Abstract
In this issue of Molecular Cell, Weinberger et al. (2012) find that particular histone deacetylases (HDACs) regulate distinct stages of transcription, implicating chromatin dynamics in the generation of gene-specific noise within populations of genetically identical cells.
Gene expression in genetically identical cells growing in common environments can show stochastic variations over time, a phenomenon known as gene expression noise. While early investigations in this area focused on identifying and distinguishing between different sources of noise within a cell (Blake et al., 2006; Blake et al., 2003; Elowitz et al., 2002; Ozbudak et al., 2002), more recent work has made clear that noise levels can vary widely between individual genes (Suter et al., 2011). Promoter sequence, nucleosome occupancy, and the presence of chromatin regulators have all been identified as factors contributing to or buffering gene expression fluctuations and phenotypic variability (Blake et al., 2006; Suter et al., 2011; Whitelaw et al., 2010). In this issue of Molecular Cell, Weinberger et al. (2012) build on these findings and show that certain chromatin regulators can act to increase noise levels at their target genes.
Eukaryotic genes are exquisitely compacted into chromatin by interactions with histone proteins, which condenses them by many orders of magnitude through organization into nucleosomes and higher-order structures. Transcription of these genes occurs in staccato bursts, in which multiple messenger RNA molecules are synthesized from a template DNA strand, and comprises multiple steps including recruitment of the pre-initiation complex, initiation of transcription, and elongation by RNA polymerase. All of these steps represent potential points of transcriptional regulation, and chromatin modifiers that control the packaging and accessibility of DNA play a key role in regulating these steps. By screening for the effect of deleting particular chromatin regulators in the yeast Saccharomyces cerevisiae and measuring the effect on gene expression noise and levels of reporter constructs driven by a library of promoters, Weinberger et al. (2012) identified chromatin regulators that can modulate expression levels and noise by controlling transcription at particular steps.
The authors screened individual deletions of 137 chromatin factors using flow cytometry of a library of promoter constructs spanning a range of expression levels, and used a theoretical model to infer regulatory mechanism from the relationship between noise and mean expression. By obtaining single-cell measurements of reporter gene expression in the individual deletion constructs, they were able to deconvolute the effect of loss of a given gene on transcriptional bursting of the reporter, and identify independent modulators of burst size and burst frequency. Deletions that affected transcriptional burst size changed mean expression levels without altering noise levels. By contrast, deletions that altered the frequency of bursting resulted in changes to both mean expression and noise levels (Figure 1).
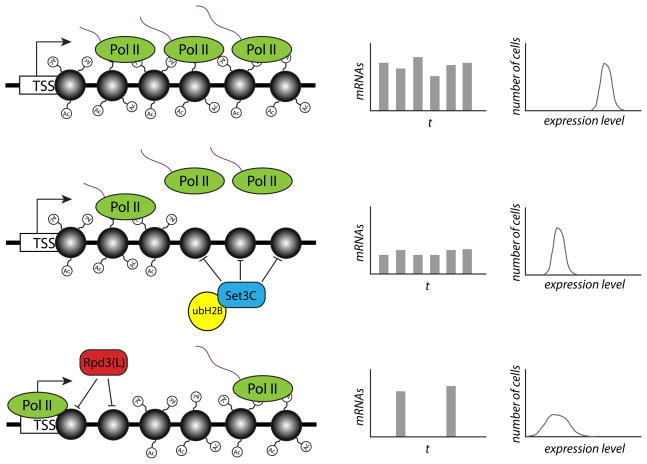
Figure 1. The HDACs Set3C and Rpd3(L) act at distinct steps of transcription to modulate either transcriptional burst size or burst frequency.
The figure illustrates the effect of histone deacetylation by either Set3C or Rpd3(L) on the expression and noise levels of a target gene. In the top row, RNA polymerase II (Pol II) transcribes RNA (purple strands) from a gene with heavily acetylated nucleosomes (grey circles), leading to high-frequency transcriptional bursts of relatively large size. When Set3C and ubH2B act to deacetylate nucleosomes in the body of the gene (middle row), it causes a decrease in Pol II processivity and an increased likelihood of abortive transcription. This affects the average size of each transcriptional burst but not the frequency, resulting in a decrease in mean expression levels but the same level of noise. Rpd3(L), by contrast, acts to deacetylate nucleosomes just downstream of the transcription start site (TSS), leading to a reduced frequency of transcriptional initiation and bursts (bottom row). This lower burst frequency results in both lower mean expression levels and increased noise levels, as seen by the increased width of the expression distribution.
Perturbations to both histone H2B mono-ubiquitination (ubH2B) and the histone deacetylase (HDAC) complex Set3C resulted in increases in transcriptional burst size, implicating these pathways as repressors of burst size (Weinberger et al. 2012). Deletions of histone acetyltransferases (HATs) decreased the frequency of transcriptional bursts; while removing other HDACs, including members of the Rpd3(L) complex, increased burst frequency (Figure 1). While both Set3C and Rpd3(L) seemed to act as transcriptional repressors, the distinct noise phenotypes of knocking out particular HDAC complexes implied that these factors are specialized to regulate distinct steps of transcription, which the authors went on to investigate further. By examining published genome-wide occupancy profiles of RNA polymerase II (PolII) subunits and elongation factors, they found that ubH2B and Set3C targets were associated with low levels of PolII processivity, suggesting that these factors act during elongation to increase the chances of abortive transcription. Correlation of ubH2B occupancy with low levels of histone acetylation, along with an increase in acetylation at Set3C targets in strains deleted for H2B ubiquitinating enzymes, support a model in which ubH2B acts to recruit Set3C to target genes. By mapping levels of the H3K9 acetylation (H3K9ac) mark, which tends to be positively associated with gene expression, in strains knocked out for Set3C or Rpd3(L) components, the authors defined a role for Rpd3(L) in deacetylating the +2 nucleosome downstream of the transcriptional start site. Earlier work had found an association between nucleosome occupancy at genes and high noise levels (Cairns, 2009). Deacetylation of the +2 nucleosome by Rpd3(L) presumably decreases the probability of transcriptional initiation, following recruitment of the transcription apparatus to the promoter.
Functional specialization of HDACs provides an opportunity for evolution to fine-tune and sculpt distinct parameters of gene expression programs. Modulating Set3C or ubH2B activity at a given gene could alter the probability of full-length transcription and change mean expression levels, while changing Rpd3(L) activity could instead adjust the frequency of transcriptional bursting and increase or decrease noise levels. Frequency-modulated signal encoding has been recognized as a regulatory strategy used by biological systems (Locke et al., 2011), and it is conceivable that transcriptional bursting frequency could play a broader role in controlling other biological processes. Regulation of transcriptional elongation is an emerging theme in many areas of biology, from embryonic fly development to mammalian stem cell fate choice (Core and Lis, 2008). Control at this post-initiation step of transcription may be advantageous to biological systems as it offers the potential for faster and more synchronous regulation in response to signaling factors through control of pause release. Surprisingly, the principal modulators of burst size identified by Weinberger et al. (2012) act at the level of elongation rather than at earlier steps of transcription, indicating that PolII processivity may be an actively regulated process. Importantly, the approach taken here allows for dissection of regulation at distinct steps of transcription.
While it is clear that different genes can exhibit unique noise profiles and transcriptional bursting kinetics, the functional consequences of gene-specific noise remain largely unknown. Might there be particular genes, environmental conditions, or lifecycle stages at which regulation of noise levels is particularly advantageous? Just as it is widely recognized that mean gene expression levels respond to environmental stimuli and developmental cues, noise levels, burst size, and burst frequency also represent distinct parameters that could potentially be manipulated to govern cell behavior (Blake et al., 2006; Locke et al., 2011; Weinberger et al., 2005). In the post-genome age, a great deal of effort has gone into understanding the changes in mean gene expression levels that occur as cells change state. Deciphering the structure of gene expression noise and dynamics in the context of biological programs represents an emerging frontier in understanding how evolution shapes transcriptional programs, and it will be interesting to see how these concepts extend to metazoan cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Mol Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Blake WJ, MKA, Cantor CR, Collins JJ. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Cairns BR. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Locke JC, Young JW, Fontes M, Hernandez Jimenez MJ, Elowitz MB. Science. 2011;334:366–369. doi: 10.1126/science.1208144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- Weinberger L, Voichek Y, Tirosh I, Hornung G, Amit I, Barkai N. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.05.008. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Whitelaw NC, Chong S, Whitelaw E. Dev Cell. 2010;19:649–650. doi: 10.1016/j.devcel.2010.11.001. [DOI] [PubMed] [Google Scholar]