SUMMARY
Comparative sequence analyses of bacterial genomes are revealing many structured RNA motifs that function as metabolite-binding riboswitches. We have identified an RNA motif frequently positioned in the 5′ UTRs of folate transport and biosynthesis genes in Firmicute genomes. Biochemical experiments confirm that representatives of this newfound RNA class selectively bind derivatives of the vitamin folate, including di- and tetrahydrofolate coenzymes. In addition, representatives of this aptamer class occasionally reside upstream of RNA structures that are predicted to control translation initiation in response to ligand binding. These findings expand the number of coenzymes that are directly sensed by RNA and reveal possible riboswitch-controlled regulons that respond to changes in single-carbon metabolism.
INTRODUCTION
Metabolite-binding riboswitches are gene control elements constructed entirely of RNA (Roth and Breaker, 2009). Each usually is composed of an aptamer domain that binds a specific metabolite and an expression platform that transforms an aptamer binding event into an adjustment in the level of expression of target genes. Riboswitches are most commonly found in the 5′ untranslated regions (UTRs) of bacterial mRNAs, where they usually control gene expression by influencing transcription termination or translation initiation (Barrick and Breaker, 2007; Dambach and Winkler, 2009). Riboswitches also have been shown to act in trans by base pairing with mRNAs transcribed elsewhere in the genome (Loh et al., 2009). Additionally, there are experimentally validated examples of eukaryotic riboswitches in both fungi (Cheah et al., 2007) and plants (Bocobza et al., 2007; Croft et al., 2007; Wachter et al., 2007) that modulate protein levels by controlling mRNA splicing.
Currently, the most productive methods for finding novel riboswitch classes make use of computer-aided searches to identify intergenic regions that are highly conserved in primary and secondary structure among several bacterial genomes or in metagenomic DNA sequences (Rodionov et al., 2003; Barrick et al., 2004; Weinberg et al., 2007; Yao et al., 2007; Rodionov et al., 2007). In the current study, we describe the discovery and characterization of a novel riboswitch candidate that is frequently associated with genes whose protein products transport folate or catalyze reactions related to single-carbon metabolism.
RESULTS AND DISCUSSION
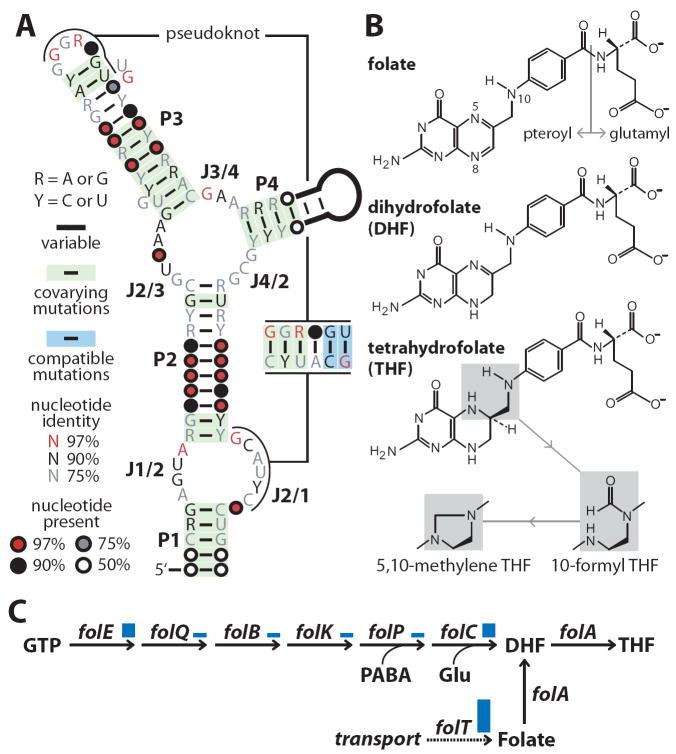
Our bioinformatics pipeline identified 57 distinct representatives (Figure S1) of a novel RNA motif (Figure 1A) consisting of four base-paired regions (P1 through P4) and an additional base-paired pseudoknot structure. The motif is frequently located immediately upstream of bacterial folate (Figure 1B) uptake transporter folT genes (Figure 1C; Figure S2), and in some cases upstream of the folate biosynthesis genes folE, folC, and folQPBK (Table S1). The RNA motif was found both in complete genomes (Clostridiales and Lactobacillales orders) and in metagenome (human gut) data. We hypothesized that the RNA may be a riboswitch that binds to a compound related to folate.
Figure 1. Conserved RNA motif associated with genes for folate metabolism.
(A) Consensus sequence and secondary structure model for the candidate riboswitch aptamer. See Supplemental Information for the alignment and methods used to establish nucleotide conservation and covariation. (B) Chemical structures of folate, DHF, THF and other folate derivatives used in this study. (C) Folate biosynthesis and uptake pathway. Genes under control of a THF riboswitch are highlighted with relative frequency of regulatory interaction reflected by the heights of the blue bars. GTP is guanosine-5′-triphosphate; PABA is p-aminobenzoic acid; Glu is L-glutamine.
Folates are composed of three moieties: pterin, p-aminobenzoate, and glutamate (Figure 1B). An active form of folate, tetrahydrofolate (THF), is an essential cofactor in one-carbon transfer reactions of central metabolism. De novo THF biosynthesis starts from GTP and proceeds in seven consecutive steps (de Crécy-Lagard et al., 2007) (Figure 1C). Alternatively, Gram-positive bacteria salvage folate from the environment utilizing a unique energy-coupling factor (ECF) transport system composed of a folate-binding protein (FolT) and a common ECF module (Rodionov et al., 2009). The identified riboswitch candidate is predicted to control expression of folT in many Firmicutes, including Lactobacillus species which are auxotrophic for folate. There is a single example, occurring in Ruminococcus obeum, in which the RNA motif us associated with the folate biosynthesis folEQPBK operon.
A 106-nucleotide RNA construct termed 106 folT (Figure 2A) that encompasses a representative motif located in the 5′ UTR of the folT gene from Alkaliphilus metalliredigens was prepared for analysis. This RNA was subjected to in-line probing (Soukup and Breaker, 1999; Regulski and Breaker, 2008) to assess ligand binding and structural modulation characteristics. In-line probing assays take advantage of the inherent instability of RNA phosphoester bonds located in unstructured regions to reveal folding changes caused by ligand binding. The 106 folT RNA undergoes extensive changes in RNA structure (Figure 2B) as it binds THF with an apparent dissociation constant (KD) of ~70 nM (Figure 2C). Two other folate derivatives, dihydrofolate (DHF) and tetrahydrobiopterin, also bind with KD values of ~300 nM (Figure S3). However, no structural modulation was observed when the RNA was incubated with folate, pterin, p-aminobenzoate, glutamate, or p-aminobenzoyl glutamate at concentrations as high as 1 mM.
Figure 2. Evidence for THF riboswitch function.
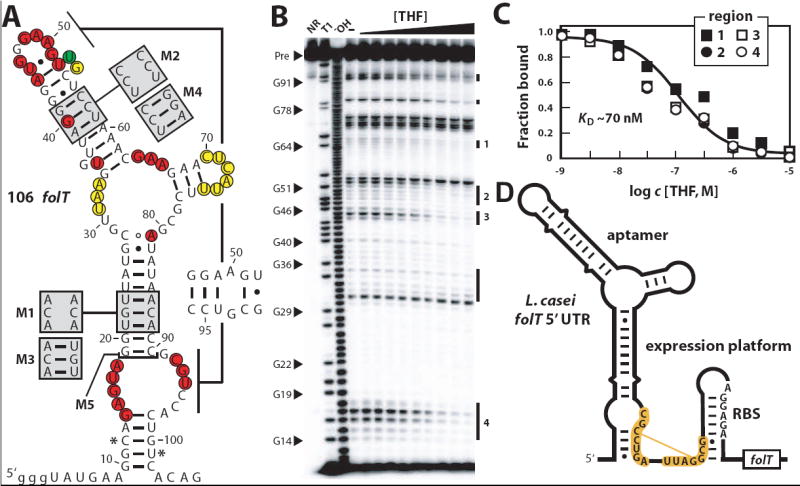
(A) Sequence and predicted secondary structure of the 106 folT RNA from A. metalliredigens. Nucleotides shaded yellow, red or green identify internucleotide linkages that undergo constant, decreasing or increasing scission when THF is added to an in-line probing assay (see gel image in B). Asterisks identify the 5′ and 3′ boundaries of the in-line probing annotations, which are restricted due to limitations of product separation by polyacrylamide gel electrophoresis. Shaded boxes designate sequence changes present in mutants M1 through M4 as indicated. M5 construct is truncated at the bracket. Lowercase letters identify nucleotides added to the construct to facilitate efficient in vitro transcription. (B) In-line probing analysis of the A. metalliredigens 106 folT RNA. NR, T1 and −OH designate precursor (Pre) RNAs subjected to no reaction, partial digestion with RNase T1 (cleaves after G residues), and partial digest under alkaline conditions (cleaves at every position). Other lanes include Pre RNAs subjected to in-line probing reactions without THF (–) or with increasing concentrations of THF (1 nM to 10 μM). Vertical lines identify cleavage products whose yields are altered by THF addition, and numbered regions were quantified and used to estimate KD values. (C) Plot of the fraction of RNAs bound to ligand versus the logarithm of the concentration of THF. The solid line represents a theoretical binding curve for a one-to-one interaction with a KD of 70 nM. Fraction bound values were generated by quantifying band intensities from the regions identified in B. (D) A possible THF riboswitch expression platform is evident in the L. casei folT mRNA. The nucleotides shaded in orange can form an alternative base pairing interaction in the absence of THF that may interfere with the formation of the putative anti-RBS hairpin.
Similar in-line probing results with THF and DHF were observed using several other representatives from this RNA class (data not shown). Additional assays revealed that these RNAs also bind to various N5- and N10-modified THF derivatives, including 5-formyl THF, 5-methyl THF, 10-formyl THF, 5,10-methenyl THF, and 5,10-methylene THF. These results indicate that the newfound RNAs function as selective aptamers for specific reduced forms of the vitamin folate. The ability of the aptamers to discriminate between folate and the reduced derivatives THF and DHF could be achieved simply by positioning a hydrogen bond acceptor to recognize the hydrogen at the N8 position of the pterin moiety pyrazine ring (Figure 1B). Furthemore, the reduced forms of folate are no longer aromatic and take on a puckered configuration (Poe and Hoogsteen, 1978) that may also serve as a molecular recognition feature. Since these aptamers tolerate modifications at the N5 and N10 positions of the ligand, they are unlikely to be important molecular recognition contacts to these positions. Given the preferential affinity for the coenzyme forms of folate, we speculate that THF, or perhaps one or more of its single-carbon derivatives, is the biologically relevant ligand for these aptamers.
Regions of the RNA that are particularly susceptible to spontaneous cleavage during in-line probing reactions (Figure 2B) largely correspond to nucleotides predicted to reside in single-stranded regions (Figure 2C). Furthermore, sites that undergo ligand-dependent reduction in spontaneous cleavage carry highly conserved nucleotides. These results are consistent with our secondary structure model for the aptamer and suggest that ligand binding stabilizes a tertiary structure formed in part by the nucleotides whose base identities are well conserved.
To further test the validity of our model, RNA constructs containing three consecutive mutations that disrupt the P2 (construct M1) or P3 (M2) stems of the 106 folT RNA (Figure 2A) were examined for THF binding using in-line probing assays. Both M1 and M2 constructs are less structured than the wild-type (WT) construct, as indicated for M1 by an increase in spontaneous cleavage at phosphodiester linkages of the nucleotides that comprise the stems (Figure S4). Furthermore, the KD of M1 for THF is ~1000-fold poorer compared to WT, whereas the M2 mutant does not bind to THF at concentrations as high as 3.16 mM. In contrast, RNA constructs carrying an additional three mutations that restore base pairing to the P2 (M3) or P3 (M4) stems (Figure 2A) exhibit KD values comparable to that observed for WT.
Riboswitch aptamer binding pockets are often composed of highly conserved residues residing in regions predicted to be single-stranded. These nucleotides participate in forming the structure necessary to bind the ligand, and some participate in forming direct contacts with the ligand. There are two sizable groups of single-stranded nucleotides evident from the consensus secondary structure model (Figure 1A). These include the junction between P1 and P2 (J1/2, nucleotides 15-18), and the central bulge area connecting the P2, P3, and P4 stems (J2/3, nucleotides 29-33; J3/4, nucleotides 64-66; J4/2, nucleotides 78-80). Note that nucleotides 91-96 that form J2/1 have the potential to form a pseudoknot structure with nucleotides 47-52, and therefore may be base-paired in the ligand-bound state. In the ligand-bound structure, it is unlikely that the nucleotides in the central single-stranded region contact nucleotides from the single-stranded region near P1 because they reside on opposite ends of the ~10 base-pair P2 stem.
Although this structural arrangement may prevent the aptamer from forming a ligand binding pocket by simultaneously using residues from both of these single-stranded regions, ligand binding induces structural changes in both single-stranded regions. Thus, we cannot identify the nucleotides that most likely form the binding pocket using only these observations. To further address this issue, we designed a truncated construct M5 wherein P1 and the nucleotides forming J1/2 and J2/1 were deleted. Specifically, this construct spans nucleotides 19 through 90 of the 106 folT RNA (Figure 2A), and carries three additional G residues at the 5′ terminus to facilitate preparation by in vitro transcription. The RNA retains THF binding function, albeit with a substantially reduced affinity (KD ~10 μM) (data not shown).
The fact that THF is still bound by the M5 construct indicates that nucleotides forming P1 and the junction linking this stem to P2 are not likely to be involved in forming molecular recognition contacts with the ligand. Furthermore, the contribution of the J2/1 nucleotides in forming the putative pseudoknot structure with nucleotides in the loop of P3 is also not essential for ligand binding. Rather, these nucleotides are more likely to support folding of the ligand binding pocket, which is probably formed at least in part using nucleotides from the three-stem junction. Interestingly, the central bulge is not conserved as well as other previously characterized riboswitch binding pockets. However, most individual THF aptamer sequences match the consensus well with a small number of representatives that deviate substantially. These outliers each may form a distinct ligand binding pocket that still recognizes THF or that selectively binds one of the several natural derivatives of this coenzyme (Figure 1B).
If these natural THF aptamers are components of cofactor-responsive riboswitches, then at least some aptamers should be associated with expression platforms whose structures are indicative of their gene control mechanism. Evidence of expression platform structures was sought by manually examining the nucleotides located immediately downstream of several aptamers. For example, in the organism Lactobacillus casei, there is the potential for formation of a strong hairpin involving nucleotides predicted to serve as the ribosome binding site (RBS) (Figure 2D). Formation of the consensus THF aptamer structure in the presence of ligand should permit formation of this anti-RBS stem and thereby preclude ribosome binding and translation initiation. However, in the absence of THF, alternative folding is possible because the J2/1 nucleotides and the right shoulder of P1 are complementary to nucleotides residing upstream and partially overlapping those required to form the anti-RBS structure. Formation of this putative anti-anti-RBS stem in the absence of THF should permit translation. Numerous examples of genetic “OFF” riboswitches that use this mechanism for translation control have been identified previously (Barrick and Breaker, 2007).
While the most common riboswitch architectures involve the integration of a single aptamer with a single expression platform, there are a number of examples wherein multiple aptamers are juxtaposed to achieve more sophisticated gene regulation (Stoddard and Batey, 2006; Breaker, 2008). The most common tandem arrangements involve two aptamers of the same class that use ligand-binding cooperativity (Mandal et al., 2004) or dual transcription termination (Welz and Breaker, 2007) to yield more digital gene control. In rare instances, riboswitches from two different ligand classes are present in the same mRNA where they function as a two-input Boolean logic gate (Sudarsan et al., 2006). Alternatively, two riboswitches that bind identical ligands can act to control both transcription and translation (Poiata et al., 2009). Interestingly, we observe a THF aptamer in Clostridium kluyveri located immediately downstream of a pfl RNA representative, which is a newfound riboswitch candidate whose putative ligand remains unknown (Weinberg et al., 2010). Thus, the pfl motif and THF aptamer may co-localize to produce another tandem riboswitch arrangement that yields genetic responses to two different ligands. However, since representatives of the pfl motif commonly are associated with genes for formyl-THF biosynthesis, the candidate riboswitch also may respond to THF or one of its natural derivatives.
The THF riboswitch represents the ninth verified riboswitch architecture that senses a coenzyme or a coenzyme derivative. The fact that nearly half of the validated riboswitch classes sense essential coenzymes highlights important sensory and regulatory roles entrusted to structured RNAs by modern cells. Furthermore, some of the coenzyme-sensing aptamer classes are very common and widespread in bacteria, suggesting an early evolutionary origin. Most coenzymes, including THF, are believed to have emerged in an RNA World (White 1976). Therefore some coenzyme-sensing aptamers, including the class of THF-binding RNAs described herein, may reflect ancient structures used by the earliest life forms to sense or use coenzymes as signaling compounds or as chemical catalysts.
Two previously described coenzyme-sensing riboswitch classes selective for thiamin pyrophosphate (Sudarsan et al, 2005) and flavin mononucleotide (Lee et al., 2009; Ott et al., 2009), have been shown to be targeted by coenzyme analogs that exhibit antimicrobial activity. Considering the importance of folate metabolism to bacterial pathogens, THF riboswitches might represent a novel drug target wherein folate analogs could be used to deceive bacteria into misregulating the import and production of the vitamin and its derivatives.
SIGNIFICANCE
Efforts to discover structured noncoding RNAs in bacteria are revealing the existence of numerous riboswitch candidates. Since the first reports of metabolite-binding riboswitches, we and others have speculated that all the major nucleotide-derived coenzymes could have corresponding riboswitches in bacteria. Justification for this speculation is based on the likelihood that many bacteria will need to sense changes in coenzyme concentrations and regulate certain metabolic pathways in response to coenzyme abundance or deficiency. Also, the existence of so many coenzyme-sensing riboswitches would be consistent with an RNA World origin for some riboswitch classes (Breaker, 2009).
Regardless of the origin of riboswitches, THF and its derivatives serve as key coenzymes for single-carbon chemistry in all forms of life, and the discovery of a riboswitch class for folate derivatives helps reveal how cells sense and respond to changing concentrations of these compounds. Folate metabolism enzymes currently serve as major antibacterial drug targets, and the co-localization of THF riboswitches with coding regions of unknown function may allow researchers to identify additional genes whose protein products are involved in folate metabolism. Furthermore, THF riboswitches could directly serve as a new series of drug targets for the manipulation of this coenzyme in bacteria.
Supplementary Material
Acknowledgments
We thank Nick Carriero and Rob Bjornson for assisting our use of the Yale Life Sciences High Performance Computing Center (NIH grant RR19895-02). We also thank Sam Kapelle for conducting preliminary binding assays, and members of the Breaker laboratory for helpful discussions. This work was supported in part by NIH grant DK070270 to R.R.B. D.A.R. is supported by the ‘Molecular and Cellular Biology’ program of the Russian Academy of Sciences and Russian President’s grant MK-422.2009.4. R.R.B. is also a Howard Hughes Medical Institute Investigator.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Data including four figures, one table, and Supplemental experimental procedures can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Complex riboswitches. Science. 2008;319:1795–1797. doi: 10.1126/science.1152621. [DOI] [PubMed] [Google Scholar]
- Breaker RR. Riboswitches: from ancient gene-control systems to modern drug targets. Future Microbiol. 2009;4:771–773. doi: 10.2217/fmb.09.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci USA. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambach MD, Winkler WC. Expanding roles for metabolite-sensing regulatory RNAs. Curr Opin Microbiol. 2009;12:161–169. doi: 10.1016/j.mib.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crécy-Lagard V, El Yacoubi B, de la Garza RD, Noiriel A, Hanson AD. Comparative genomics of bacterial and plant folate synthesis and salvage : predictions and validations. BMC Genomics. 2007;8:245. doi: 10.1186/1471-2164-8-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6:187–194. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Ott E, Stolz J, Lehmann M, Mack M. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol. 2009;6:276–280. doi: 10.4161/rna.6.3.8342. [DOI] [PubMed] [Google Scholar]
- Poe M, Hoogsteen K. 5,6,7,8-Tetrahydrofolic acid: conformation of the tetrahydropyrazine ring. J Biol Chem. 1978;253:543–546. [PubMed] [Google Scholar]
- Poiata E, Meyer MM, Ames TD, Breaker RR. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA. 2009;15:2046–2056. doi: 10.1261/rna.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- Rodionov DA. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem Rev. 2007;107:3467–3497. doi: 10.1021/cr068309+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Hebbeln P, Eudes A, ter Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD, et al. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol. 2009;191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res. 2003;31:6748–6757. doi: 10.1093/nar/gkg900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CD, Batey RT. Mix-and-match riboswitches. ACS Chem Biol. 2006;1:751–754. doi: 10.1021/cb600458w. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes. Genome Biol. 2010;11:R38. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HB., 3rd Coenzymes as fossils of an earlier metabolic state. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- Yao Z, Barrick J, Weinberg Z, Neph S, Breaker R, Ruzzo WL. A computational pipeline for high-throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLoS Comput Biol. 2007;3:e126. doi: 10.1371/journal.pcbi.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.