fig. 7B). These data provided us the first clinical evidence to support the notion that reduction of SUV3 expression may participate in human tumorigenesis, which can be resulted from the loss of mtDNA integrity.
Figure 7.
Reduced SUV3 expression in human tumors. (A) The relative expression level of SUV3 in tumor sample was compared to that of the normal adjacent tissue. Relative expression level of SUV3 gene normalized by β-actin. (B) Comparison of the expression level of SUV3 in normal breast tissue and breast tumor (Student's t-test was used to assess P value).
Discussion
In this communication, we demonstrated that inactivating one allele of mSuv3 in mice shortens the lifespan and predisposes the animals to tumor formation with 90% penetrance. Heterozygote offsprings derived from crossing mSuv3+/− male with C57/BL6 female live as long as wild type mice whereas those from cronssing mSuv3+/− female to C57/BL6 male exhibit shortened lifespan. Moreover, shortened lifespan, reduced fecundity, and multiple tumor formations are seen in mSuv3+/+ mice with mitochondria derived from mSuv3+/− mother, suggesting the disease phenotypes follow patterns of maternal mitochondrial inheritance. Findings such as higher mutation rates and reduced mtDNA copy numbers are observed in the offsprings with mitochondria derived from mSuv3+/− mother, suggesting that SUV3 is a novel tumor suppressor that functions through maintainance of the mitochondrial genome integrity.
Genomic instability is a hallmark of human cancer. Many of the tumor suppressor genes identified so far are critical for maintaining genome stability. For example, the prototypic RB gene has been shown to be essential in monitoring chromosome segregation fidelity and cell cycle entrance (17). p53, and BRCA1/2, participate in maintaining genome stability by regulating DNA damage repair response pathways in additional to other functions (18, 19). For this group of tumor suppressor genes, both alleles are inactivated in tumor cells, but reintroduction of one wild type allele can suppress tumor growth by inducing cell cycle arrest or tumor cell apoptosis (20). On the other hand, another group of tumor susceptibility genes, such as Mismatch repair genes (MMR), Werner syndrome gene (WNS) and Bloom’s syndrome gene (BLM), directly participate in DNA repair (21, 22). Defects in MMR genes can lead to mutations in the type II TGF-beta receptor, causing Hereditary Non-polyposis Colorectal Cancer (HNPCC) (23). However, different from the previous group of tumor suppressor genes like RB, reintroducing MMR gene back to HNPCC cells cannot suppress the tumor progression. The other two genes in this group, WNS and BLM, both encode RecQ helicases, and inactivating both copies of the genes results in premature ageing and cancer (22). Interestingly, inactivation of one allele of SUV3 helicase in our mouse model appears to trigger an increase of mtDNA mutations and a reduction in the mtDNA copy numbers, an analogy of mitochondrial genomic instability, leading to increased tumor incidences. These mutated and dysfunctional mitochondria are sufficient to enhance tumor formation even when both copies of wild type mSuv3 were present. The mSuv3 knockout mice described here provides a unique genetic model to study mitochondrial genomic instability in cancer predisposition.
It is noted that the protein level of SUV3 in mouse is critical for maintaining mitochondria genomic stability since the expression from just one copy of wild type allele is insufficient (Fig. 1). This haploinsufficiency principle appears to be the common mechanism seen in many other transgenic mouse tumor models that inactivating both copies of critical genes, such as AML1 (24), p27KIP1 (25), Lkb1 (26), CtIP (27), Smad4 (28), and RINT1 (29), usually leads to cell death and embryonic lethality, while inactivation of one allele prediposes the animal to tumorigenesis. More intriguingly, tumor cells derived from these animals frequently keep the remaining wild type allele, instead of losing it as in the case of RB or p53. If such principle also applies to human cancer, reduction of mtDNA as well as SUV3 protein should be observed in at least some human cancers. Indeed, when we surveyed two cohorts of human breast tumor specimens, the expression of SUV3 mRNA in tumors is also significantly lower than in normal tissues. These results implicate that a reduction of SUV3 expression may be involved in the tumorigenesis in a portion of human cancers. Thus, any genetic or epigenetic event that reduces the expression of SUV3 may contribute to mitochondria genomic instability and promote tumor formation.
How does inactivating one allele of mSuv3 lead to mitochondrial genomic instability? SUV3 is known as a component of the mitochondrial RNA degradosome for decades (10, 12, 13). In yeast, we and others have demonstrated that inactivation of the ySuv3 gene leads to respiratory deficiency and compromised mtDNA integrity (12, 30). In mammalian system, we also observed an accumulation of truncated mitochondrial RNA transcripts and reduction in mtDNA copy number in SUV3 knockdown cells (14). In fact, in addition to its role in RNA degradation (31), mammalian SUV3 has been shown to participate in several DNA processes, including unwinding double stranded DNA substrates (32), and associating with purified mitochondrial nucleoids (33). These potential biochemical activities of SUV3 suggest that SUV3 may be involved in preventing mtDNA mutagenesis and/or facilitating replication. Consistant with this notion, we have demonstrated that SUV3 is associated with actively replicating mitochondrial nucleoids in yeast (30), although a direct biochemical link remains to be demonstrated. Recently, an increase of mtDNA mutations has been observed in mice carrying a mutant POLG, a mtDNA mutator (34, 35, 36). However, the mtDNA copy numbers stayed at the same level as wild type animals (37), suggesting a potentially different mechanism from the role of SUV3 in maintaining mitochondrial genomic stability. It is noted that a small amount of SUV3 localizes to nucleus, where it interacts with BLM helicase (15). However, the precise function of such interaction remains to be established. Although we cannot completely rule out the potential nuclear role of SUV3 impacting on these phenotypes, our backcrossing data (Fig. 3) clearly indicated the maternal effect of SUV3. Regardless the mechanism, mitochondrial dysfunctions resulted from SUV3 insufficiency clearly enhances tumorigenesis, strongly supporting the notion that mitochondrial integrity is important during development and progression of cancers (2, 38). It has been reported that mtDNA mutations are prevalent in colon cancer (39, 40). Therefore, loss of mtDNA integrity may be a general feature for cancer, as the accumulation of mtDNA mutations may increase mitochondrial ROS production, which is a potent mitogen (41, 42). SUV3 as a tumor suppressor operating on mitochondria is conforming with the growing data that both tumor suppressors and oncogenes can target to mitochondrial functions. For example, SIRT3, a newly identified tumor suppressor, is nuclear-encoded but localized to mitochondria (43). In addition, deregulation of metabolic enzymes such as NADP+-dependent isocitrate dehydrogenase enzymes (IDH1 and IDH2) have been linked to human glioma (44, 45). Thus, this new paradigm offers a potential mechanism for a group of human cancers with maternal inheritance otherwise recognized as sporadic cancer.
Experimental procedures and Materials
Construction of mSuv3 targeting vector
The Mus musculus Suv3 (mSuv3) gene was isolated by screening a mouse 129/Sv genomic library (kindly provided by Dr. Tom Doetschman, University of Cincinnati) using a 2.4-kb fragment of human SUV3 cDNA as the probe. A 5-kb BglII fragment of the mouse mSuv3 gene was subcloned into the pBluescript SK vector (Stratagene, CA), yielding a pBg5 plasmid. The internal HindIII site, corresponding to amino acid residue 684 of human SUV3, was restricted and converted to SalI by ligation with a SalI linker. A pgkneopA cassette (46) in antisense orientation was inserted into the engineered SalI site and the resulting plasmid was designated as pmSuv3-neo. The BglII fragment of mSuv3-neo was then subcloned into a p2TK vector (47) to produce the final targeting vector designated as mSuv3-KO.
Selection of ES clones
SalI-linearized mSuv3-KO targeting vector was electroporated into an E14.1 ES cells and selected in medium containing G418 (250µg/ml) and 2’-fluoro-2’-deoxy-1-β-D-arabinofuranosyl-5-iodo-uracil (FIAU) (1µM). DNA extracted from the resistant ES cell clones was digested with BamHI-SalI, and analyzed by Southern blotting. A 0.8-kb BamHI-BglII fragment, designated as probe A, corresponding to a region at 5' end of the genomic sequence in the targeting vector, was used to identify bands of 10-kb and 11.6-kb, that correspond to germ line wild type mSuv3 and the homologous recombinant allele, respectively. An additional DNA probe corresponding to a fragment in the neo gene was also used to identify the neo-containing restriction fragment.
Embryo collection, PCR genotyping, and histology of embryos from mSuv3 heterozygotes intercrosses
Mice heterozygous (F1) for the mSuv3 mutant allele were mated and toes from F2 progeny were cut for genotype analysis. Embryos from timed pregnancies were dissected from maternal decidua. For embryos older than E9.5, the visceral yolk sac was collected and genotyped by PCR (48). For E4.5 to E8.5 embryos, uterine embryos were excised and fixed in 4% paraformaldehyde. After embedded in paraffin, the embryos were sectioned and stained with Mayer’s hematoxylin and eosin (H&E). Genotypings were performed on the microdissected cells from stained slides. E3.5 blastocysts were flushed from maternal uterus and genotyped by PCR. The targeted allele, a 236 bp product within the pgkneopA cassette, was assayed using primers: forward: 5’-TGA TAT TGC TGA AGA GCT TGG CGG C-3’; reverse: 5’-TGG GAG TGG CAC CTT CCA GGG TCA A-3’. The wild type allele, 149 bp product within the targeted exon of the mouse mSuv3 gene, was assessed using the primers: forward: 5’-TCC AAG AAG GCG TGC ACA ACA TCA C-3’; reverse: 5’-ACT TTT GGT GCC ACG TGT CCT TCT C-3’.
Breeding of mSuv3 heterozygous mice
The F1 heterozygous mSuv3+/− mice were derived from crossing two chimeric mice with C57/BL6 mice, generating a hybrid between the 129SvJ and C57/BL6 inbred backgrounds. The F1 mSuv3+/− mice were then interbred to generate mice with two mSuv3 genotypes (mSuv3+/+ and mSuv3+/−). The F2 mSuv3+/− mice were interbred to generate F3 mSuv3 +/+ and mSuv3+/− mice. Similarly, F3 mSuv3+/− mice were interbred to generate F4 mSuv3 +/+ and mSuv3+/− mice. Since the female F3 and F4 mSuv3+/− mice were not very fertile, the mSuv3 knockout allele was maintained by crossing male F4 mSuv3+/− mice to C57/BL6 females. The F1 to F4 mice, which were on a mixed C57/BL6 and 129SvJ backgrounds, were analyzed (Figure 2).
Breeding of mSuv3 heterozygous Mice into C57/BL6 predominant background
The F3 heterozygous mSuv3+/− mice were crossed to C57BL/6 wild type mice, and the progeny were analyzed (Figure 3B). Subsequent crosses for two additional generations were analyzed in Figure 5D.
Fecundity Determination
Female mSuv3+/− or mSuv3+/+ mice were housed together with C57/BL6 male starting at eight weeks of age. The birth date and number of pups of each delivery were recorded. Pups were weaned at 21 days.
Histology and Immunohistochemistry
Tissues were fixed in 4% paraformaldehyde, paraffin embedded, and stained with H&E. Immunostaining was performed using homemade SUV3 specific mouse monoclonal antibody 18C5 and Vectastain Elite ABC kit (Vector Laboratories, CA).
mtDNA Sequence analysis
Genomic DNA including mtDNA was isolated from aged mSuv3+/− mice by the Puregene DNA purification kit (Gentra System Inc, MN). The region surrounding the L-strand origin of DNA replication (OL fragment nt 4418–6409) was PCR-amplified using PfuUltra DNA polymerase (Agilent, CA) and primers: OL forward: 5’-TTT CAT AGG GGC ATG AGG AG-3’; OL reverse: 5’-AAG CAC GAT GTC AAG GGA TG-3’. The PCR products were subcloned into the Zero Blunt PCR cloning vectors (Invitrogen, CA). Forty-eight bacterial clones were isolated for plasmid DNA extraction. Sequencing were performed using primers: 4418F: 5’-TTT CAT AGG GGC ATG AGG AG-3’; 4698F: 5’-ACC ACT AAC AGG ATT CTT ACC-3’; 5262F: 5’-TAT CAC CTT AAG ACC TCT GG-3’; and 5865F:5’-CCC AGC CAT AAC ACA GTA TC-3’.
mtDNA copy number determination
The ratio of mtDNA to nDNA was determined by quantitative real time PCR using the cycling parameters: 95 °C for 5 min, 95 °C for 30s, 58 °C for 45s, for 40 cycles (49, 50). Primer sequences included: mitochondrial 12S rDNA: 12S forward: 5’-ACC GCG GTC ATA CGA TTA AC-3’; 12S reverse: 5’-CCC AGT TTG GGT CTT AGC TG-3’; nuclear 18S rDNA: 18S forward: 5’-CGC GGT TCT ATT TTG TTG GT-3’; 18S reverse: 5’-AGT CGG CAT CGT TTA TGG TC-3’ Determinations of samples and standards were performed in triplicate. The 12S/18S ratios were calculated from the means.
Mitochondrial enzymatic assays
OXPHOS enzyme analysis (51) was performed on isolated mitochondria, which were lyzed by three cycles of freeze-and-thaw in liquid N2 and 37°C water bath, respectively. OXPHOS complexes I, II+III and IV were assayed immediately using spectrophotometric assays. Complex I activity was measured as the rotenone-sensitive NADH oxidation rate at 340 nm. Complex II+III activity was measured as the cytochrome c (cyt c) reduction rate at 550 nm and complex IV activity as the reduced cytochrome c oxidation rate at 550 nm. Citrate synthase activity was used as a standard to normalize for mitochondria amount.
Isolation and culturing of mouse embryonic fibroblasts (MEFs)
Primary MEF cultures were isolated from individual E12.5 embryos obtained from crossing heterozygous male and female mSuv3+/− mice with C57/BL6 mice. The MEFs were expanded for one passage and genotyped by PCR analysis. Complete DMEM with high glucose containing 10% fetal bovine serum, 25 µM/ml sodium pyruvate, 100 units/ml penicillin, and 100 µg of streptomycin was used for culturing MEF cells. For the prolifereation asssays, the MEFs were propagated according to the 3T3 protocol (16), in which the cells were harvested, counted, and 3 × 105 cells were seeded into a new 60-mm dish every three days,. Cell lines were propagated until all MEF cultures entered crisis.
Western blot analysis
Proteins were separated by SDS-PAGE, blotted to Immobilon-P membranes (Milipore, MA) and probed with antibodies against p48 (GeneTex, CA) or SUV3, using a mouse anti-human SUV3 polyclonal antibody generated against the GST-SUV3 fusion protein (amino acids 1–122).
Quantitative real-time PCR assay
Quantitative real-time PCR was performed using Fast SYBR-Green master mix according to the manufacturer's instruction and analyzed on a StepOnePlus Real-Time PCR system (Applied Biosystems, CA). Human SUV3 mRNA levels were calculated according to the relative ΔCt method using β actin mRNA as an internal control. Primers used are: human β actin: ACTB forward: 5’-ATC TGG CAC CAC ACC TTC TAC A-3’; ACTB reverse: 5’-TCA CCG GAG TCC ATC ACG AT-3’; human SUV3: SUV3 forward: 5’-ATC AGG GAG CCA GTC ACG AT-3’; SUV3 reverse: 5’-GCT GGG TGG CTC AGT AGC TT-3’.
Statistical analysis
Except for the quantifications of specific immunoblots, mouse tumorigenesis assay and cell growth assay, all data are presented as means±standard deviation. Student's t-test was used to compare control and treatment groups. Asterisk (*) indicates statistic significance with P-value <0.05.
Acknowledgements
We thank Dennis Wang for his critical reading, Dr. Tom Doetschman for providing 129/Sv mouse genomic library, Andrea Nikitin, Suna Cai, and Weiwei Fan for their excellent assistance. This study was supported by grant NIH AG027877 to WHL and PLC, and NS21328 and AG13154 to DCW. Based on the UCI’s policy, it is declared that WHL serves as a member of Board of Directors of GeneTex, Inc. This arrangement has been reviewed and approved by UCI COI committee.
Footnotes
Contributions
P.L.C. and W.H.L. conceptualized experiments, prepared figures and drafted the manuscript. C.F.C. performed majority of the experiments. Y.C. performed the time pregnancy analysis. X.E.G. performed cell experiments. J.Y.S. and C.K.H analyzed of the SUV3 expression level in human tumor specimens, R.L.R. analyzed histology experiments. D.C.W. provided scientific advice and critical comments on manuscript.
Competing financial interests
The authors declare no competing financial interests.
Reference
- 1.Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 4.Golden TR, Melov S. Mitochondrial DNA mutations, oxidative stress, and aging. Mech. Ageing Dev. 2001;122:1577–1589. doi: 10.1016/s0047-6374(01)00288-3. [DOI] [PubMed] [Google Scholar]
- 5.de Grey AD. The reductive hotspot hypothesis of mammalian aging: membrane metabolism magnifies mutant mitochondrial mischief. Eur J Biochem. 2002;269:2003–2009. doi: 10.1046/j.1432-1033.2002.02868.x. [DOI] [PubMed] [Google Scholar]
- 6.Yui R, Ohno Y, Matsuura ET. Accumulation of deleted mitochondrial DNA in aging Drosophila melanogaster. Genes Genet Syst. 2003;78:245–251. doi: 10.1266/ggs.78.245. [DOI] [PubMed] [Google Scholar]
- 7.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 9.Golik P, Szczepanek T, Bartnik E, Stepien PP, Lazowska J. The S. cerevisiae nuclear gene SUV3 encoding a putative RNA helicase is necessary for the stability of mitochondrial transcripts containing multiple introns. Curr Genet. 1995;28:217–224. doi: 10.1007/BF00309780. [DOI] [PubMed] [Google Scholar]
- 10.Margossian SP, Li H, Zassenhaus HP, Butow RA. The DExH box protein Suv3p is a component of a yeast mitochondrial 3'-to-5' exoribonuclease that suppresses group I intron toxicity. Cell. 1996;84:199–209. doi: 10.1016/s0092-8674(00)80975-7. [DOI] [PubMed] [Google Scholar]
- 11.Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, et al. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- 12.Stepien PP, Margossian SP, Landsman D, Butow RA. The yeast nuclear gene suv3 affecting mitochondrial post-transcriptional processes encodes a putative ATP-dependent RNA helicase. Proc Natl Acad Sci U S A. 1992;89:6813–6817. doi: 10.1073/pnas.89.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stepien PP, Kokot L, Leski T, Bartnik E. The suv3 nuclear gene product is required for the in vivo processing of the yeast mitochondrial 21s rRNA transcripts containing the r1 intron. Curr Genet. 1995;27:234–238. doi: 10.1007/BF00326154. [DOI] [PubMed] [Google Scholar]
- 14.Khidr L, Wu G, Davila A, Procaccio V, Wallace D, Lee WH. Role of SUV3 helicase in maintaining mitochondrial homeostasis in human cells. J Biol Chem. 2008;283:27064–27073. doi: 10.1074/jbc.M802991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira M, Mason P, Szczesny RJ, Maddukuri L, Dziwura S, Jedrzejczak R, et al. Interaction of human SUV3 RNA/DNA helicase with BLM helicase; loss of the SUV3 gene results in mouse embryonic lethality. Mech Ageing Dev. 2007;128:609–617. doi: 10.1016/j.mad.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng L, Lee WH. Retinoblastoma tumor suppressor and genome stability. Adv Cancer Res. 2002;85:13–50. doi: 10.1016/s0065-230x(02)85002-3. [DOI] [PubMed] [Google Scholar]
- 18.Chen PL, Chen Y, Bookstein R, Lee WH. Genetic mechanisms of tumor suppression by the human p53 gene. Science. 1990;250:1576–1580. doi: 10.1126/science.2274789. [DOI] [PubMed] [Google Scholar]
- 19.Ting NS, Lee WH. The DNA double-strand break response pathway: becoming more BRCAish than ever. DNA Repair (Amst) 2004;3:935–944. doi: 10.1016/j.dnarep.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 20.Riley DJ, Lee EY, Lee WH. The retinoblastoma protein: more than a tumor suppressor. Annu Rev Cell Biol. 1994;10:1–29. doi: 10.1146/annurev.cb.10.110194.000245. [DOI] [PubMed] [Google Scholar]
- 21.Rowley PT. Inherited susceptibility to colorectal cancer. Annu Rev Med. 2005;56:539–554. doi: 10.1146/annurev.med.56.061704.135235. (2005) [DOI] [PubMed] [Google Scholar]
- 22.Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 23.Markowitz SD, Wang JY, Myeroff L, Parsons R, Sun L, Lutterbaugh J, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 24.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 25.Fero ML, Randel E, Gurley KE, Roberts JM, Kemp CJ. The murine gene p27Kip1 is haplo-insufficient for tumour suppression. Nature. 1998;396:177–180. doi: 10.1038/24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardeesy N, Sinha M, Hezel AF, Signoretti S, Hathaway NA, Sharpless NE, et al. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 2002;419:162–167. doi: 10.1038/nature01045. [DOI] [PubMed] [Google Scholar]
- 27.Chen PL, Liu F, Cai S, Lin X, Li A, Chen Y, et al. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol. 2005;25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alberici P, Jagmohan-Changur S, De Pater E, Van Der Valk M, Smits R, Hohenstein P, et al. Smad4 haploinsufficiency in mouse models for intestinal cancer. Oncogene. 2006;25:1841–1851. doi: 10.1038/sj.onc.1209226. [DOI] [PubMed] [Google Scholar]
- 29.Lin X, Liu CC, Gao Q, Zhang X, Wu G, Lee WH. RINT-1 serves as a tumor suppressor and maintains Golgi dynamics and centrosome integrity for cell survival. Mol Cell Biol. 2007;27:4905–4916. doi: 10.1128/MCB.02396-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo XE, Chen CF, Wang DD, Modrek AS, Phan VH, Lee WH, et al. Uncoupling the Roles of the SUV3 Helicase in Maintenance of Mitochondrial Genome Stability and RNA Degradation. J Biol Chem. 2011;286:38783–38794. doi: 10.1074/jbc.M111.257956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang DD, Shu Z, Lieser SA, Chen PL, Lee WH. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3'-to-5' directionality. J Biol Chem. 2009;284:20812–20821. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu Z, Vijayakumar S, Chen CF, Chen PL, Lee WH. Purified human SUV3p exhibits multiple-substrate unwinding activity upon conformational change. Biochemistry. 2004;43:4781–4790. doi: 10.1021/bi0356449. [DOI] [PubMed] [Google Scholar]
- 33.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 34.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 35.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, et al. Mitochondrial DNA Mutations, Oxidative Stress, and Apoptosis in Mammalian Aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 36.Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, et al. Mitochondrial point mutations do not limit the natural lifespan of mice. Nature Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- 37.Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, et al. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci U S A. 2005;102:17993–17998. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zanssen S, Schon EA. Mitochondrial DNA Mutations in Cancer. PLoS Med. 2005;2:e401. doi: 10.1371/journal.pmed.0020401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nature Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 40.He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burdon RH. Superoxide and hydrogen peroxide in relation to mammalian cell proliferation. Free Rad Biol Med. 1995;18:775–794. doi: 10.1016/0891-5849(94)00198-s. [DOI] [PubMed] [Google Scholar]
- 42.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of Éø-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;18:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 47.Lee EY-HP, Chang C-Y, Hu N, Wang Y-CJ, Lai C-C, Herrup K, et al. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 48.Liu CY, Flesken-Nikitin A, Li S, Zeng Y, Lee W-H. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 1996;10:1835–1843. doi: 10.1101/gad.10.14.1835. [DOI] [PubMed] [Google Scholar]
- 49.Rodríguez-Santiago B, Casademont J, Nunes V. Is mitochondrial DNA depletion involved in Alzheimer's disease? Eur J Hum Genet. 2001;9:279–285. doi: 10.1038/sj.ejhg.5200629. [DOI] [PubMed] [Google Scholar]
- 50.Coskun PE, Beal MF, Wallace DC. Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101:10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]