Summary
The domestication of animals, plants and microbes fundamentally transformed the lifestyle and demography of the human species [1]. Although the genetic and functional underpinnings of animal and plant domestication are well understood, little is known about microbe domestication [2–6]. We systematically examined genome-wide sequence and functional variation between the domesticated fungus Aspergillus oryzae, whose saccharification abilities humans have harnessed for thousands of years to produce sake, soy sauce and miso from starch-rich grains, and its wild relative A. flavus, a potentially toxigenic plant and animal pathogen [7]. We discovered dramatic changes in the sequence variation and abundance profiles of genes and wholesale primary and secondary metabolic pathways between domesticated and wild relative isolates during growth on rice. Through selection by humans, our data suggest that an atoxigenic lineage of A. flavus gradually evolved into a “cell factory” for enzymes and metabolites involved in the saccharification process. These results suggest that whereas animal and plant domestication was largely driven by Neolithic “genetic tinkering” of developmental pathways, microbe domestication was driven by extensive remodeling of metabolism.
Keywords: primary metabolism, secondary metabolism, saccharification, selective sweep, functional genomics, proteomics
Results and Discussion
Examination of several plants and animals suggests that domestication was driven by genetic changes in diverse developmental pathways that ultimately led to large fruits, naked grains, small brains and big bodies [1, 8, 9]. Although the molecular genetics and phenotypic outcomes of crop and livestock domestication have been extensively studied [8–10], the evolutionary paths traversed by domesticated microbes remain poorly understood [2–6]. In China, evidence for a fermented beverage based on rice mixed with honey and fruit dates back to 7,000 B.C. [11]. Over the millennia that followed, the gradual development of the saccharification process, in which filamentous fungi break down the starch-rich rice to sugars that yeast ferments, morphed the beverage into the high-alcohol rice wine known as sake [11–15]. The filamentous fungus used in saccharification for making sake, as well as other traditional Japanese products such as soy sauce and miso, is Aspergillus oryzae (class Eurotiomycetes, phylum Ascomycota). For sake making, A. oryzae spores (koji-kin) are first spread onto steamed rice. After a ~ 2-day growth period, the resulting A. oryzae-rice (koji) is mixed with additional steamed rice and water and fermented by Saccharomyces cerevisiae, such that the breakdown of the rice starch by A. oryzae occurs in parallel with the conversion of sugars to alcohol by S. cerevisiae [16]. However, the saccharific and more generally proteolytic and metabolic, activities of A. oryzae do not only fuel the yeast, but they also contribute metabolites that influence the flavor and aroma of sake [16].
A. oryzae is closely related to the wild species A. flavus [17, 18], the two species sharing 99.5% genome-wide nucleotide similarity [19]. However, A. oryzae is an atoxigenic domesticate recognized by the U.S. Department of Agriculture as a Generally Regarded As Safe (GRAS) organism [7], whereas A. flavus is a destructive agricultural pest of several seed crops and producer of the potent natural carcinogen aflatoxin [20]. This striking contrast between genomic and phenotypic variation makes the A. oryzae – A. flavus lineage an excellent microbe domestication model for the study of the functional changes associated with microbe domestication and the impact of the process on genome variation [6, 7, 21, 22].
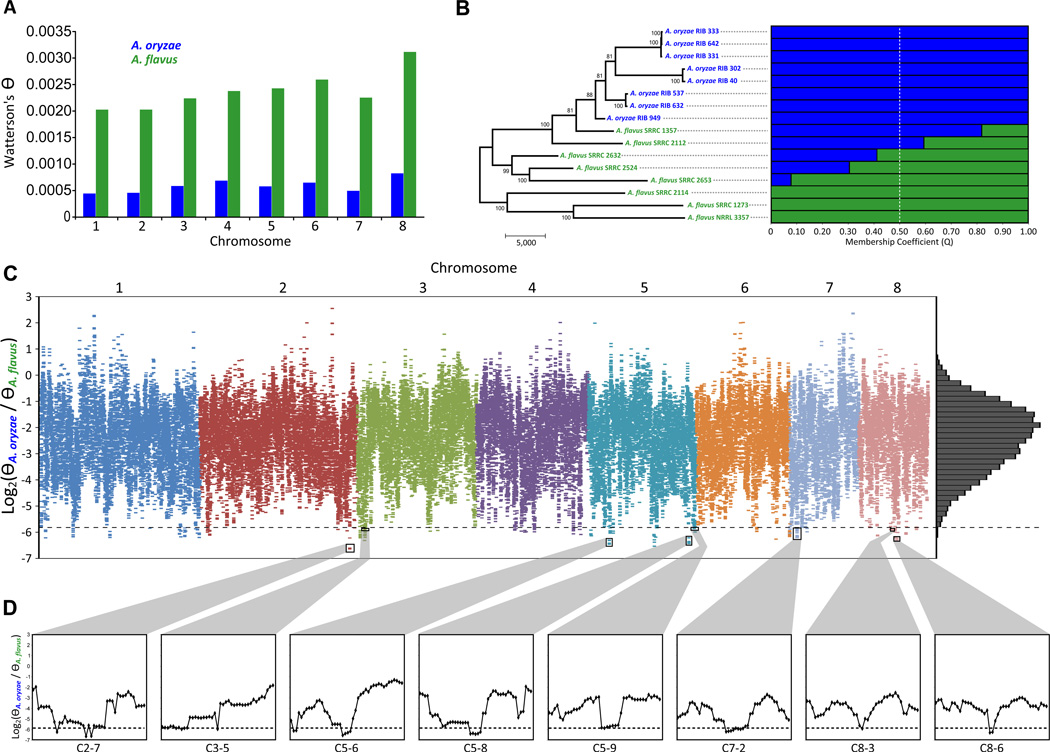
Domesticated organisms have typically been selected for beneficial traits conferred by certain genetic loci and have undergone several rounds of population bottlenecks. Although we previously did not find evidence that the A. oryzae genome exhibited a relaxation of selective constraints, a common characteristic accompanying plant an animal domestication [6], whether the A. oryzae genome has experienced positive selection during the domestication process remains an open question. To address this question, we Illumina sequenced 14 geographically and industrially diverse isolates from A. oryzae and A. flavus and jointly analyzed them with the two species’ reference genomes [7, 23] (A. oryzae RIB 40 and A. flavus NRRL 3357; Tables S1 and S2). Analysis of the genome-wide nucleotide diversity across the 16 isolates showed that the genetic diversity of the A. oryzae isolates is ~25% of that found in the A. flavus isolates (chromosome average nucleotide variation ΘA. oryzae = 0.0006 versus ΘA. flavus = 0.0024; T-test, P = 4.1e-7), consistent with previous gene-level estimates [17, 24, 25] (Figure 1a). Evolutionary analysis of 100,084 high quality SNPs (see Supplemental Methods) suggested that the A. oryzae isolates are monophyletic, in agreement with the previous hypotheses that A. oryzae originated via a single domestication event [17, 25], and do not group by geography or ecology (Figure 1b). Interestingly, two A. flavus isolates (SRRC 1357 and SRRC 2112) show closer affinity to A. oryzae than to other A. flavus isolates (Figure 1b), suggesting that A. oryzae originated from within A. flavus.
Figure 1. Phylogenetic relationship and genomic patterns of variation in A. oryzae and A. flavus.
(A) Chromosomal levels of nucleotide variation (Θ) in A. oryzae (blue) and A. flavus (green). (B) Left panel: Parsimony-inferred phylogeny of the 16 A. oryzae (blue) and A. flavus (green) isolates from the 100,084 high quality genome-wide variant sites. Values near internodes indicate bootstrap support, generated by 1,000 replicates. The scale bar represents the number of changes. Right panel: structure-based membership coefficient for each isolate (population number K = 2). The A. oryzae and A. flavus genetic backgrounds are shown in blue and green, respectively. (C) Relative nucleotide diversity scores (ΘOF) for 5-kb windows (65,894 windows) with a 500 bp step size scanning the eight chromosomes. Points below the dotted line represent genomic regions below the empirical 0.25% quantile (164 windows) and comprise the candidate Putative Selective Sweep Regions (PSSRs). The right panel shows the distribution of ΘOF scores. (D) Close-ups of representative PSSRs and flanking regions.
One of the footprints of recent selection on the genome is the reduction in variation of regions that are close to the variants under selection [26]. When a beneficial allele is rapidly driven toward fixation, nearby neutral variants are likely to also become fixed as a result of the low rate of recombination between closely linked sites [27]. By estimating the relative genome-wide nucleotide diversity ΘOF = log2 (ΘA. oryzae / ΘA. flavus) we identified 61 putative selective sweep regions (PSSRs) (Figure 1c, d and Table S3; see Supplemental Methods). Examination of PSSR gene content indicates that the main targets of selection were genes and pathways involved in primary metabolism (PM) and secondary metabolism (SM). For example, the 148 PSSR genes were significantly overrepresented for SM (Fisher’s Exact Test (FET), P = 0.0004), whereas five PSSRs contained SM gene clusters, including one for the biosynthesis of the tremorgenic mycotoxin aflatrem (PSSR C5-9; Figure 1c,d) [28]. These results were particularly noteworthy as SM gene families are thought to have expanded and be located in unique genomic regions of the A. oryzae - A. flavus lineage compared to the far more distantly related species A. fumigatus and A. nidulans [7]. Furthermore, several PSSR genes are involved in protein and peptide degradation (genes in PSSRs C2-7 and C5-8) and carbohydrate metabolism (C3-5, C5-6) (Figure 1 c, d). One of the strongest supported PSSRs (C8-6) contained a glutaminase (Figure 1 c, d), which catalyzes the hydrolysis of carbon-nitrogen bonds of L-glutamine to glutamic acid, a widely used food flavor enhancer found at considerable levels in sake [29]. Strikingly, whereas there are six polymorphic sites within the A. oryzae isolates (2 promoter, 4 intron), A. flavus isolates are polymorphic at 86 sites (14 synonymous, 2 nonsynonymous, 18 promoter region and 52 intron) (Figure S1).
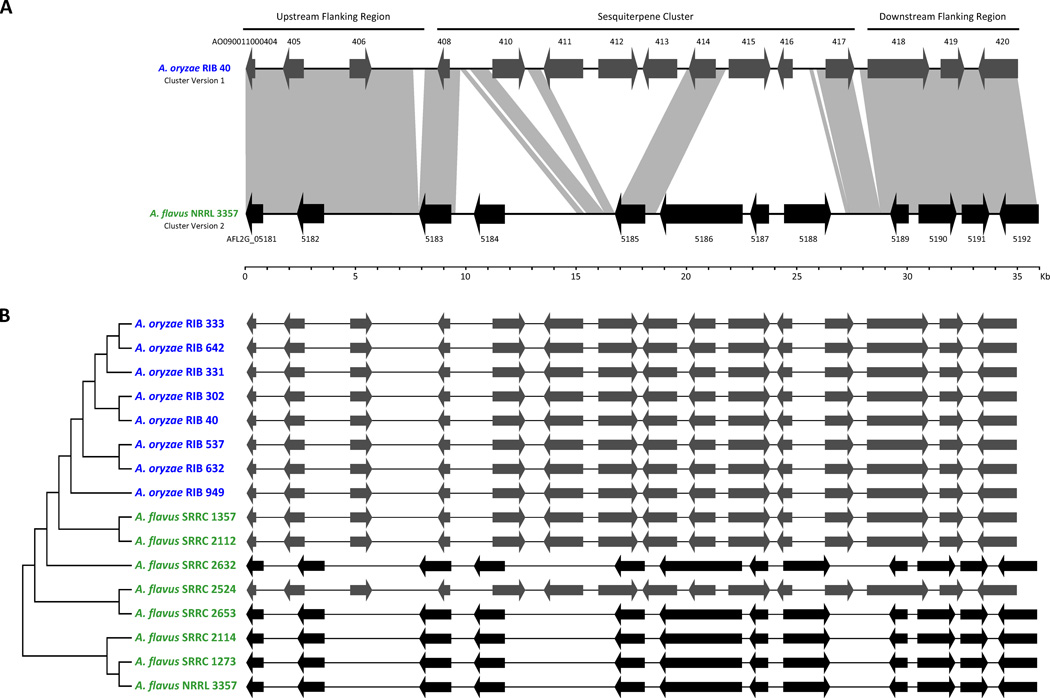
We also examined the isolate genome data to identify differences in genome architecture between the two species (see Supplemental Methods). Although our search identified only five genes shared uniquely by all A. oryzae isolates and none by A. flavus isolates (Table S4), it did also identify a locus that contains a 9-gene cluster in the A. oryzae genome, but contains a 6-gene cluster in the A. flavus NRRL 3357 genome (Figure 2a). Interestingly, the 9-gene cluster is very similar to the sesquiterpene gene cluster in Trichoderma virens [30, 31], whose product belongs to a class of food flavoring aromatic compounds [32], whereas the 6-gene cluster comprises of a terpene cyclase and GAPDH from the 9-gene cluster together with four other unrelated genes (Figure S2). Remarkably, although A. oryzae is fixed for the 9-gene cluster, A. flavus is polymorphic; three isolates contain the 9-gene cluster, while the other five contain the alternative 6-gene cluster (Figure 2b). Furthermore, the genes contained in the two alternative cluster “alleles” at this locus have different evolutionary histories (Figure S2). Most unique genes of the 9-gene cluster group with sequences from A. clavatus and very divergent fungi related to T. virens, consistent with horizontal transfer, whereas most A. flavus unique genes of the alternative cluster group with sequences from A. aculeatus, suggesting a very different history.
Figure 2. The variable genome architecture of the sesquiterpene cluster locus.
(A) Microsynteny of the locus harboring the sesquiterpene encoding gene cluster and its flanking regions in A. oryzae RIB 40 and A. flavus NRRL 3357 isolates. Gray blocks represent genomic regions exhibiting significant sequence similarity between species. Genes, and the direction of transcription, are symbolized by arrows and labeled. The A. oryzae RIB 40 genme contains a 9-gene cluster “allele”, whereas the A. flavus NRRL 3357 genome contains a 6-gene cluster “allele”. Only the terpene cyclase (AO090011000408) and the GAPDH (AO090011000414), as well as a few non-coding regions are homologous between the two “alleles”. (B) A graph showing the allele present in each of the 16 isolates. Note that all eight A. oryzae contain the 9-gene cluster “allele”, whereas A. flavus is polymorphic.
A. oryzae has been grown continually on starch-rich grains, such as rice and soy, for thousands of years [7, 14]. To identify functional differences and putative adaptations to this starch-rich diet, we examined the transcriptome profiles of three phylogenetically distinct isolates of sake-derived A. oryzae, as well as the proteome profiles of the reference isolate of each species, during growth on rice. Similar to the analyses of the PSSR gene content, comparison of the transcriptome and proteome profiles between A. oryzae and A. flavus identified several differentially abundant transcripts, proteins and pathways involved in PM and SM.
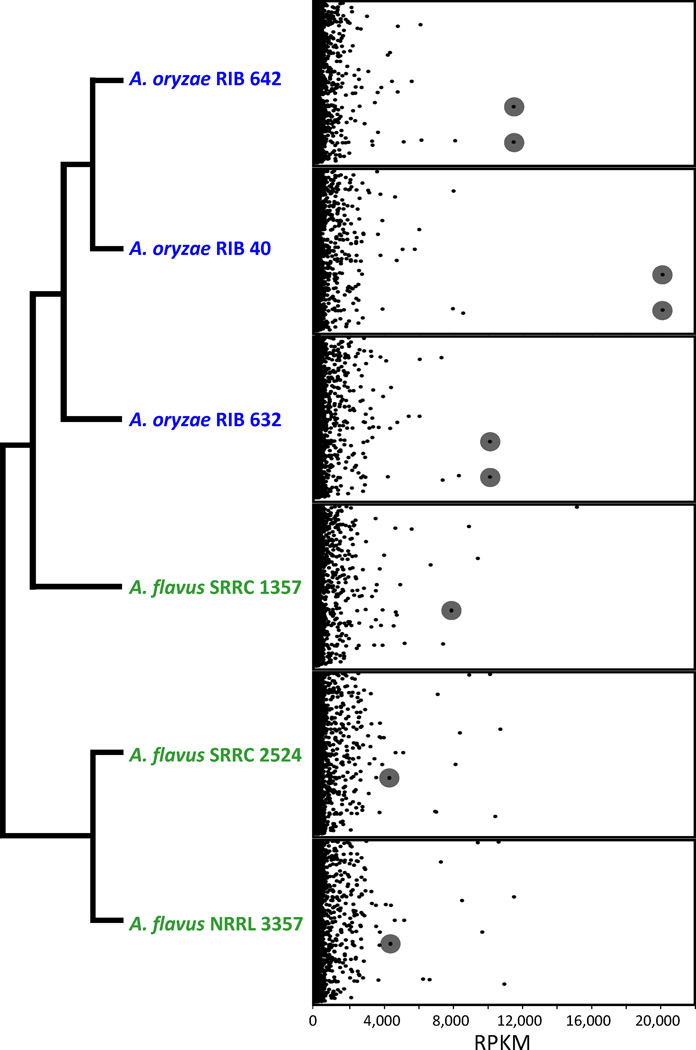
All A. oryzae isolates possess two or three copies of α-amylase [7, 21], the enzyme that hydrolyzes the α-D-glycosidic bonds of starch to produce dextrin, compared to a single copy in A. flavus. We found that the transcript and protein abundance of α-amylase was the highest of any A. oryzae gene or protein and was significantly up-regulated compared to A. flavus (gene expression: FET; P < 1e-300 and protein abundance: >30-fold, FET, P = 2.15e-51) (Figure 3 and Tables S5 to S8). Several other A. oryzae up-regulated genes are involved in carbohydrate PM, including the genome neighbors amylolytic transcriptional activator amyR [33] (FET; P = 1.68e-97) and saccharide metabolizing enzyme maltase glucoamylase (FET; P = 1.79e-17), as well as the glucose metabolizing enzyme sorbitol dehydrogenase (FET; P = 8.22e-252) (Figures S3 and S4, and Tables S6 and S8). Importantly, comparison of the transcriptional profile of the two species showed that both the up-regulated and down-regulated gene sets in A. oryzae were overrepresented for carbohydrate PM (FET; P = 6.24e-5 and P = 4.22e-12, respectively), suggesting that differential regulation of PM is a key functional difference between the two species.
Figure 3. α-amylase is the most highly expressed transcript in A. oryzae.
Expression levels (RPKM) (X-axis) of all genes (Y-axis) for each of the six isolates organized by their phylogenetic relatedness. The two α-amylase paralogs are highlighted in gray. Expression levels for the two paralogs are depicted as equal because they have identical coding sequences and differentiation of their expression levels is not possible.
A. oryzae is also equipped with an arsenal of secreted enzymes that break down the proteins and complex polysaccharides of the grain outer layers, providing access to the starch-rich interior layers [7, 13, 16, 34]. Several protease-encoding genes are located in PSSRs (e.g., the methionine aminopeptidase located in the PSSR C5-8), or are up-regulated (e.g., extracellular cellulase celA), or both (e.g., the up-regulated proteinase located in PSSR C2-7) (Figure 1 c, d, and Tables S3 and S5 to S8). In contrast, 16 of the 27 plant polysaccharide degrading genes were down-regulated (a few of them are also located in PSSRs, e.g., endoglucanase and feruloyl esterase in PSS C3-5 and endo-1,4-β-xylanase in PSS C5-6) (Tables S5, S7, and S8). The broad down-regulation of this subset of genes likely reflects differences between A. oryzae and A. flavus.
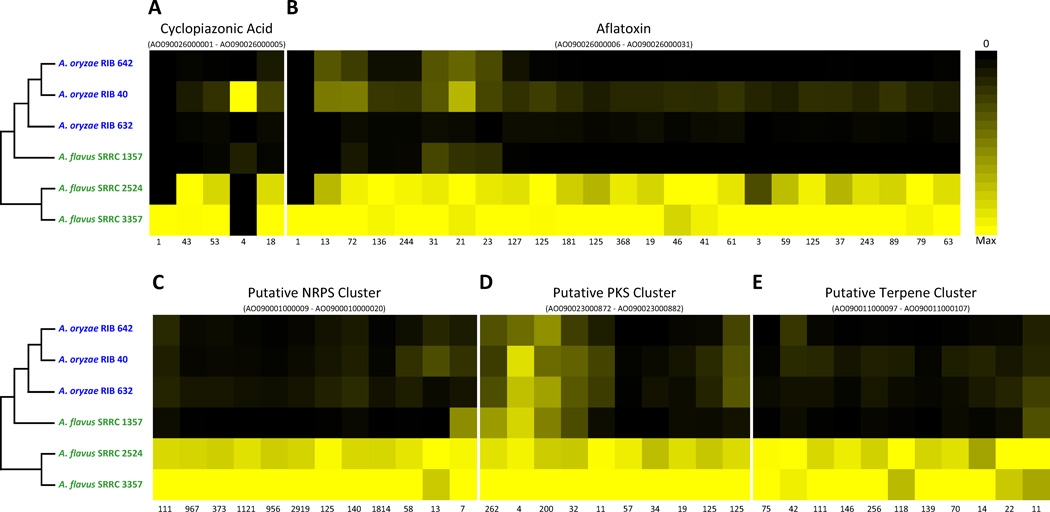
Comparison of the gene expression profiles of 610 genes in all 55 predicted SM gene clusters [35] against background genes in the two species showed that another general characteristic of the A. oryzae transcriptome during growth on rice is SM down-regulation (FET, P = 7.3e-10). This is consistent with the wholesale down-regulation of five SM gene clusters in A. oryzae (Figure 4). Importantly, both the cyclopiazonic acid and the aflatoxin SM pathway in A. oryzae were down-regulated (Figure 4a, b), explaining a key phenotypic difference between A. oryzae and A. flavus, which is the inability of the first to produce either of the two toxins [7, 22, 36]. We further investigated sequence variation in the isolates with expression data with respect to five previously characterized types of mutations observed at the aflatoxin gene cluster locus: (i) transcription binding site mutations in the aflR promoter [37], (ii) a ~250 bp 3’ deletion in the aflT coding region [37], (iii) a frameshift mutation in the norA coding region [37], (iv) multiple nonsynonymous mutations in the verA coding region [37], and (v) ~40 Kb deletion from norB to norA genes [38]. This analysis revealed mutation v in A. oryzae RIB 632 and mutations i – iv in A. oryzae RIB 632 and RIB 40 [37] when compared to A. flavus NRRL 3357. Furthermore, the A. oryzae-like isolate A. flavus SRRC 1357, contained 5 and 13 nonsynonymous mutations in the aflT (ii) and verA (iv) genes respectively, while A. flavus SRRC 2524 was nearly identical to A. flavus NRRL 3357 (3 and 1 synonymous mutations in the norA (iii) and verA (iv) genes). Interestingly, aflatoxin is genotoxic to S. cerevisiae [39], suggesting that the atoxicity of A. oryzae might have been driven by its impact on yeast survival and, as a consequence, fermentation for making sake.
Figure 4. The A. oryzae secondary metabolism transcriptome is widely down-regulated during growth on rice.
Expression levels of five down-regulated secondary metabolism biosynthesis gene clusters for the six isolates for: (A) cyclopiazonic acid, (B) aflatoxin, (C) putative nonribosomal peptide metabolite, (D) putative polyketide synthase metabolite, and (E) putative terpene. The range of genes included in each gene clusters is given under each cluster’s name. For each gene, the color of the heat map cell corresponds to its expression level (in RPKM units), where black is zero expression and yellow is the maximum RPKM for that gene (listed below each gene).
A. flavus natural isolates show substantial variation in SM production and several are known to be atoxigenic [20, 22, 36, 38, 40–42]. Interestingly, the SM expression profile of the atoxigenic A. flavus SRRC 1357, the isolate most closely related to A. oryzae (Figure 1a), was more similar to A. oryzae than to those of the other A. flavus isolates (Figure 4a–e, Figure 4d (C7-2) and Table S6), consistent with the hypothesis that A. oryzae was domesticated from an atoxigenic clade of A. flavus.
During malt rice (koji) production A. oryzae also produces a variety of aromatic, flavor-producing volatile compounds and associated enzymes [16, 43, 44]. In addition to the sequence and genome architecture differences observed in the glutaminase and sesquiterpene loci, we also detected functional differences in other industrially-associated genes. Two particularly interesting examples of up-regulated genes include a glycosyl transferase (FET; P = 1.75e-237), a member of a broad sugar modifier family involved in the making of many sweeteners [45] (Tables S6 and S8), and an asparaginase (gene expression: FET; P = 1.29e-15 and protein abundance: FET, P = 0.006), an enzyme used commercially to reduce acrylamide levels in starch rich foods, such as rice [46] (Tables S6 to S8). Surprisingly however, of the more than 500 genes annotated as MFS or ABC transporters, only 6 were up-regulated in all A. oryzae isolates when compared to all A. flavus isolates and an additional 6 were up-regulated in the A. oryzae isolates and the closely related A. flavus isolate when compared against all other A. flavus isolates (Figure S3).
In summary, our systematic comparison of sequence, gene expression, and protein abundance variation in the A. oryzae – A. flavus lineage indicates that A. oryzae domestication was accompanied by dramatic changes in primary and secondary metabolism. In a span of a few millennia, unintentional human breeding of predominantly segregating variation present in A. flavus resulted, through the gradual accumulation of small (e.g., Figure 2 c, d and Figure S1) and large scale (e.g., Figures 2 and 3) genetic and functional changes (e.g., Figures 3 and 4), to the evolution of the saccharific and proteolytic A. oryzae “cell factory”. Although alterations in metabolic pathways were also likely targets of selection during both plant and animal domestication [47], the majority of changes was primarily driven by modifications in developmental pathways that affect growth and form. In stark contrast, these and previous [4, 48–53] findings argue that the molecular foundations of microbe domestication largely rested in the restructuring of metabolism.
Experimental Procedures
Please see Supplementary Information for a full description of methods.
Sequencing and proteomics
gDNA and mRNA libraries were prepared as previously described [54, 55] and sequenced on an Illumina GA II. We used the Multi-Dimensional Protein Identification Technology (MudPIT) to examine the proteomic profile of A. oryzae RIB40 and A. flavus NRRL 3357. For mRNA and proteomics samples, isolates were grown on rice at 30°C for 24 hours to mimic sake-making conditions. Raw Illumina sequence reads were submitted to the NCBI Sequence Read Archive (SRA) (Accession Numbers: A. oryzae gDNA: SRA0502658, A. flavus gDNA: SRA052664, A. oryzae RNAseq, SRA0502666 and A. flavus RNAseq: SRA052667). Raw proteomics data was submitted to Tranche and can be downloaded from the Vanderbilt MSRC Bioinformatics Data page (http://www.mc.vanderbilt.edu/root/vumc.php?site=msrc/bioinformatics&doc=21164).
SNP detection and evolutionary analysis
We used the Maq package [56] to identify SNPs for each isolate by mapping genomic reads against the A. oryzae RIB40 reference genome. We required that SNP sites had ≥ 5× coverage, an average quality score ≥ 20 and have no ambiguously called in any isolate. We then extracted the nucleotide from each variants site in all isolates. The alignment of variant sites was used to infer the phylogenetic relationships and population structure of our isolates.
Selective sweep detection
Using Variscan [57], we measured relative nucleotide diversity ΘOF = log2(ΘA. oryzae / ΘA. flavus) in 5 kb windows, with a 500 bp step-size to detect regions of the A. oryzae genome with relatively reduced levels of variation. We considered the lower 0.25% quantile of ΘOF values as putative selective sweep regions (PSSR).
Gene expression and protein abundance analysis
Using the rSeq package [58], mRNA reads were mapped against the A. oryzae RIB40 reference transcriptome and gene expression was quantified in terms of Reads Per Kilobase per Million mapped reads (RPKM) [59]. We identified differentially expressed genes and differentially abundant proteins between the reference strains of each species by comparing the proportion of mapped reads using Fisher’s exact tests. Species-level and clade-level gene expression up-regulation was further identified where all isolates of a group were expressed ≥ 10 RPKM and up-regulated by at least 1.5-fold vs. all isolates of the other group.
Supplementary Material
Highlights.
Aspergillus oryzae was likely domesticated from an atoxigenic A. flavus ancestor
Domestication was driven by wholesale genetic and functional changes in metabolism
Secondary metabolic pathways were targets of selection and down-regulation
α-amylase is the most abundant A. oryzae transcript and protein during rice growth
Acknowledgments
We thank members of Rokas lab, Chris Hittinger, Shannon Beltz, David Geiser, Kathy Friedman, David Friedman, Jim Patton, Julian Hillyer, Kamya Rajaram, Abby Olena, Scott Egan, Jonathan Flowers, David McCauley, Travis Clark, Chelsea Baker, and Dr. Osamu Yamada and the National Research Institute of Brewing of Japan. JGG is funded by the Graduate Program in Biological Sciences at Vanderbilt University and the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH, NIAID: F31AI091343-01). Research in A.R.'s lab is supported by the Searle Scholars Program and the National Science Foundation (DEB-0844968).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 2.Hyma KE, Saerens SM, Verstrepen KJ, Fay JC. Divergence in wine characteristics produced by wild and domesticated strains of Saccharomyces cerevisiae. Fems. Yeast. Res. 2011;11:540–551. doi: 10.1111/j.1567-1364.2011.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legras JL, Merdinoglu D, Cornuet JM, Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- 4.Libkind D, Hittinger CT, Valerio E, Goncalves C, Dover J, Johnston M, Goncalves P, Sampaio JP. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. USA. 2011;108:14539–14544. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rokas A. The effect of domestication on the fungal proteome. Trends Genet. 2009;25:60–63. doi: 10.1016/j.tig.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 8.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- 10.Andersson L, Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nature Rev. Genet. 2004;5:202–212. doi: 10.1038/nrg1294. [DOI] [PubMed] [Google Scholar]
- 11.McGovern PE, Zhang JH, Tang JG, Zhang ZQ, Hall GR, Moreau RA, Nunez A, Butrym ED, Richards MP, Wang CS, et al. Fermented beverages of pre- and proto-historic China. Proc. Natl. Acad. Sci. USA. 2004;101:17593–17598. doi: 10.1073/pnas.0407921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abe K, Gomi K, Hasegawa F, Machida M. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia. 2006;162:143–153. doi: 10.1007/s11046-006-0049-2. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T, Abe K, Asai K, Gomi K, Juvvadi PR, Kato M, Kitamoto K, Takeuchi M, Machida M. Genomics of Aspergillus oryzae. Biosci. Biotech. Bioch. 2007;71:646–670. doi: 10.1271/bbb.60550. [DOI] [PubMed] [Google Scholar]
- 14.Machida M, Yamada O, Gomi K. Genomics of Aspergillus oryzae: Learning from the History of Koji Mold and Exploration of Its Future. DNA Res. 2008;15:173–183. doi: 10.1093/dnares/dsn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teramoto Y, Hano T, Ueda S. Production and characteristics of an ancient form of sake made with shitogi. J.I. Brewing. 2000;106:95–99. [Google Scholar]
- 16.Yoshizawa K. Sake: Production and flavor. Food Rev. Int. 1999;15:83–107. [Google Scholar]
- 17.Geiser DM, Pitt JI, Taylor JW. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc. Natl. Acad. Sci. USA. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurtzman CP, Smiley MJ, Robnett CJ, Wicklow DT. DNA relatedness among wild and domesticated species in the Aspergillus flavus Group. Mycologia. 1986;78:955–959. [Google Scholar]
- 19.Rokas A, Payne G, Fedorova ND, Baker SE, Machida M, Yu J, Georgianna DR, Dean RA, Bhatnagar D, Cleveland TE, et al. What can comparative genomics tell us about species concepts in the genus Aspergillus? Stud. Mycol. 2007;59:11–17. doi: 10.3114/sim.2007.59.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami H, Takase S, Ishii T. Non-productivity of aflatoxin by Japanese industrial strains of Aspergillus. J. Gen. Appl. Microbiol. 1967;13 323-&. [Google Scholar]
- 21.Hunter AJ, Jin B, Kelly JM. Independent duplications of alpha-amylase in different strains of Aspergillus oryzae. Fungal Genet. Biol. 2011;48:438–444. doi: 10.1016/j.fgb.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Kato N, Tokuoka M, Shinohara Y, Kawatani M, Uramoto M, Seshime Y, Fujii I, Kitamoto K, Takahashi T, Takahashi S, et al. Genetic safeguard against mycotoxin cyclopiazonic acid production in Aspergillus oryzae. Chembiochem. 2011;12:1376–1382. doi: 10.1002/cbic.201000672. [DOI] [PubMed] [Google Scholar]
- 23.Payne GA, Nierman WC, Wortman JR, Pritchard BL, Brown D, Dean RA, Bhatnagar D, Cleveland TE, Machida M, Yu J. Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 2006;44:S9–S11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- 24.Chang PK, Ehrlich KC. What does genetic diversity of Aspergillus flavus tell us about Aspergillus oryzae? Int. J. Food Microbiol. 2010;138:189–199. doi: 10.1016/j.ijfoodmicro.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 25.Geiser DM, Dorner JW, Horn BW, Taylor JW. The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae. Fungal Genet. Biol. 2000;31:169–179. doi: 10.1006/fgbi.2000.1215. [DOI] [PubMed] [Google Scholar]
- 26.Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 27.Smith JM, Haigh J. Hitch-hiking effect of a favorable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 28.Nicholson MJ, Koulman A, Monahan BJ, Pritchard BL, Payne GA, Scott B. Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl. Environ. Microbiol. 2009;75:7469–7481. doi: 10.1128/AEM.02146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujisawa K, Yoshino M. Formation of inosinic acid as the taste compound in the fermentation of Japanese sake. Volume 40. Elsevier; 1998. pp. 227–231. [Google Scholar]
- 30.Mukherjee M, Horwitz BA, Sherkhane PD, Hadar R, Mukherjee PK. A secondary metabolite biosynthesis cluster in Trichoderma virens: evidence from analysis of genes underexpressed in a mutant defective in morphogenesis and antibiotic production. Curr. Genet. 2006;50:193–202. doi: 10.1007/s00294-006-0075-0. [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee PK, Horwitz BA, Kenerley CM. Secondary metabolism in Trichoderma - a genomic perspective. Microbiology. 2011;158:35–45. doi: 10.1099/mic.0.053629-0. [DOI] [PubMed] [Google Scholar]
- 32.Janssens L, Depooter HL, Schamp NM, Vandamme EJ. Production of flavors by microorganisms. Process Biochem. 1992;27:195–215. [Google Scholar]
- 33.Gomi K, Akeno T, Minetoki T, Ozeki K, Kumagai C, Okazaki N, Iimura Y. Molecular cloning and characterization of a transcriptional activator gene, amyR involved in the amylolytic gene expression in Aspergillus oryzae. Biosci. Biotech. Bioch. 2000;64:816–827. doi: 10.1271/bbb.64.816. [DOI] [PubMed] [Google Scholar]
- 34.Kitamoto K. Molecular biology of the Koji molds. Adv. Appl. Microbiol. 2002;51:129–153. doi: 10.1016/s0065-2164(02)51004-2. [DOI] [PubMed] [Google Scholar]
- 35.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rank C, Klejnstrup M, Petersen L, Kildgaard S, Frisvad J, Gotfredsen C, Larsen T. Comparative chemistry of Aspergillus oryzae (RIB40) and A. flavus (NRRL 3357) Metabolites. 2012;2:36–56. doi: 10.3390/metabo2010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tominaga M, Lee YH, Hayashi R, Suzuki Y, Yamada O, Sakamoto K, Gotoh K, Akita O. Molecular analysis of an inactive aflatoxin biosynthesis gene cluster in Aspergillus oryzae RIB strains. Appl. Environ. Microbiol. 2006;72:484–490. doi: 10.1128/AEM.72.1.484-490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang PK, Horn BW, Dorner JW. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet. Biol. 2005;42:914–923. doi: 10.1016/j.fgb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Keller-Seitz MU, Certa U, Sengstag C, Wurgler FE, Sun MZ, Fasullo M. Transcriptional response of yeast to aflatoxin B-1: Recombinational repair involving RAD51 and RAD1. Mol. Biol. Cell. 2004;15:4321–4336. doi: 10.1091/mbc.E04-05-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amaike S, Keller NP. Aspergillus flavus. Annu. Rev. Phytopathol. 2011;49:107–133. doi: 10.1146/annurev-phyto-072910-095221. [DOI] [PubMed] [Google Scholar]
- 41.Georgianna DR, Fedorova ND, Burroughs JL, Dolezal AL, Bok JW, Horowitz-Brown S, Woloshuk CP, Yu JJ, Keller NP, Payne GA. Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol. Plant Pathol. 2010;11:213–226. doi: 10.1111/j.1364-3703.2009.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kusumoto K, Nogata Y, Ohta H. Directed deletions in the aflatoxin biosynthesis gene homolog cluster of Aspergillus oryzae. Curr. Genet. 2000;37:104–111. doi: 10.1007/s002940050016. [DOI] [PubMed] [Google Scholar]
- 43.Ito K, Yoshida K, Ishikawa T, Kobayashi S. Volatile compounds produced by the fungus Aspergillus oryzae in rice koji and their changes during cultivation. J. Ferment. Bioeng. 1990;70:169–172. [Google Scholar]
- 44.Yoshizaki Y, Yamato H, Takamine K, Tamaki H, Ito K, Sameshima Y. Analysis of volatile compounds in Shochu Koji, sake Koji, and steamed rice by gas chromatography-mass spectrometry. J.I. Brewing. 2010;116:49–55. [Google Scholar]
- 45.Scott D. Specialty enzymes and products for the food-industry. Acs Sym. Ser. 1989;389:176–192. [Google Scholar]
- 46.Friedman M. Chemistry, biochemistry, and safety of acrylamide. A reviewJAgr. Food Chem. 2003;51:4504–4526. doi: 10.1021/jf030204+. [DOI] [PubMed] [Google Scholar]
- 47.Aharoni A, Giri AP, Verstappen FWA, Bertea CM, Sevenier R, Sun ZK, Jongsma MA, Schwab W, Bouwmeester HJ. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell. 2004;16:3110–3131. doi: 10.1105/tpc.104.023895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bachmann H, Starrenburg MJC, Molenaar D, Kleerebezem M, Vlieg JETV. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 2012;22:115–124. doi: 10.1101/gr.121285.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelly WJ, Ward LJH, Leahy SC. Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures. Genome Biol. and Evol. 2010;2:729–744. doi: 10.1093/gbe/evq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, et al. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makarova KS, Koonin EV. Evolutionary genomics of lactic acid bacteria. J. Bacteriol. 2007;189:1199–1208. doi: 10.1128/JB.01351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M, Chambers PJ. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. Plos Genet. 2011;7 doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Douglas GL, Klaenhammer TR. Genomic evolution of domesticated microorganisms. Annu. Rev. Food Sci. Technol. 2010;1:397–414. doi: 10.1146/annurev.food.102308.124134. [DOI] [PubMed] [Google Scholar]
- 54.Hittinger CT, Goncalves P, Sampaio JP, Dover J, Johnston M, Rokas A. Remarkably ancient balanced polymorphisms in a multi-locus gene network. Nature. 2010;464:54-U61. doi: 10.1038/nature08791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibbons JG, Janson EM, Hittinger CT, Johnston M, Abbot P, Rokas A. Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol. Biol. Evol. 2009;26:2731–2744. doi: 10.1093/molbev/msp188. [DOI] [PubMed] [Google Scholar]
- 56.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vilella AJ, Blanco-Garcia A, Hutter S, Rozas J. VariScan: Analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics. 2005;21:2791–2793. doi: 10.1093/bioinformatics/bti403. [DOI] [PubMed] [Google Scholar]
- 58.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics. 2009;25:1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.