Abstract
Yin Yang 1 (YY1) is a ubiquitously expressed transcription factor that performs numerous functions including transcriptional regulation, cell growth control, apoptosis, large-scale chromosomal dynamics, and X-chromosome inactivation. YY1 clearly is able to control cell functions, including proliferation, by acting as a transcription factor either to activate or repress specific genes. Based on its ability to regulate cell growth control genes, it has been argued that YY1 can function as an oncogene that initiates oncogenesis. Although this is an attractive hypothesis, no reports indicate that YY1 can acutely transform cells in culture or form tumors within animals when overexpressed. Thus, it remains unclear whether YY1 is a “classic” oncogene. However, YY1 controls many diverse cell functions, and these functions may provide clues to its role in oncogenesis. We propose that in many cases YY1 may function in oncogenesis and disease progression through “indirect” effects by virtue of its role in either recruiting Polycomb group proteins to DNA, regulating mutator protein accumulation, controlling large-scale chromosomal dynamics or genomic integrity. Disruption of these functions may causally initiate cancer or may contribute to disease progression. Targeting YY1 functions provides possible avenues for clinical intervention.
Keywords: transcription, Polycomb, DNA repair, mutagenesis, genome stability
I. INTRODUCTION
Transcription factor Yin Yang (YY) 1 is a ubiquitous and multifunctional zinc-finger transcription factor that mediates multiple diverse functions. YY1 plays roles in a large array of diverse cell functions including transcriptional regulation, cell growth control, apoptosis, large-scale chromosomal dynamics, X-chromosome inactivation, and DNA repair. YY1 can act as a transcriptional activator, repressor, or initiator protein depending upon DNA binding site context or cell type.1–3 It is implicated in lineage differentiation and cell growth control,4–6 as well as in oncogenesis (discussed here) and other diseases such as dystrophic muscle disease.4,7–9 A number of excellent reviews have covered some of the basic functions of YY1 in relation to function in transcription and possible role in disease.2,3,10 In this review, we will briefly cover YY1 structure, gene regulation functions, role in development and chromosomal structure, and role in epigenetic phenomena. We then will assess what we perceive to be potential roles of YY1 in certain malignancies and possible therapeutic interventions targeting YY1.
YY1 originally was purified and cloned based upon its ability to bind to the adeno-associated virus P5 promoter and to interact physically with adenoviral E1A protein. This interaction converts YY1 from a transcriptional repressor into an activator.11 The YY1 complementary DNAwas cloned simultaneously based on its ability to bind to and repress the immunoglobulin kappa 3´ enhancer and termed nuclear factor (NF) E1,12 or to bind to and activate the ribosome protein L32 promoter and named delta.13 Shortly thereafter, YY1 was cloned based on its ability to bind to retroviral long terminal repeat sequences and named upstream control region binding protein.14 The biphasic role of YY1 in transcriptional activation and repression, however, best matched the name YY1, and this name persists.
YY1 is greatly conserved in multiple species (the human and mouse proteins are 96% identical) and has functional orthologs even in insects (flies and honey bees).15 Mammals have 2 additional YY1-related proteins, YY2 and Rex1, that are thought to have originated from YY1 via retrotransposition events.16,17 YY2 is 56% identical to YY1 and shows overlapping DNA binding site specificity.16 YY2 shows a more restricted expression pattern than YY1 and seems to regulate a subset of genes also regulated by YY1. YY1 also shows homology within its DNA binding domain with transcription factor Rex1.17 The retention of YY1 and related proteins YY2 and Rex1 in mammals suggests functional divergence mediated by differences in DNA binding affinities or sites and specific protein interactions that may regulate distinct genes.17
II. YIN YANG 1 STRUCTURE
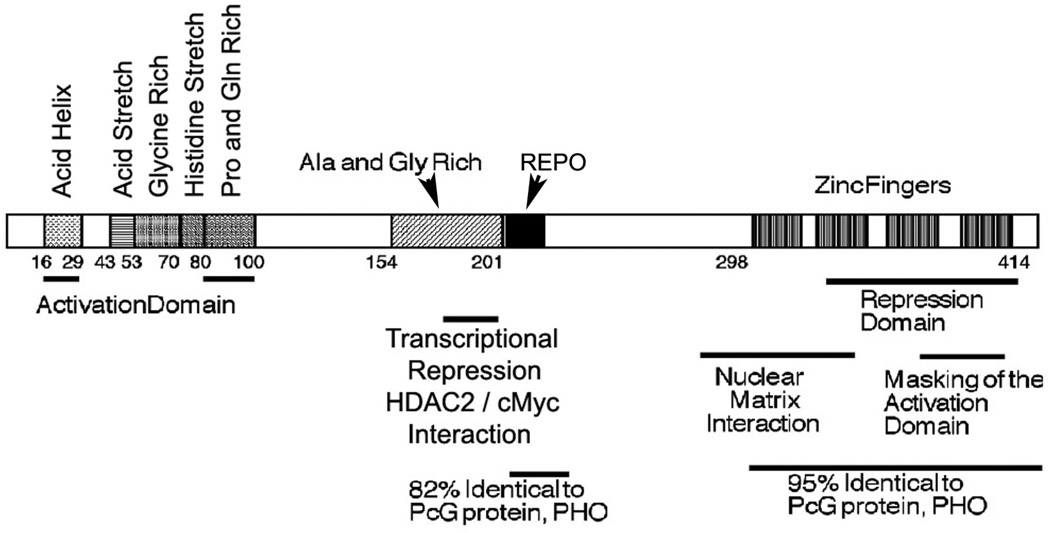
YY1 contains 4 zinc fingers at its carboxyl terminus (amino acids 298–414) and a region rich in alanine and glycine between amino acids 154 and 201. The amino terminal 100 amino acids of YY1 encode several notable features. Sequences 43 to 53 contain 11 consecutive acidic residues whereas amino acids 70 to 80 consist of 11 consecutive histidine residues. These 2 segments are separated by a region rich in glycine (residues 54–69). In addition, sequences 16 to 29 have the potential to form an amphipathic negatively charged helix, and sequences 80 to 100 are rich in proline and glutamine. Function of the histidine and acidic stretches are still unknown, but transcriptional activation function maps to sequences 16 to 29 and 80 to 100.18–21
Sequences near the YY1 carboxyl terminus (333–397) that overlap the zinc finger region, and sequences 170 to 200, have been reported to be involved in transcriptional repression.1,11,18–20,22–24 These sequences are known to interact physically with a variety of transcriptionally important proteins including TATA binding protein, p300, c-Myc, and histone deacetylase (HDAC) 2.2 The YY1 zinc-finger region also is capable of interacting with the nuclear matrix,24,25 and C-terminal sequences seem to interact with and mask the transcriptional activation domain; YY1 transactivation function increases upon their deletion if YY1 is linked to a heterologous DNA binding domain.20
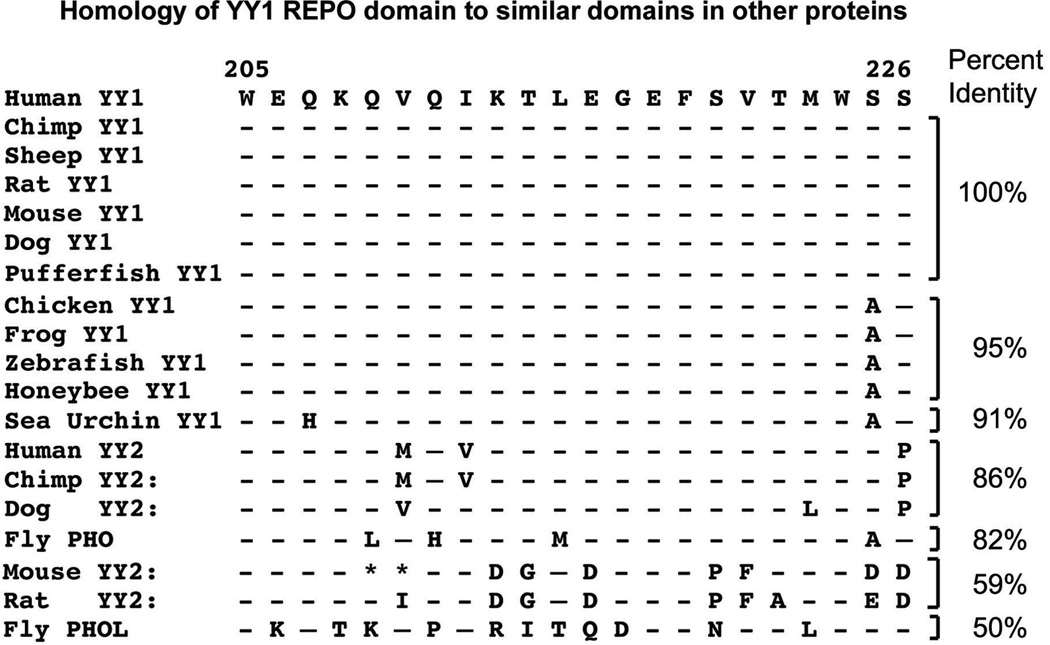
Interesting regions of homology exist between YY1 and its apparent Drosophila ortholog, pleiohomeotic (PHO). PHO is highly homologous to YY1 in 2 regions, which include YY1 sequences 296 to 414 and 205 to 226 (the corresponding segments in PHO are residues 357–475 and 148–169, respectively). Sequences 298 to 414 constitute the 4 YY1 zinc fingers. The homology over this region is extraordinary for organisms as diverse as flies and humans (112 identities out of 118; 95%). Within this segment, zinc fingers 2 and 3 are 100% identical. The 205 to 226 segment is also highly homologous. YY1 sequences 205 to 225 are 82% identical to the corresponding sequences in PHO and this region has been coined the “recruitment of Polycomb” (REPO) domain.15 Other than the zinc-finger region, this is the most highly conserved segment of YY1 homologs in other species (see Fig. 1). This domain will be discussed below with reference to its function in Polycomb group (PcG) protein DNA recruitment. Outside of these regions of high similarity (i.e., the zinc fingers and REPO domain), YY1 and PHO show no discernible similarity. PHO does not contain an obvious transcriptional activation domain and lacks YY1 structural features such as acid and histidine stretches. However, the 2 regions of high similarity between YY1 and PHO, and their similar spatial locations within the proteins, suggest that they might carry out some of the same functions in vertebrates and flies, respectively (discussed later). Notable sequence features and functional domains of YY1 are shown in Fig. 2.
FIGURE 1.
The Yin Yang (YY) 1 recruitment of Polycomb (REPO) domain is highly conserved. Shown is the REPO domain from a variety of species. Dashes indicate identity. Amino acid replacements are shown and deletions are represented by asterisks. Accession numbers are as follows: human YY1 (AAA59926); chimpanzee YY1 (XP_510162); sheep YY1 (AAT74924); rat YY1 (AAR14688); mouse YY1 (AAH55899); dog YY1 (XP_854514); chicken YY1 (NP_510162); frog YY1 (CAA54777); zebra fish YY1 (AAH71351); puffer fish YY1 (CAG01508); sea urchin YY1 (XP_790188); honeybee YY1 (XP-397280); fly pleiohomeotic (AAL48765); fly PHOL (NP_648317); human YY2 (AAS68634); chimpanzee YY2 (XP_529235); dog YY2 (XP_868531); rat YY2 (AAZ38710). The mouse YY2 sequence was taken from Luo et al.159
FIGURE 2.
Map of Yin Yang (YY) 1 functional domains and regions of similarity between YY1 and pleiohomeotic. Specific YY1 amino acid domains are listed above the map. Functional segments are shown below.
III. GENES REGULATED BY YIN YANG 1
Because YY1 was first identified as a transcription factor, initial studies focused primarily on its ability to activate or repress transcription. Numerous genes are either repressed or activated by YY1, and it has been estimated that perhaps 7% of mammalian genes are regulated by this transcription factor.26–28 In addition, some promoter sequences can bind to YY1 where it functions as an initiator protein.29 Thus, YY1 mediates diverse roles in transcription and regulates a surprising array of genes. Genes regulated by YY1 include housekeeping genes and genes involved in cell cycle control, apoptosis, chromatin structure, cytokinesis, development, and differentiation.1,2,10,30 YY1 binding sites are present in numerous promoters and enhancers, indicating its importance for the control of many genes. YY1-regulated genes have been reviewed previously2,3,10,30 and will not be elaborated upon here.
IV. YIN YANG 1 FUNCTION IN DEVELOPMENT
The ubiquitous expression of YY1 and the large number of genes regulated by this protein suggest that YY1 might be crucial for cellular function and for development. Indeed, the Shi laboratory showed that YY1 is crucial for embryonic development because homozygous mutation of the YY1 gene in mice results in peri-implantation lethality.31 Heterozygous knockout mice show growth retardation and some neurological defects, suggesting a YY1 dosage effect.31 In addition, heterozygous mutant mice showed homeotic transformations of axial skeleton in mice,32 consistent with YY1 PcG function (described more fully below). YY1 also is involved in eye development because ablation of YY1 enhances an anterior eye developmental abnormality caused by Ring1 knockout.32
Interestingly, reduction of YY1 levels impairs embryonic growth and viability in a dose-dependent manner.33 The mechanism of this dosage effect is not clear, but there is a tight correlation between YY1 dosage and cell proliferation, with deletion of the YY1 gene resulting in cytokinesis failure and cell cycle arrest.33 Cellular levels of YY1 can determine whether YY1 functions as an activator or a repressor, indicating the YY1 levels in the cell can have a profound effect on function.20
YY1 also is crucial for B-cell development and immune function. B-cell development involves progression from Lin−Sca-1+c-kit+ (LSK) progenitor cells through pro-B, pre-B, immature B, mature B, and plasma cell stages. The early stages of B-cell development can be delineated by the rearrangement status of the immunoglobulin (Ig) heavy and light chain genes. Both heavy and light chain genes are produced during early, antigen-independent B-cell development by a somatic rearrangement process that links together either V, D, and J segments (heavy chain), or V and J segments (light chain), to produce functional Ig genes.34–36 The Ig loci are huge (2.4–3.2 Mb), and for rearrangement of distal variable region genes to occur, the loci must go through a physical contraction process. Although IgH D through J and proximal V to D rearrangements can occur without contraction, the distal V genes require locus contraction and looping for rearrangement.37,38
YY1 has long been believed to play some role in Ig gene regulation and B-cell biology because it associates with multiple Ig enhancer elements including the heavy chain intron and 3´ enhancers, as well as the Ig kappa 3´ enhancer.12,39 The Shi laboratory at Harvard University provided some insight into the role of YY1 in B-cell development by demonstrating that conditional knockout of YY1 in the B-cell lineage (using mb1-CRE recombinase; expressed early after B lineage commitment) results in arrest at the pro-B cell stage.40 Pro-B cells lacking YY1 have normal DH-JH recombination but reduced frequency of VH-DHJH recombination, with the defect being most severe for more distal VH genes.40 These knockout pro-B cells also show a defect in Ig locus contraction. Thus, conditional knockout of YY1 using mb1-CRE results in arrest at the pro-B cell stage, lost Ig locus contraction, and reduced rearrangement of distal V genes.
V. YIN YANG 1 EXPRESSION IN CANCER
Considerable data exist on the relative expression levels of YY1 in various cancers. The majority of this data is based on transcript analyses, although some include protein expression data. Collectively the data are somewhat diverse, with some cancers showing increased YY1 expression and some showing lowered expression. For instance, increased YY1 RNA or protein expression has been observed in cancers of the prostate,41–45 colon,42,46 ovary,42,43 breast,47–49 bone,50,51 liver,42 lung,42 bladder,42,43 cervix,43 skin,43,49 and blood (diffuse large B-cell lymphoma, acute myeloid leukemia, chronic myeloid leukemia, B and T acute lymphoblastic leukemia, Hodgkin lymphoma, Burkitt lymphoma, mantle cell lymphoma, chronic lymphocytic leukemia, and follicular lymphoma).9,43,52,53 However, YY1 expression was found to be reduced in some melanomas, pediatric osteosarcomas, and urothelial carcinomas.43 In addition, conflicting data exist on the survival outcome with relation to YY1 expression. In some cases, high YY1 expression correlated with poor prognosis (prostate, breast, and bone cancers),47,51 whereas in other situations YY1 expression correlated with positive outcomes (ovarian cancer, colon cancer, follicular lymphoma).46,54–56 In the case of ovarian cancer, small interfering RNA knockdown of YY1 resulted in reduced cell proliferation, reduced expression of cell division cycle6, reduced cell motility, and reduced growth in soft agar.55 On the contrary, YY1 knockdown led to resistance to paclitaxel and docetaxel treatments in ovarian cancer cell lines.55
YY1 overexpression correlates with aggressive phenotype in osteosarcoma and this might be partially due to cooperation between Myc and YY1 silencing specific target genes.50,57 To determine whether overexpression of YY1 was involved directly in osteosarcoma, the Napoli group used YY1 small interfering RNA treatment of sarcoma osteogenic 2 cells. YY1 knockdown caused dramatic growth differences including reduced cell growth, lack of growth in soft agar, reduced cell invasion in Matrigel filters (BD Biosciences, Sparks, MD), and reduced formation of new blood vessels.51 On the other hand, YY1 can upregulate an invasion suppressor to inhibit cancer cell invasion.58 Thus the data are complex and the role of YY1 in cancer is unclear.
VI. MECHANISMS OF YIN YANG 1 FUNCTION IN ONCOGENESIS
A. Identification of the Polycomb Group Function of Yin Yang 1
More than a decade ago, the Kassis laboratory cloned a Drosophila protein, PHO. We immediately were intrigued by homologies between the mammalian YY1 protein and the Drosophila PHO59 (Figs. 1 and 2). Girton and Jeon60 demonstrated that PHO is a PcG protein, a family of proteins involved in hematopoietic development, epigenetic chromosomal condensation, stable transcriptional repression, control of cell proliferation, as well as stem cell self-renewal. This raised the exciting possibility that YY1 can function as a vertebrate PcG protein.
PcG proteins generate stable, heritable repression complexes on DNA.61–64 The complexes assemble on Polycomb response element (PRE) sequences and repress transcription of nearby genes. Upon recruitment of complexes to DNA, the histones in the vicinity become deacetylated on H3 lysines 9 and 14 and become methylated on H3 lysines 9 and 27.65–73 It is speculated that these modifications are part of the repression mechanism, but their precise functions are unknown. The molecular details of how PcG complexes are recruited to DNA are uncertain. The complexes bind to PREs apparently via interaction with sequence-specific DNA binding proteins. The current best candidates for these proteins are PHO (in Drosophila) and YY1 (in mammals). Many Drosophila PRE sequences contain PHO/YY1 binding sites, suggesting that this could be a common mechanism for PcG recruitment to DNA.74
Prompted by the possibility that YY1 functions as a PcG protein, we tested this hypothesis using a Drosophila in vivo transcription system as well as a phenotypic correction assay. Our results showed that human YY1 does indeed function as a PcG protein in vivo.75–77 We found that YY1 can repress transcription in a PcG-dependent fashion, can phenotypically correct pho mutant flies, and can recruit PcG proteins to specific DNA sequences.75–77 A number of mammalian DNA sequences have been identified that bind to PcG proteins in vivo.78,79 Some of these DNA sequences also bind to YY1, and knockdown of YY1 expression in mammals can lead to loss of PcG DNA binding and loss of methylation on histone H3 lysine 27.78 The demonstration that YY1 is a mammalian PcG protein with high affinity, sequence-specific DNA binding activity is particularly exciting because PcG proteins are known to contribute to a number of malignancies.80–83 The ability of YY1 to control PcG DNA occupancy in mammals suggests that it could be a potential target for therapies against cancers that rely on PcG function (discussed below).84,85 Thus, it would be advantageous to better understand YY1 PcG function and the YY1 sequences involved in PcG DNA recruitment.
B. The Yin Yang 1 Recruitment of Polycomb Domain
Using a fly transgenic approach, our laboratory identified the YY1 sequences involved in PcG function.15 We found that a 26 amino acid segment (amino acid residues 201–226), when fused to a heterologous GAL4 DNA binding domain, was necessary and sufficient for PcG-dependent transcriptional repression. Amazingly, this small 26 amino acid segment also was necessary and sufficient for recruitment of PcG proteins to DNA.15 Therefore, we named YY1 sequences 201 through 226 the REPO domain for their ability to recruit Polycomb.zxsV Cxxxxxxc A REPO domain YY1 mutant (Δ201–226) can mediate nearly all YY1 functions such as DNA binding, transcriptional activation, transient transcriptional repression, and interaction with histone acetyltransferase (HAT) and HDAC proteins. However, this mutant fails to carry out YY1 PcG functions and fails to recruit PcG proteins to DNA.15
We also identified biochemical interactions that link the YY1 REPO domain with PcG proteins in vivo. Using a yeast 2-hybrid screen approach, we found that the YY1 REPO domain interacts with the PcG protein YY1-associated factor 2 (Yaf2) and can recruit Yaf2 to DNA.86 In turn, Yaf2 can recruit other PcG proteins to DNA, leading to transcriptional repression. As expected, loss of the Drosophila Yaf2 homolog, Drosophila Ring and YY1 binding protein, results in reduced PcG recruitment. Therefore, Yaf2 may provide a bridging function between YY1/PHO and other PcG complex proteins, and this too could be a target for therapy against malignancies caused by PcG dysfunction.
C. Polycomb Group Proteins in Hematopoietic Stem Cell Development
PcG proteins are involved in hematopoietic stem cell self-renewal and YY1 may play a role in this process.80,87 PcG protein Bmi-1 is known to be necessary for hematopoietic stem cell self-renewal and can control cell proliferation.88,89 Similarly, the PcG protein EZH2 can prevent hematopoietic stem cell exhaustion,90 whereas Mel18 negatively regulates hematopoietic stem cell self-renewal.91 Other PcG proteins implicated in hematopoietic development include Mph1/Rae 28 and extraembryonic ectoderm.92–94 Based on the above PcG functions in stem cell biology, we hypothesize that YY1 also may be important for hematopoietic stem cell biology and may be involved in stem cell function.
Polycomb Group D. Proteins in Cancer
PcG proteins have long been associated with cancer biology and stem cell function.80 The PcG protein Bmi-1 originally was identified as a protein that cooperates with c-Myc to promote B-cell and T-cell lymphomas.95 EZH2 and extraembryonic ectoderm are involved in cell proliferation, and EZH2 provides proliferative advantage on cells, with high levels associating with breast cancer aggressiveness.48,96 EZH2 also is overexpressed in hormone-refractory metastatic prostate cancer, leukemia, and breast cancer, and it directly contributes to the control of prostate cell proliferation and malignant behavior.44,48,49,97–99 EZH2 or SUZ12 are overexpressed in a large variety of cancers,83 and the high level of PcG gene expression in prostate, breast, and hepatocellular cancers can be considered a prognostic indicator for these diseases.44,48,100 In some cases, PcG expression can be linked directly to disease phenotype. For instance, overexpression of EZH2 can cause anchorage-independent growth of breast epithelial cells.48 In addition, EZH2 overexpressing cells can cause tumors in mice.101 Similarly, high levels of EZH2 are observed in T-cell lymphoma/leukemia, and patients overexpressing YY1 have a worse prognosis.102 The Oncomine database indicates overexpression of 4 PcGs (EZH2, Suz12, YY1, and RBB7) in brain tumors, and high levels of EZH2 correlate with the severity of the tumor.103 Therefore, levels of PcG expression can be used as prognostic markers for cancer progression.
Although a definitive mechanism is not established for how PcG proteins contribute to oncogenesis, many PcG proteins regulate cell proliferation by negative regulation of the INK4A-ARF locus that encodes p15INK4B, p16INK4A, and p19ARF genes.84,88,104–106 The PcG protein Bmi-1 also can regulate cell senescence88 and can inhibit c-Myc–induced apoptosis.107 In addition, PcG proteins can regulate p53 function, leading to cell cycle control.108,109 It is not clear if the contribution of PcG proteins to cancer is directly due to their PcG repressive function, or if it is due to simple transcriptional repression, which is more transient in nature. However, it is clear in some cases that specific PcG proteins are directly involved in the malignant phenotype. The general assumption is that elevation of PcG proteins results in hyperrepression of a group of target genes involved in cancer, including tumor suppressor genes. Indeed, a group of PcG repressed genes correlates with poor outcome in prostate cancer patients.110 Consistent with a role in cancer mechanism, reducing PcG protein EZH2 levels through RNA interference approaches reduces prostate cancer cell proliferation.44 The role of EZH2 seems to depend upon its histone methyltransferase activity because pharmacologic disruption of PcG methyltransferase activity using 3-Deazaneplanocin A selectively induces apoptosis in breast cancer cells.111 This is quite exciting because specifically targeting PcG function seems to be a viable approach for therapies against cancers dependent on aberrant PcG activity. Thus, inhibition of PcG function represents an important potential target for therapy against cancer.
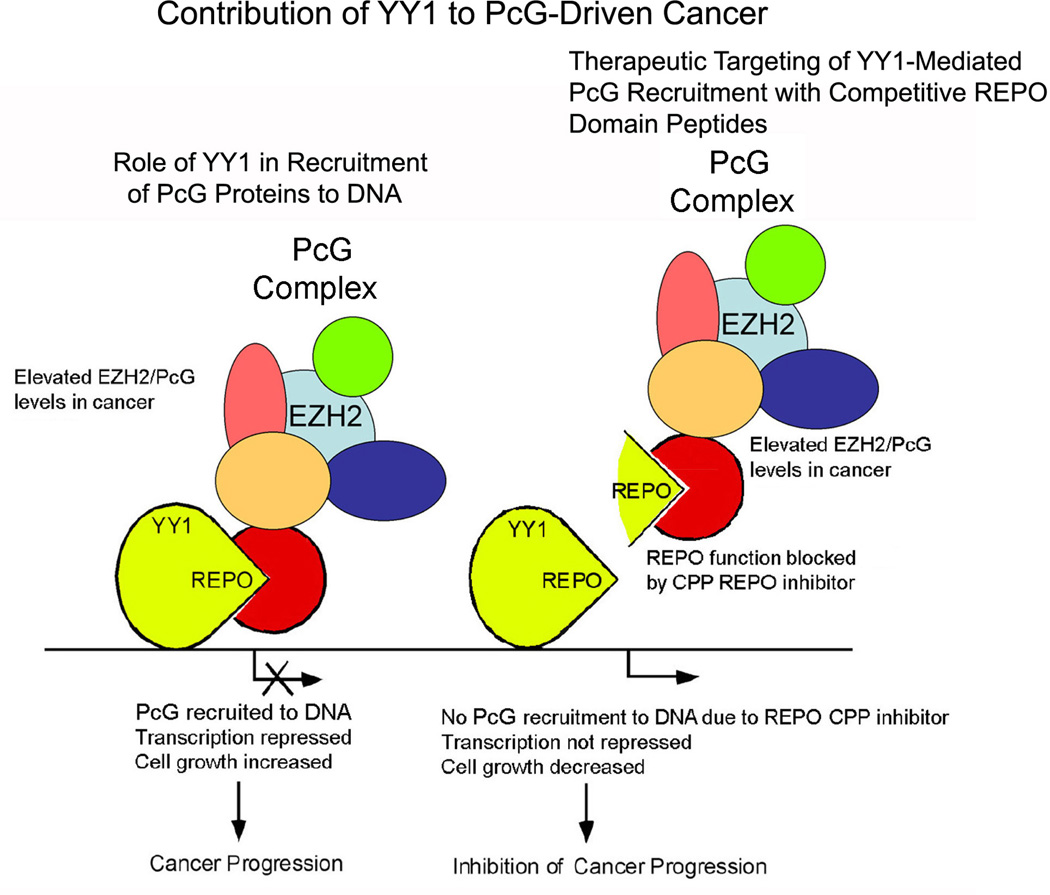
Unfortunately, 3-Deazaneplanocin A is not specific for PcG methyltransferase activity because it inhibits S-adenosylhomocysteine hydrolase, resulting in accumulation of S-adenosylhomocysteine, which causes by-product inhibition of all S-adenosyl-L-methionine–dependent methyltransferases. A more specific inhibitor for PcG function would be advantageous for treatment. Because most PcG proteins do not recognize specific DNA sequences, but instead require recruitment molecules such as YY1, targeting PcG DNA recruitment is a potential therapeutic target. As described above, we showed that the 26 amino acid YY1 REPO domain is necessary and sufficient for the recruitment of PcG complexes to DNA, for methylation of histone H3 on lysine 27, and for PcG-mediated silencing.15 We reasoned that disruption of REPO domain function might prove to be an effective approach to therapy against cancer. In this case, a REPO peptide could be used as a competitive inhibitor of YY1 recruitment of PcG proteins to DNA. We used 26 amino acid small peptide inhibitors (cell-penetrating peptides; cell-penetrating peptide inhibitors) that cross the cell membrane to show that the YY1 REPO domain peptide can indeed inhibit prostate cancer cell growth, as well as growth of an Abelson-transformed pro-B cell line.112 We found that this inhibition is at least in part due to inducing cell death by stimulating apoptosis.112 We propose that the REPO domain peptide functions to inhibit cancer cell growth by competitively inhibiting YY1 interaction with PcG proteins, thus ablating PcG-dependent repression (Fig. 3).
FIGURE 3.
Model of Polycomb group (PcG)–induced oncogenesis and the strategy of using recruitment of Polycomb (REPO) domain peptides as competitive inhibitors to reduce PcG DNA recruitment and to inhibit cancer growth. The left panel shows the Yin Yang (YY) 1 REPO domain recruiting PcG proteins to DNA, resulting in repression of gene expression. The right panel shows a REPO peptide acting as a competitive inhibitor to interfere with the ability of YY1 to recruit PcG proteins to DNA. Loss of PcG recruitment to DNA leads to de-repression of regulatory genes and inhibition of cancer cell growth. Adapted from Basu et al.112
E. Cell Growth Control
YY1 clearly is able to control cell functions, including proliferation, by acting as a transcription factor either to activate or repress specific genes. Based on its ability to regulate cell growth control genes, it has been argued that YY1 likely can function as an oncogene that initiates oncogenesis.3,10 For instance, YY1 can activate c-Myc P1 promoter activity in Burkitt lymphoma113 and can enhance murine double minute 2-mediated p53 inactivation, thus potentiating cellular proliferation and tumorigenesis.114 YY1 can control human immunodeficiency virus gene expression and viral titers, and deletion of YY1 binding sites in human papilloma viruses correlates with increased viral gene expression and the development of cervical cancer.115–123 Thus, YY1 function is related to transcriptional regulation, embryonic development, oncogenesis, viral gene expression, and a growing list of diseases.
On the contrary, in human basal cell carcinoma, YY1 shows repressive activity at the glutathione S-transferase locus and may prevent tumor progression caused by the glutathione S-transferase M3 genotype.124 Indeed, Lichy et al125 found a marked decrease in YY1 binding in malignant HeLa/fibroblast somatic cell hybrids when compared to nontumor cells, whereas Austen and colleagues126 showed that YY1 is a negative regulator of cell growth via potent inhibition of c-Myc transforming activity and possibly functions as a tumor suppressor. These diverse YY1 functions probably result from its ability to interact with numerous proteins and complexes.
A long list of proteins can interact physically with YY1 to control gene expression. It has long been known that YY1 interacts with various HATs and HDACs (reviewed by Thomas and Seto,2 Gordon et al,3 and Sui10). These proteins no doubt contribute to the role of YY1 in activating and repressing transcription, and YY1 itself is a target of acetylation.127 For instance, YY1 interaction with the HAT protein p300 overcomes transcriptional repression,19 and YY1 recruits the HDAC protein mRPD3 to DNA to repress expression.23 YY1 also recruits histone H4 methyltransferase protein arginine N-methyltransferase to the GRP78 gene to activate transcription.128 On the other hand, YY1 recruits histone methyltransferase protein EZH2 to a variety of genes to repress transcription.78,79 YY1 also interacts with proteins in the ubiquitinylation pathway and thus can control protein stability to regulate function.114 YY1 also can bind to the retinoblastoma (Rb) protein to accelerate cell cycle progression to the S phase.6,19 Hypophosphorylated Rb interacts with YY1, and this disrupts YY1-DNA association. The YY1-Rb interaction is observed in resting cells, but not in serum- or bacterial lipopolysaccharide-stimulated cells.6,39 In addition, YY1 interacts with murine double minute 2 and tumor suppressor p53, leading to enhanced p53 ubiquitinylation and subsequent degradation. As discussed above, overexpression of YY1 might therefore predispose cells to a transformed phenotype by inhibiting p53 tumor suppressor activity.
Although it is an attractive hypothesis that YY1 can directly participate in oncogenesis by regulating expression of growth control genes, no reports indicate that YY1 can acutely transform cells in culture (viz. 3T3 transformation assays; growth in soft agar) or form tumors within animals when overexpressed. Thus, it remains unclear whether YY1 is a “classic” oncogene. However, YY1 controls cell functions involved in DNA repair and genomic stability. These functions may provide clues to the role of YY1 in oncogenesis.
F. Role of Yin Yang 1 in Genomic Stability
YY1 can regulate large portions of the genome through imprinting mechanisms. Clusters of YY1 binding sites reside at imprinting control regions.129,130 Knockdown of YY1 results in upregulation of many imprinted genes within the Peg3 region and a resultant loss of DNA methylation.130 YY1 can function as a cofactor with CCCTC binding factor within the imprinting domain of Tsix. Loss of YY1 causes aberrant expression of Tsix as well as Xist.131 Strikingly, YY1 also tethers Xist RNA to the inactive X nucleation center during X-chromosome inactivation.132 How this relates to the role on YY1 in cancer biology is uncertain, but clearly YY1 can regulate large-scale chromosomal structures.
YY1 also physically interacts with the chromatin remodeling complex INO80, related to the yeast switch/sucrose nonfermentable complex.133 As might be expected for a complex involved in chromatin remodeling, this interaction can result in access of YY1 to promoter sequences, leading to changes in gene expression.133 However, this interaction also seems to be crucial for genomic stability because YY1-INO80 interactions seem to control homologous recombination-based repair.134 Knockdown of YY1 leads to genomic instability and aneuploidy, and knockdown of INO80 subunits yields a similar phenotype.134 The role of YY1 in genome and epigenetic functions is particularly intriguing. Mouse embryo fibroblasts isolated from YY1f/f mice are particularly sensitive to chromosomal aberrations after transfection with adenoviral-driven CRE recombinase to delete the endogenous YY1 gene.134 These cells also are highly sensitized to DNA damaging agents, suggesting that YY1 plays an important role in DNA repair and genomic integrity. Mouse embryo fibroblast knockout and knockdown studies indicate that YY1 and INO80 together are involved in homology-directed DNA repair, and YY1 can bind directly to recombination intermediates.134 Therefore, YY1 loss or overexpression may disrupt genomic stability and predispose cells to acquiring mutations that eventually may lead to oncogenesis or to malignant progression. The ability of YY1 to interact with and stimulate Poly (adenosine diphosphate-ribose) function in DNA repair is consistent with this idea.135
Interestingly, we recently observed that YY1 can interact physically with activation-induced cytidine deaminase (AID).136 Both Ig class switch recombination (CSR) and somatic hypermutation (SHM) require AID function,137,138 and AID deficiency leads to loss of CSR and SHM. Both processes require that AID deaminate cytidine to uracil, followed by either mutagenic processing by error prone repair mechanisms (SHM) or double strand breaks leading to rearrangement (CSR).139 AID function must be tightly regulated to avoid deleterious mutagenic activity because, in addition to diversifying the immune response, AID catalyzed cytidine deamination is believed to be involved in generation of lymphomagenic chromosome translocations, and overexpression of AID in transgenic animals leads to T-cell lymphomas and tumors in lung epithelium.140–142 An increasing number of non-Ig genes also have been revealed to be hypermutated by AID in wild-type B cells in which efficient, error-free repair masks most AID mutations.143
AID expression levels directly correlate with the frequency of AID-dependent DNA remodeling events and incidence of c-Myc/IgH translocations.141,144–147 Limiting AID levels in the nucleus protects the B-cell genome from mistargeted mutations and is regulated by multiple mechanisms. Upon stimulation of B cells, AID expression is dramatically upregulated in germinal center B cells.138 However, most AID is retained in the cytoplasm and only a small fraction translocates to the nucleus to mediate CSR and SHM.148–151
Factors that control AID subcellular localization are only now being defined. However, we found that YY1 can interact physically with AID, leading to increased AID nuclear levels. This YY1-AID interaction and increased nuclear AID accumulation can regulate AID function in Ig CSR.136 This suggests that if YY1 is overexpressed in B lymphoid or myeloid (chronic myelogenous leukemia) cancers that also express AID (diffuse large B-cell lymphoma, chronic myelogenous leukemia, acute lymphoblastic leukemia, Burkitt lymphoma, follicular lymphoma, chronic lymphoblastic leukemia), it would lead to increased levels of nuclear AID, increased mutagenic activity, and increased incidence of B-cell lymphomas or augmented disease progression. Indeed, patients with germinal center–derived diffuse large B-cell lymphoma show elevated YY1 expression.52 In addition, AID expression seems to promote B lymphoid blast crisis and drug resistance in chronic myeloid leukemia.152 Thus, high levels of YY1 could elevate nuclear AID levels, leading to accumulation of mutations that could contribute to disease progression from chronic to acute stages.
G. Yin Yang 1 and Fas-Induced Apoptosis
High levels of YY1 seem to contribute to the failure of therapies that induce apoptosis. CH11 is a monoclonal antibody against Fas/apoenzyme 1 that can induce apoptosis in Fas/apoenzyme 1–expressing cells.153,154 Some tumors acquire resistance to CH-11-induced apoptosis apparently because of down-regulation of the Fas gene. The Bonavida group found that YY1 binds to a silencer in the Fas gene to mediate silencing.155 Knockdown of YY1 causes elevated Fas expression, resulting in responsiveness to CH11-induced apoptosis.155 Similar results are obtained by inhibiting YY1 DNA binding using the nitric oxide donor DETANONOate.156 An additional approach is to down-regulate factors that control YY1 expression. Transcription factor NF-κB activates YY1 expression, and reduced levels of NF-κB results in reduced YY1 levels. In addition, the monoclonal antibody rituximab against cell surface protein CD20 can increase Fas expression.156,157 Interestingly, rituximab reduces p38 signaling and constitutive NF-κB activity, leading to reduced YY1 expression and concomitant upregulation of Fas.156,157 This increased Fas expression restores CH11-induced apoptosis in both non-Hodgkin lymphoma cells and prostate cancer cells.156–158 Therefore, rituximab treatment of CH11-resistant cancers can be used to treat B lymphoid cancers that express CD20 on the cell surface. The general strategy of reducing YY1 expression can be used in other cancers via blocking tumor necrosis factor α activity that activates NF-κB and YY1, leading to reduced Fas expression.
VII. CONCLUSION
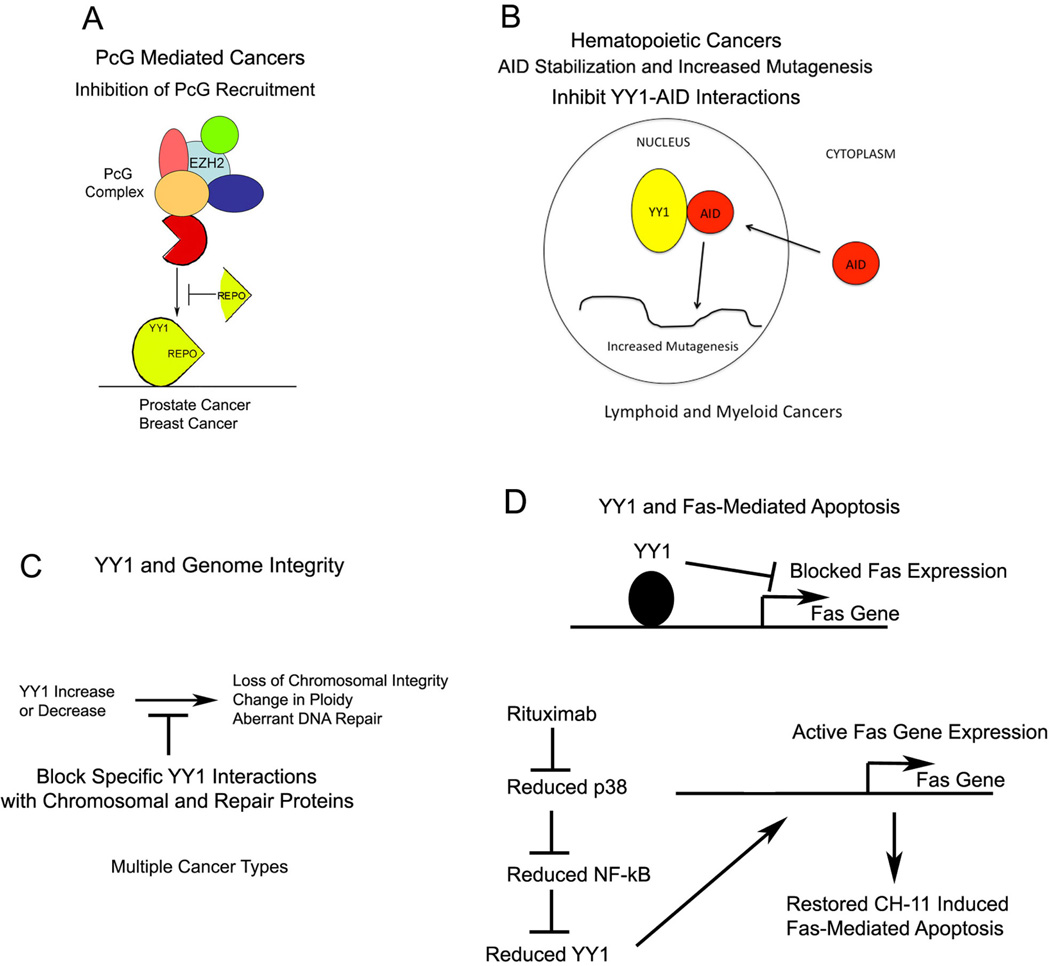
Despite the myriad functions performed by YY1 and its direct role in regulating growth control genes and protein functions, it is still unclear whether it directly acts as an oncoprotein. We propose that many of YY1’s functions in oncogenesis and disease progression are “indirect” effects of its role in recruiting PcG proteins to DNA, regulating mutator protein accumulation (AID), controlling large-scale chromosomal dynamics (such as imprinting and X-inactivation), or controlling genomic integrity (DNA repair) (Fig. 4). Disruption of these functions may causally initiate cancer or may contribute to disease progression. In any event, YY1 provides some possible avenues for clinical intervention:
PcG recruitment. Substantial evidence indicates that PcG proteins are expressed at higher levels in various cancers and are intimately involved in the disease phenotype. Disruption of PcG recruitment to DNA by targeting YY1 REPO domain function is a promising potential therapy (Fig. 4A).
Regulation of mutator proteins. AID is a mutator enzyme expressed during antibody maturation phases of B-cell development. It is now clear that AID can contribute to a number of hematopoietic malignancies, and off target effects of AID can result in gene mutations and chromosomal translocations. Our observation that YY1 can control levels of AID in the nucleus suggest that disruption of AID-YY1 interactions, or reduction of YY1 levels, could be used to reduce AID nuclear accumulation, thus protecting against AID-induced mutations (Fig. 4B). This is a new area of investigation that will require more work to determine effectiveness as a therapy.
Control of genome integrity. It is now clear that reducing YY1 levels results in significant alterations in chromosomal integrity. The ability to maintain diploid status and to repair double stranded DNA breaks is greatly reduced. Modulation of this function of YY1 could be used to sensitize cancer cells to chemotherapeutic agents (Fig. 4C).
Fas-induced apoptosis. Overexpression of YY1 leads to loss of Fas gene expression and loss of CH11-induced apoptosis. On the contrary, reduction in YY1 results in elevated Fas expression and a restoration of CH11-induced apoptosis. Therefore, agents that regulate YY1 expression (such as rituximab) may be used in combination with agents that cause Fas-induced apoptosis (Fig. 4D). Although the precise role of YY1 in oncogenesis is not yet clear, our current knowledge enables us to develop therapies that may be useful in cancer treatment.
FIGURE 4.
Models of Yin Yang (YY) 1 function in cancer development and potential strategies for developing therapeutic agents to control YY1 function.
ACKNOWLEDGMENTS
This material was funded in part by National Institutes of Health grants AI-079002 and GM071830, awarded to Dr. Michael Atchison of the University of Pennsylvania School of Veterinary Medicine.
ABBREVIATIONS
- YY1
Yin Yang 1
- YY2
Yin Yang 2
- PHO
pleiohomeotic
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- PcG
Polycomb Group
- REPO
Recruitment of Polycomb
- Ig
immunoglobulin
- VDJ
variable diversity joining segments
- NF-E1
nuclear factor E1
- NF-κB
nuclear factor kappa B
- UCRBP
upstream control region binding protein
- AID
activation induced cytidine deaminase
- CSR
class switch recombination
- SHM
somatic hypermutation
- MEF
mouse embryo fibroblasts
- CTCF
CCCTC binding factor
- Swi/Snf
Switch/Sucrose nonfermentable
- DLBCL
diffuse large B cell lymphoma
- CML
chronic myelogenous leukemia
- ALL
acute lymphoblastic leukemia
- TNFα
tumor necrosis factor alpha
- Mdm2
mouse double minute 2
- TBP
TATA binding protein
- PRMT1
protein arginine N-methyltransferase
- Rb
retinoblastoma protein
- DZNep
3-Deazaneplanocin A
- GST
glutathione S-transferase
- CDC6
cell division cycle 6
- SaOS2
sarcoma osteogenic 2
- PRE
Polycome response element
- Yaf2
YY1 associated factor 2
- dRYBP
Drosophila Ring and YY1 binding protein
REFERENCES
- 1.Galvin KM, Shi Y. Multiple mechanisms of transcriptional repression by YY1. Mol Cell Biol. 1997;17:3723–3732. doi: 10.1128/mcb.17.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 3.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 4.Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walowitz JL, Bradley ME, Chen S, Lee T. Proteolytic regulation of the zinc finger transcription factor YY1, a repressor of muscle-restricted gene expression. J Biol Chem. 1998;273:6656–6661. doi: 10.1074/jbc.273.12.6656. [DOI] [PubMed] [Google Scholar]
- 6.Petkova V, Romanowski MJ, Sulijoadikusumo I, Rohne D, Kang P, Shenk T, Usheva A. Interaction between YY1 and the Retinoblastoma protein. J Biol Chem. 2001;276:7932–7936. doi: 10.1074/jbc.M007411200. [DOI] [PubMed] [Google Scholar]
- 7.Gabellini D, Greeen MR, Tupier R. Inappropriate gene activation in FSHD: A repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Shen H, Himmel KL, Dupuy AJ, Largaespada DA, Nakamura T, Shaughnessy JD, Jr, Jenkins NA, Copeland NG. Leukaemia disease genes: large scale cloning and pathway predictions. Nature Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- 9.Erkeland S, Valkhof M, Heijmans-Antonissen C, Delwel R, Valk PJM, Hermans MHA, Touw IP. The gene encoding transcriptional regulator Yin Yang 1 (YY1) is a myeloid transforming gene interfering with neutrophilic differentiation. Blood. 2003;101:1111–1117. doi: 10.1182/blood-2002-04-1207. [DOI] [PubMed] [Google Scholar]
- 10.Sui G. The regulation of YY1 in tumorigenesis and its targeting potential in cancer therapy. Mol Cell Pharmacol. 2009;1:157–1176. [Google Scholar]
- 11.Shi Y, Seto E, Chang L-S, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 12.Park K, Atchison ML. Isolation of a candidate repressor/activator, NF-E1 (YY-1, δ), that binds to the immunoglobulin κ 3′ enhancer and the immunoglobulin heavy-chain μE1 site. Proc Natl Acad Sci USA. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hariharan N, Kelley DE, Perry RP. δ, trancription factor that binds to downstream elements in several polymerase II promoters, is a functionally diverse zinc finger protein. Proc Natl Acad Sci USA. 1991;88:9799–9803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flanagan JR, Becker KG, Ennist DL, Gleason SL, Driggers PH, Levi B-Z, Appella E, Ozato K. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney Murine Leukemia Virus. Mol Cell Biol. 1992;12:38–44. doi: 10.1128/mcb.12.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci USA. 2006;103:19296–19301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen N, Zhang X, Olashaw N, Seto E. Molecular cloning and functional characterization of the transcription factor YY2. J Biol Chem. 2004;279:25927–25934. doi: 10.1074/jbc.M402525200. [DOI] [PubMed] [Google Scholar]
- 17.Kim JD, Faulk C, Kim J. Retroposition and evolution of the DNA-binding motifs of YY1, YY2, and REX1. Nucleic Acids Res. 2007;35:3442–3452. doi: 10.1093/nar/gkm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee T-C, Zhang Y, Schwartz RJ. Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene. 1994;9:1047–1052. [PubMed] [Google Scholar]
- 19.Lee J-S, Galvin KM, See RH, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by EIA-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 20.Bushmeyer S, Park K, Atchison ML. Characterization of functional domains within the multifunctional transcription factor, YY1. J Biol Chem. 1995;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- 21.Austen M, Luscher B, Luscher-Firzlaff JM. Characterization of the transcriptional regulator YY1. J Biol Chem. 1997;272:1709–1717. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BA, Tullis G, Seto E, Horikoshi N, Weinmann R, Shenk T. Adenovirus E1A protein interact with the cellular YY1 transcription factor. J Virol. 1995;69:1628–1636. doi: 10.1128/jvi.69.3.1628-1636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W-M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bushmeyer SM, Atchison ML. Identification of YY1 sequences necessary for association with the nuclear matrix and for transcriptional repression functions. J Cell Biochem. 1998;68:484–499. [PubMed] [Google Scholar]
- 25.McNeil S, Guo B, Stein J, Lian JB, Bushmeyer S, Seto E, Atchison ML, Penman S, van Wijnen AJ, Stein GS. Targeting of the YY1 transcription factor to the nucleolus and the nuclear matrix in situ: the C-terminus is a principal determinant for nuclear trafficking. J Cell Biochem. 1998;68:500–510. [PubMed] [Google Scholar]
- 26.Hyde-DeRuyscher RP, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nuc Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schug J, Schuller W-P, Kappen C, Salbaum JM, Bucan M, Stoeckert CJJ. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi H, Yu Y, Fu Y, Foley J, Halees A, Weng Z. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res. 2009;17:798–806. doi: 10.1101/gr.5754707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Usheva A, Shenk T. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- 30.Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulatory Yin Yang-1. Nuc Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Molec Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorente M, Perez C, Sanchez C, Donohoe M, Shi Y, Vidal M. Homeotic transformations of the axial skeleton of YY1 mutant mice and genetic interaction with the Polycomb group gene Ring1/Ring1A. Mech Develop. 2006;123:312–320. doi: 10.1016/j.mod.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Affar EB, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. Essential dosage-dependent functions of the transcription factor Yin Yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Ann Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 35.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Ann Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 36.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 37.Roldán E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, Skok JA. Locus “decontraction” and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nature Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sayegh C, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon SJ, Saleque S, Birshtein BK. Yin Yang 1 is a lipopolysaccharide-inducible activator of the murine 3′ Igh enhancer, hs3. J Immunol. 2003;170:5549–5557. doi: 10.4049/jimmunol.170.11.5549. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Schmidt-Supprain M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, Shi Y. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seligson D, Horvath S, Huerta-Yepez S, Hanna S, Garban H, Roberts A, Shi T, Liu X, Chia D, Goodglick L, Bonavida B. Expression of transcription factor Yin Yang 1 in prostate cancer. Int J Oncol. 2005;1:131–141. [PubMed] [Google Scholar]
- 42.Pilarsky C, Wenzig M, Specht T, Saeger HD, Grutzmann R. Identification and validation of commonly overexpresssed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–750. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaravinos A, Spandidos DA. Yin yang 1 expression in human tumors. Cell Cyle. 2010;9:512–522. doi: 10.4161/cc.9.3.10588. [DOI] [PubMed] [Google Scholar]
- 44.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RGAB, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 45.Varambally S, Yu J, Laxman B, Rhodes DR, Hehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM. Integrative genomic and proteom analysis of protstate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Chinnappan D, Xiao D, Ratnasari A, Andry C, King TC, Weber HC. Transcription factor YY1 expression in human gastrointestinal cancer cells. Int J Oncol. 2009;34:1417–1423. [PubMed] [Google Scholar]
- 47.Allouche A, Nolens G, Tancredi A, Delacroix L, Mardaga J, Fridman V, Vinkler R, Boniver J, Delvenne P, Begon DY. The combined immunodetection of AP-2alpha and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res. 2008;10:R9. doi: 10.1186/bcr1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RGAB, Otte AP, Hayes DR, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggresisive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bachmann I, Halvorsen O, Collett K, Stefansson IM, Straume O, Haukaas SA, Salvesen HB, Otte AP, Akslen LA. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 50.de Nigris F, Botti C, de Chiara A, Rossiello R, Apice G, Fazioli F, Fiorito C, Sica V, Napoli C. Expression of transcription factor Yin Yang 1 in human osteosarcomas. Eur J Cancer. 2006;42:2420–2424. doi: 10.1016/j.ejca.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 51.de Nigris F, Rossiello R, Schiano C, Arra C, Williams-Ignarro S, Barbieri A, Lanza A, Balestrieri A, Giuliano MT, Ignarro LJ, Napoli C. Deletion of Yin Yang 1 protein in osteosarcoma cells on cell invasion and CXCR4/angiogenesis and metastasis. Cancer Res. 2008;68:1797–1808. doi: 10.1158/0008-5472.CAN-07-5582. [DOI] [PubMed] [Google Scholar]
- 52.Castellano G, Torrisi E, Ligresti G, Nicoletti F, Malaponte G, Travali S, McCubrey JA, Canevari S, Libra M. Yin Yang 1 overexpression in diffuse large B-cell lymphoma is associated with B-cell transformation and tumor progression. Cell Cycle. 2010;9:557–563. doi: 10.4161/cc.9.3.10554. [DOI] [PubMed] [Google Scholar]
- 53.Grubach L, Juhl-Christensen C, Rethmeier A, Olesen LH, Aggerholm A, Hokland P, Ostergaard M. Gene expression profiling of Polycomb, Hox, and Meis genes in patients with acute myeloid leukaemia. Eur J Haematol. 2008;81:112–122. doi: 10.1111/j.1600-0609.2008.01083.x. [DOI] [PubMed] [Google Scholar]
- 54.Berchuck A, Iversen ES, Lancaster JM, Pittman J, Luo J, Lee PS, Murphy SK, Dressman HK, Febbo PG, West M, Nevins JR, Marks JR. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res. 2005;11:3686–3704. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
- 55.Matsumura N, Huang Z, Baba T, Lee PS, Barnett JC, Mori S, Chang JT, Kuo W-L, Gusberg AH, Whitaker RS, Gray JW, Fujii S, Berchuck A, Murphy SK. Yin Yang 1 modulates taxane response in epithelial ovarian cancer. Mol Cancer Res. 2009;7:210–220. doi: 10.1158/1541-7786.MCR-08-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naidoo K, Clay V, Hoyland JA, Swindell R, Linton K, Illidge T, Radford JA, Beyers RJ. YY1 expression predicts favourable outcome in follicular lymphoma. J Clin Pathol. 2011;64:125–129. doi: 10.1136/jcp.2010.078188. [DOI] [PubMed] [Google Scholar]
- 57.de Nigris F, Botti C, Rossiello R, Crimi E, Sica V, Napoli C. Cooperation between Myc and YY1 provides novel silencing transcriptional targets of α3β1-integrin in tumour cells. Oncogene. 2007;26:382–394. doi: 10.1038/sj.onc.1209804. [DOI] [PubMed] [Google Scholar]
- 58.Wang C-C, Tsai M-F, Hong T-M, Chang G-C, Chen C-Y, Yang W-M, Chen JJW, Yan P-C. The transcriptional factor YY1 upregulates the novel invasion suppressor HLJ1 expression and inhibits cancer cell invasion. Oncogene. 2005;24:4081–4093. doi: 10.1038/sj.onc.1208573. [DOI] [PubMed] [Google Scholar]
- 59.Brown JL, Mucci D, Whiteley M, Dirksen M-L, Kassis JA. The Drosophila polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 60.Girton JR, Jeon SH. Novel embryonic and adult homeotic phenotypes are produced by pleiohomeotic mutations in Drosophila. Dev Biol. 1994;161:393–407. doi: 10.1006/dbio.1994.1040. [DOI] [PubMed] [Google Scholar]
- 61.Pirrotta V. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 1997;13:314–318. doi: 10.1016/s0168-9525(97)01178-5. [DOI] [PubMed] [Google Scholar]
- 62.Pirrotta V. PcG complexes and chromatin silencing. Curr Opin Genet Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- 63.Satijn DP, Otte AP. Polycomb group protein complexes: do different complexes regulate distinct target genes? Biochim Biophys Acta. 1999;1447:1–16. doi: 10.1016/s0167-4781(99)00130-x. [DOI] [PubMed] [Google Scholar]
- 64.Schumacher A, Magnuson T. Murine polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- 65.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 66.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 67.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 69.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Min J, Zhang Y, Xu R-M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nature Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 72.Tie F, Prasad-Sinha J, Birve A, Rasmuson-Lestander A, Harte PJ. A 1-megadalton ESC/E(Z) complex from Drosophila that contains Polycomblike and RPD3. Mol Cell Biol. 2003;23:3352–3362. doi: 10.1128/MCB.23.9.3352-3362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128:275–286. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- 74.Mihaly J, Mishra RK, Karch F. A conserved sequence motif in Polycomb response elements. Mol Cell. 1998;1:1065–1066. doi: 10.1016/s1097-2765(00)80107-0. [DOI] [PubMed] [Google Scholar]
- 75.Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. The YY1 transcription factor functions as a PcG protein in vivo. EMBO J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Srinivasan L, Atchison ML. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 2004;18:2596–2601. doi: 10.1101/gad.1228204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Srinivasan L, Pan X, Atchison ML. Transient requirements of YY1 expression for PcG transcriptional rescue and phenotypic rescue. J Cell Biochem. 2005;96:689–699. doi: 10.1002/jcb.20562. [DOI] [PubMed] [Google Scholar]
- 78.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valk-Lingbeek ME, Bruggeman SWM, van Lohuizen M. Stem cells and cancer: The polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 81.van Lohuizen M. The trithorax-group and Polycomb-group chromatin modifiers: implications for disease. Curr Opin Genet Dev. 1999;9:355–361. doi: 10.1016/s0959-437x(99)80053-7. [DOI] [PubMed] [Google Scholar]
- 82.Lin Y-W, Chen H-M, Fang J-Y. Gene silencing by the Polycomb Group proteins and associations with cancer. Cancer Invest. 2011;29:187–195. doi: 10.3109/07357907.2010.512605. [DOI] [PubMed] [Google Scholar]
- 83.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mut Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 84.Jacobs JJL, van Lohuizen M. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602:151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- 85.van Lohuizen M. Functional analysis of mouse Polycomb group genes. Cell Mol Life Sci. 1998;54:71–79. doi: 10.1007/s000180050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilkinson F, Pratt H, Atchison ML. PcG recruitment by the YY1 REPO domain can be mediated by Yaf2. J Cell Biochem. 2009;109:478–486. doi: 10.1002/jcb.22424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jacobs JJL, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycombgroup gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 89.Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, Ema H, Kamijo T, Katoh-Fukui Y, Koseki H, van Lohuizen M, Nakauchi H. Enhanced self-renewal of hematopoietic stem cells mediated by the Polycomb gene product Bmi-1. Immunity. 2004;21:843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 90.Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E, Dontje B, de Haan G. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107:2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kajiume T, Ninomiya Y, Ishihara H, Kanno R, Kanno M. Polycomb group gene mel-18 modulates the self-renwal activity and cell cycle status of hematopoietic stem cells. Exp Hematol. 2004;32:571–578. doi: 10.1016/j.exphem.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 92.Ohta H, Sawada A, Kim JY, Tokimasa S, Nishiguchi S, Humphries RK, Hara J, Takihara Y. Polycomb group gene rae28 is required for sustaining activity of hematopoietic stem cells. J Exp Med. 2002;195:759–770. doi: 10.1084/jem.20011911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lessard J, Schumacher A, Thorsteinsdottir U, van Lohuizen M, Magnuson T, Sauvageau G. Functional antagonism of the Polycomb Group genes eed and Bmi1 in hematopoietic cell proliferation. Genes Dev. 1999;13:2691–2703. doi: 10.1101/gad.13.20.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:252–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 95.van Lohuizen M, Verbeek S, Scheijen b, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in Eμ-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 96.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saramaki OR, Tammela TLJ, Martikainen PM, Vessella R, Visakorpi T. The gene for Polycomb Group protein enhancer of Zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer. 2006;45:639–645. doi: 10.1002/gcc.20327. [DOI] [PubMed] [Google Scholar]
- 98.Raaphorst FM, Otte AP, Meijer CJ. Polycomb-group genes as regulators of mammalian lymphopoiesis. Trends Immunol. 2001;22:682–690. doi: 10.1016/s1471-4906(01)02082-8. [DOI] [PubMed] [Google Scholar]
- 99.Berezovska OP, Glinskii AB, Yang Z, Li XM, Hoffman RM, Glinsky GV. Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer. Cell Cycle. 2006;5:1886–1901. doi: 10.4161/cc.5.16.3222. [DOI] [PubMed] [Google Scholar]
- 100.Sasaki M, Ikeda H, Itatsu K, Yamaguchi J, Sawada S, Minato H, Ohta T, Nakanuma Y. The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and agressive biological behavior of hepatocellular carcinoma. Lab Invest. 2008;88:873–882. doi: 10.1038/labinvest.2008.52. [DOI] [PubMed] [Google Scholar]
- 101.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorulation of EZH2 suppresses methylation of lysine 27 in histones. Science. 2005;310:305–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 102.Sasaki D, Imaizumi Y, Hasegawa H, Osaka A, Tsukasaki K, Choi YL, Mano H, Marquez VE, Hayashi T, Yanagihara K, Moriwaki Y, Miyazaki Y, Kamihira S, Yamada Y. Overexpression of enhancer of zeste homolog 2 with trimethylation of lysine 27 on histone H3 in adult T-cell leukemia/lymphoma as a target for epigenetic therapy. Haematologica. 2011;96:712–719. doi: 10.3324/haematol.2010.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Crea F, Hurt EM, Farrar WL. Clinical significance of Polycomb gene expression in brain tumors. Molec Cancer. 2010;9:265–270. doi: 10.1186/1476-4598-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maertens G, El Messaoudi-Aubert S, Racek T, Stock JK, Nicholls F, Rodriguez-Niedenfuhr M, Gil J, Peters G. Several distinct polycomb complexes regulate and co-localize on the INK4a tumor supressor locus. PLoS One. 2009;4:e6380. doi: 10.1371/journal.pone.0006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine J-C, Hansen KH, Helin K. The Polycomb group proteins bind throughout the INK4A-ARF locus and are dissociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one and one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 107.Jacobs JJL, Scheijen B, Voncken J-W, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-myc in tumorogenesis by inhibiting c-myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen D, Zhang J, Rayburn ER, Wang H, Zhang R. RYBP stabilizes p53 by modulating MDM2. EMBO Rep. 2009;10:166–172. doi: 10.1038/embor.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zeng Y, Kotake Y, Pei XH, Smith MD, Xiong Y. p53 binds to and is required for the repression of Arf tumor suppressor by HDAC and polycomb. Cancer Res. 2011;71:2781–2792. doi: 10.1158/0008-5472.CAN-10-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu J, Yu J, Rhodes DR, Tomlins SA, Cao X, Chen G, Mehra R, Wang X, Ghosh D, Shah RB, Varambally S, Pienta KJ, Chinnaiyan AM. A Polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007;67:10657–10663. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- 111.Tan J, Yang X, Zhuang L, Jian X, Chen W, Lee PL, Karuturi RKM, Tan PBO, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Basu A, Hodawadekar S, Andrews O, Knox A, Pan X, Wilkinson F, Atchison ML. YY1 PcG function as a potential cancer theraeutic target. Forum Immunopath Dis Ther. 2010;1:31–50. [Google Scholar]
- 113.Riggs KJ, Saleque S, Wong KK, Merrell KT, Lee JS, Shi Y, Calame K. Yin-yang 1 activates the c-myc promoter. Mol Biol Cell. 1993;13:7487–7495. doi: 10.1128/mcb.13.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sui G, Affar EB, Shi Y, Brignone C, Wall NR, Yin P, Donohue M, Luke MP, Calvo D, Grossman SR, Shi Y. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 115.Margolis DM, Somasundaran M, Green MR. Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J Virol. 1994;68:905–910. doi: 10.1128/jvi.68.2.905-910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moriuchi M, Moriuchi H. YY1 transcription factor down-regulates expression of CCR5, a major coreceptor for HIV-1. J Biol Chem. 2003;278:13003–13007. doi: 10.1074/jbc.M204980200. [DOI] [PubMed] [Google Scholar]
- 117.Coull JJ, He G, Rucker MC, Dervan PB, Margolis DM. Targeted derepression of the human immunodeficiency virus type 1 long terminal repeat by pyrrole-imidazole polyamines. J Virol. 2002;76:12349–12354. doi: 10.1128/JVI.76.23.12349-12354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.He G, Margolis DM. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol Cell Biol. 2002;22:2965–2973. doi: 10.1128/MCB.22.9.2965-2973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.May M, Dong XP, Beyer-Finkler E, Stubenrauch F, Fuchs PG, Pfister H. The E6/E7 promoter of extrachromosomal HPV16 DNA in cervical cancers escapes from cellular repression by mutation of target sequences for YY1. EMBO J. 1994;13:1460–1466. doi: 10.1002/j.1460-2075.1994.tb06400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dong XP, Stubenrauch F, Beyer-Finkler E, Pfister H. Prevalence of deletions of YY1-binding sites in episomal HPV16 DNA from cervical cancers. Int J Cancer. 1994;58:803–808. doi: 10.1002/ijc.2910580609. [DOI] [PubMed] [Google Scholar]
- 121.Tan SH, Baker CC, Stunkel W, Bernard HU. A transcriptional initiator overlaps with a conserved YY1 binding site in the long control region of human papillomavirus type16. Virology. 2003;305:486–501. doi: 10.1006/viro.2002.1779. [DOI] [PubMed] [Google Scholar]
- 122.Schmidt M, Kedzia W, Gozdzicka-Jozefiak A. Intratype HPV16 sequence variation within LCR of isolates from asymptomatic carriers of cervical cancers. J Clin Virol. 2001;23:65–77. doi: 10.1016/s1386-6532(01)00189-5. [DOI] [PubMed] [Google Scholar]
- 123.Veress G, Murvai M, Szarka K, Juhasz A, Konya J, Gergely L. Transcriptoinal activity of human papillomavirus type 16 variants having deletions in the long control region. Eur J Cancer. 2001;37:1946–1952. doi: 10.1016/s0959-8049(01)00222-2. [DOI] [PubMed] [Google Scholar]
- 124.Yengi L, Inskip A, Gilford J, Allderse J, Bailey L, Smith A, Lear JT, Heagerty AH, Bowers B, Hand P, Hayes JD, Jones PW, Strange RC, Fryer AA. Polymorphism at the glutathione S-transferase locus GSTM3: interactions with cytochrome P450 and glutathione S-transferase genotypes as risk factors for multiple cutaneous basal cell carcinoma. Cancer Res. 1996;56:1974–1977. [PubMed] [Google Scholar]
- 125.Lichy JH, Majidi M, Elbaum J, Tsai MM. Differential expression of the human ST5 gene in HeLa-fibrobast hybrid cell lines mediated by YY1: evidence that YY1 plays a part in tumor suppression. Nuc Acids Res. 1996;24:4700–4708. doi: 10.1093/nar/24.23.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Austen M, Cerni C, Luscher-Firzlaff JM, Luscher B. YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene. 1998;17:511–520. doi: 10.1038/sj.onc.1201968. [DOI] [PubMed] [Google Scholar]
- 127.Yao Y-L, Yang W-M, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Baumeister P, LLuo S, Skarnes WC, Sui G, Seto E, Shi Y, Lee AS. Endoplasmic retiuculum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol Cell Biol. 2005;25:4529–4540. doi: 10.1128/MCB.25.11.4529-4540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim JD, Hinz AK, Bergmann A, Huang JM, Ovcharenko I, Stubbs L, Kim J. Identification of clustered YY1 binding sites in imprinting control regions. Genome Res. 2006;16:901–911. doi: 10.1101/gr.5091406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim JD, Hinz AK, Choo JH, Stubbs L, Kim J. YY1 as a controlling factor for the Peg3 and Gnas imprinted domains. Genomics. 2007;89:262–269. doi: 10.1016/j.ygeno.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Donohoe ME, Zhang L-F, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, YY1, for the X chromosome binary switch. Mol Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 132.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, Conaway JW. YY1 functions with INO80 to activte transcription. Nature Struct Molec Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- 134.Wu S, Shi Y, Mulligan P, Gay F, Landry J, Liu H, Lu J, Qi HH, Wang W, Nickoloff JA, Wu C, Shi Y. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nature Struct Biol. 2007;14:1665–1672. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Oei SL, Shi Y. Transcription factor Yin Yang 1 stimulates Poly(ADP-Ribosyl)ation and DNA repair. Biochem Biophys Res Commun. 2001;284:450–454. doi: 10.1006/bbrc.2001.4985. [DOI] [PubMed] [Google Scholar]
- 136.Zaprazna K, Atchison M. 2011 Submitted. [Google Scholar]
- 137.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 138.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 139.Martin A, Scharff MD. AID and mismatch repair in antibody diversification. Nature Rev Immunol. 2002;2:605–614. doi: 10.1038/nri858. [DOI] [PubMed] [Google Scholar]
- 140.Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HCIII, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nature Genetics. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 141.Takizawa M, Tolarova H, Li Z, Dubois W, Lim S, Callen E, Franco S, Mosaico M, Feigenbaum L, Alt FW, Nussenzweig A, Potter M, Casellas R. AID expression levels determine the extent of cMyc oncogenic tranlocations and the incidence of B cell tumor development. J Exp Med. 2008;205:1949–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Okazaki I-M, Kotani A, Honjo T. Role of AID in tumorigensis. Adv Immunol. 2007;94:245–273. doi: 10.1016/S0065-2776(06)94008-5. [DOI] [PubMed] [Google Scholar]
- 143.Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30:173–181. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 144.Sernandez IV, de Yebenes VG, Dorsett Y, Ramiro AR. Haploinsufficiency of activation-induced deaminase for antibody diversification and chromosome translocations both in vitro an in vivo. PLoS One. 2008;3:e3927. doi: 10.1371/journal.pone.0003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de Yebenes VG, Belver L, Pisana DG, Gonzalez S, Villasante A, Croce C, He L, Ramiro AR. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai T-H, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rada C, Jarvis JM, Milstein C. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc Natl Acad Sci USA. 2002;99:7003–7008. doi: 10.1073/pnas.092160999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, Nakata M, Honjo T. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci USA. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brar SS, Watson M, Diaz M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem. 2004;279:26395–26401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- 151.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Klemm L, KLDuy C, Iacobucci I, Kuchen S, von Levetzow G, Feldhahn N, Henke N, Li Z, Hoffmann TK, Kim Y-m, Hofmann W-K, Jumaa H, Groffen J, Heisterkamp N, Martinelli G, Lieber MR, Casellas R, Muschen M. The B cell mutator AID promotes B lympoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Trauth BC, Klas C, Peters AMJ, Matzku S, Moller P, Falk W, Debatin KM, Krammer PH. Monoclonal antibody-mediate tumor regression by induction of apoptosis. Science. 1989;245:301–305. doi: 10.1126/science.2787530. [DOI] [PubMed] [Google Scholar]
- 154.Yondhara S, Ishii A, Yonehara M. A cell-killing monoclonal antibody (anti-Fas) to a cell surface antigen co-downregulated with the receoptor of tumor necrosis factor. J Exp Med. 1991;169:1747–1756. doi: 10.1084/jem.169.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Garban HJ, Bonavida B. Nitric oxide inhibts the transcription repressor Yin-Yang 1 binding activity at the silencer region of the Fas promoter: a pivotal role for nitric oxide in the up-regulation of Fas gene expression in human tumor cells. J Immunol. 2001;167:75–81. doi: 10.4049/jimmunol.167.1.75. [DOI] [PubMed] [Google Scholar]
- 156.Vega MI, Jazirehi AR, Huerta-Yepez S, Bonavida B. Rituximab-induced inhibition of YY1 and Bcl-xl expression in Ramos non-Hodgkin’s lymphoma cell line via inhibition of NF-κB activity: role of YY1 and Bcl-xl in Fas resistance and chemoresistance, respectively. J Immunol. 2005;175:2174–2183. doi: 10.4049/jimmunol.175.4.2174. [DOI] [PubMed] [Google Scholar]
- 157.Vega MI, Huerta-Yepez S, Jazirehi AR, Garban H, Bonavida B. Rituximab (chimeric anti-CD20) sensitizes B-NHL cell lines to Fas-induced apoptosis. Oncogene. 2005;24:8114–8127. doi: 10.1038/sj.onc.1208954. [DOI] [PubMed] [Google Scholar]
- 158.Huerta-Yepez S, Vega M, Garban H, Bonavida B. Involvement of the TFN-α autocrine-paracrine loop, via NF-κB and YY1, in the regulation of tumor cell resistance to Fas-induced apoptosis. Clin Immunol. 2006;120:297–309. doi: 10.1016/j.clim.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 159.Luo C, Lu X, Stubbs L, Kim J. Rapid evolution of a recently retrotransposed transcription factor YY2 in mammalian genomes. Genomics. 2006;87:348–355. doi: 10.1016/j.ygeno.2005.11.001. [DOI] [PubMed] [Google Scholar]