Abstract
While lysine acetylation in the nucleus is well characterized, comparatively little is known about its significance in cytoplasmic signaling. Here we show that inhibition of the Sirt1 deacetylase, which is primarily cytoplasmic in cancer cell lines, sensitizes these cells to caspase-2-dependent death. To identify relevant Sirt1 substrates, we developed a novel proteomics strategy, enabling the identification of a range of putative substrates, including 14-3-3ζ, a known direct regulator of caspase-2. We show here that inhibition of Sirtuin activity accelerates caspase activation and overrides caspase-2 suppression by nutrient abundance. Furthermore, 14-3-3ζ is acetylated prior to caspase activation, and supplementation of Xenopus egg extract with glucose-6-phosphate, which promotes caspase-2/14-3-3ζ binding, enhances 14-3-3ζ-directed Sirtuin activity. Conversely, inhibiting Sirtuin activity promotes 14-3-3ζ dissociation from caspase-2 in both egg extract and human cell lines. These data reveal a role for Sirt1 in modulating apoptotic sensitivity, in response to metabolic changes, by antagonizing 14-3-3ζ acetylation.
Introduction
Protein lysine acetylation is a reversible post-translational modification (PTM), controllled by several families of acetylases and deacetylases, and is known to regulate diverse cellular processes [reviewed in (Norris et al., 2009)]. While acetylation has been well studied in the context of histones, recent reports demonstrated the high prevalence of acetylation on non-histone proteins, particularly those involved in central metabolic pathways, as well as hundreds of other proteins in different cellular subcompartments (Choudhary et al., 2009; Kim et al., 2006; Lombard et al., 2007; Wang et al., 2010; Zhao et al., 2010). Notably, certain deacetylases are either expressed exclusively in the cytoplasm or in some cases, may shuttle between the nucleus and cytoplasm (e.g., Sirt1, Sirt2, HDAC6).
Sirtuin deacetylases (Sirt1–7 in mammals) have been implicated in aging and many age-related diseases including neurodegenerative disorders, heart disease, and cancer (Imai and Guarente, 2010). Sirt1, the sirtuin most commonly linked to apoptosis, is upregulated in numerous cancers and has been shown to protect tumor cells from apoptosis (Chen et al., 2005; Huffman et al., 2007; Liu et al., 2006; Liu et al., 2009). Sirt1 is overexpressed in chemoresistant leukemia, osteosarcoma, neuroblastoma, ovarian and breast cancer cells compared to their chemosensitive counterparts, and tumor biopsies from cancer patients given chemotherapeutics have been reported to exhibit higher levels of Sirt1 as compared to those from untreated patients (Chu et al., 2005). Possible mechanisms to explain the antiapoptotic effect of Sirt1 involve its modulation of histones, resulting in suppression of tumor repressor gene transcription, and its deacetylation of transcription factors that regulate cell survival, including p53, and FOXO (Brunet et al., 2004; Luo et al., 2001; Wang et al., 2006).
Motivated in part by our observation of Sirt1 cytoplasmic localization in cancer cells, we devised an unbiased proteomics approach to identify cytoplasmic substrates of Sirt1. This approach yielded several putative Sirt1-targeted proteins, mostly in the areas of glycolysis/metabolism, oxidative stress, cytoskeletal dynamics, and apoptosis. Prominent among these newly identified substrates was the small acidic phosphobinding protein, 14-3-3ζ.
This identification of 14-3-3ζ as a Sirt1 substrate was of particular interest, as we had previously observed in Xenopus eggs/oocytes that 14-3-3ζ governed the activation of the pro-apoptotic protease, caspase-2 (C2) (Nutt et al., 2009). Specifically, under nutrient replete conditions, in fresh oocytes or egg extract, high levels of pentose phosphate pathway (PPP) activity stimulated a CaMKII-dependent suppressive phosphorylation of C2 at Ser135 (Nutt et al., 2005), which was protected from dephosphorylation by 14-3-3ζ binding to C2. Indeed, the exhaustion of metabolites with time in Xenopus egg extracts led to 14-3-3ζ release from C2, exposing Ser135 to PP1-mediated dephosphorylation, and triggering C2 activation and apoptosis. Conversely, supplementation of extract with excess glucose-6-phosphate (G6P), to stimulate PPP activity, maintained the binding between 14-3-3ζ and C2 and suppressed caspase activation (Nutt et al., 2009; Nutt et al., 2005).
Extending this previous work, we show here that acetylation of 14-3-3ζ occurs as PPP activity wanes and promotes release of 14-3-3ζ from C2. Conversely, stimulation of the PPP with G6P promotes 14-3-3ζ-directed Sirtuin activity. In breast tumor cell lines, in which we observe aberrant cytoplasmic localization of Sirt1 compared to normal breast epithelial cells, Sirt1 inhibition enhanced the cells’ sensitivity to paclitaxel, while RNAi ablation of C2 abrogated this effect. Moreover, Sirt1 inhibition caused dissociation of 14-3-3ζ from C2 in cultured cells. These findings point to a critical link between Sirt1 and 14-3-3ζ acetylation to control C2-dependent cell death in response to chemotherapeutic agents.
Results
Inhibition of cytoplasmic Sirt1 in breast cancer cell lines enhances taxol- induced cell death
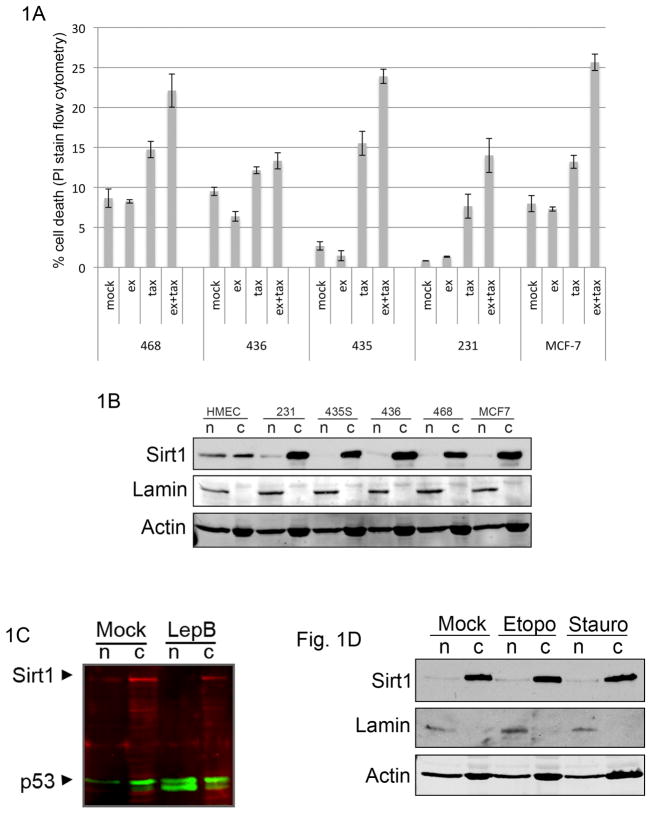
The potential role for Sirt1 in breast cancer is underscored by observations that DBC1 (deleted in breast cancer-1), a direct inhibitor of Sirt1 (Kim et al., 2008; Zhao et al., 2008), can be lost genetically in breast tumors by deletion of the 8p21 chromosomal region (Hamaguchi et al., 2002), or functionally, by DBC1 losing its ability to interact with Sirt1 (Kim et al., 2009). Since much of the effect of Sirt1 on apoptosis is attributed to its role in transcriptional regulation, we were interested in determining whether Sirt1 inhibition might affect cell sensitivity to death under conditions in which transcription is not ongoing (e.g., taxol-induced mitotic arrest). To test this, we treated a panel of breast tumor cell lines with combinations of taxol and the Sirt1-specific inhibitor ex527 (Solomon et al., 2006). As shown in in figure 1A, while we observed a modest level of cell death induced by taxol alone in different breast cancer cell lines, there was a significant increase in taxol-induced death in most cell lines when Sirt1 was inhibited. Based on previous reports demonstrating acetylation of numerous cytoplasmic proteins, we were interested in determining Sirt1 localization in these cell lines. In contrast to an even distribution of Sirt1 in the nucleus and cytoplasm of normal human mammary epithelial cells, Sirt1 was predominantly cytoplasmic in each of the breast cancer cell lines examined (figure 1B). We also observed a similar pattern of cytoplasmic Sirt1 localization in a panel of ovarian cancer cell lines (figure S1). Sirt1 has previously been reported to be capable of shuttling between the nucleus and cytoplasm in a chromosome region maintenance-1 (Crm1)-dependent manner (Tanno et al., 2007). To test whether the cytoplasmic localization of Sirt1 in breast cancer cell lines was due to an increase in Sirt1-directed Crm1 activity, we treated MDA-MB-231 cells with leptomycin B, an inhibitor of Crm1. Although leptomycin B caused nuclear accumulation of p53, it failed to induce nuclear accumulation of Sirt1 (figure 1C), suggesting that a lack of nucleocytoplasmic shuttling in breast cancer cell lines, as opposed to upregulated Crm1-dependent nuclear export, leads to Sirt1’s cytoplasmic distribution. Moreover, treatment of cells with apoptotic stimuli did not induce nuclear translocation of Sirt1 (figure 1D). Together, these data suggest that Sirt1 modulates targets in the cytoplasm that regulate sensitivity to cell death.
Figure 1. Inhibition of cytoplasmic Sirt1 enhances caspase-2-dependent cell death in breast cancer cell lines.
(A) Breast cancer cell lines (MCF-7, MDA-MB-435, MDA-MB-468, MDA-MB-231, and MDA-MB-436) were treated with 100 uM ex527 and/or 50 nM taxol. Cell death was measured 36 hours after taxol treatment by propidium idodide staining and FACS analysis. Results of three independent experiments are summarized with error bars representing standard error. (B) Breast cancer cell lines from panel A were fractionated into nuclear (n) and cytoplasmic (c) fractions. Each fraction was immunoblotted for Sirt1, lamin and actin. (C) MDA-MB-231 cells were treated with 180 nM leptomycin B or control, fractionated as in panel C then immunoblotted for Sirt1 (red) and p53 (green). Proteins visualized by Li-Cor imaging. (D) MDA-MB-231 cells were incubated with etoposide (30 h) or staurosporine (4 h), fractionated, and immunoblotted for Sirt1, lamin, and actin.
A biotin-switch based proteomics approach to identify deacetylase substrates
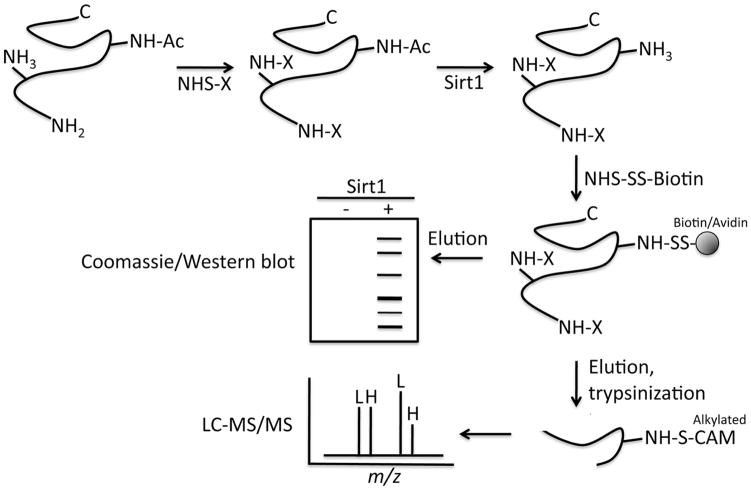
Protein acetylation/deacetylation is well characterized in the context of histone modifications and transcription (most deacetylase substrates identified to date are nuclear proteins), yet recent reports (Choudhary et al., 2009; Kim et al., 2006; Wang et al., 2010; Zhao et al., 2010) raise the possibility that the success of deacetylase inhibitors in cancer may be, in part, due to their regulation of targets outside the nucleus. This is supported by observations that cancer-implicated deacetylases show cytoplasmic localization (Byles et al., 2010; North et al., 2003; Zhang et al., 2004), and that clinical deacetylase inhibitors modulate the acetylation of numerous cytoplasmic proteins (Choudhary et al., 2009). Furthermore, our initial observation that cytoplasmic Sirt1 seemed to modulate the apopototic response to taxol, prompted our interest in identifying cytoplasmic Sirt1 substrates. Our laboratory has previously used Xenopus egg extracts as a model system to study cell cycle and apoptotic pathways. One feature of this system that makes it amenable to proteomics approaches is the ease with which one can collect grams of biologically active concentrated cytosol stably poised at a given cell cycle phase. Therefore, we devised a proteomics approach to identify deacetylase substrates that takes advantage of this and other aspects of an in vitro system.
The technique presented here, termed acetyl-bst, is based on biotin-switch (bst) methodology and N-hydrosuccinimide (NHS) chemistry, and builds upon two approaches designed for the study of s-nitrosothiols (Hao et al., 2006; Jaffrey and Snyder, 2001). Acetyl-bst is performed in cell extracts where NHS forms a stable amide bond with the ε-amine of lysine residues and protein N-termini. As depicted in figure 2, it involves three main steps: Cell extracts are treated with N-succinimidyl-N-methlycarbamate (NSMC) to block PTM-free lysines, producing a mixture of proteins with blocked lysines and lysines previously acetylated by endogenous acetylases. Following blocking, the extract is incubated with deacetylase and the acetylated lysines freed by deacetylase activity are derivatized with NHS-SS-biotin and captured on streptavidin resin. Once captured, the putative deacetylase substrates are removed from SS-biotin with a reducing agent, and the resulting free sulfhydryl is alkylated with iodoacetemide. The identity of the biotin-retrieved peptides is determined by LC-MS/MS and the alkylated lysines (those freed by deacetylase activity) are marked as putative deacetylase-targeted residues.
Figure 2.
Summary of steps in acetyl-bst proteomics approach.
Results of acetyl bst and validation of substrates in mammalian cells
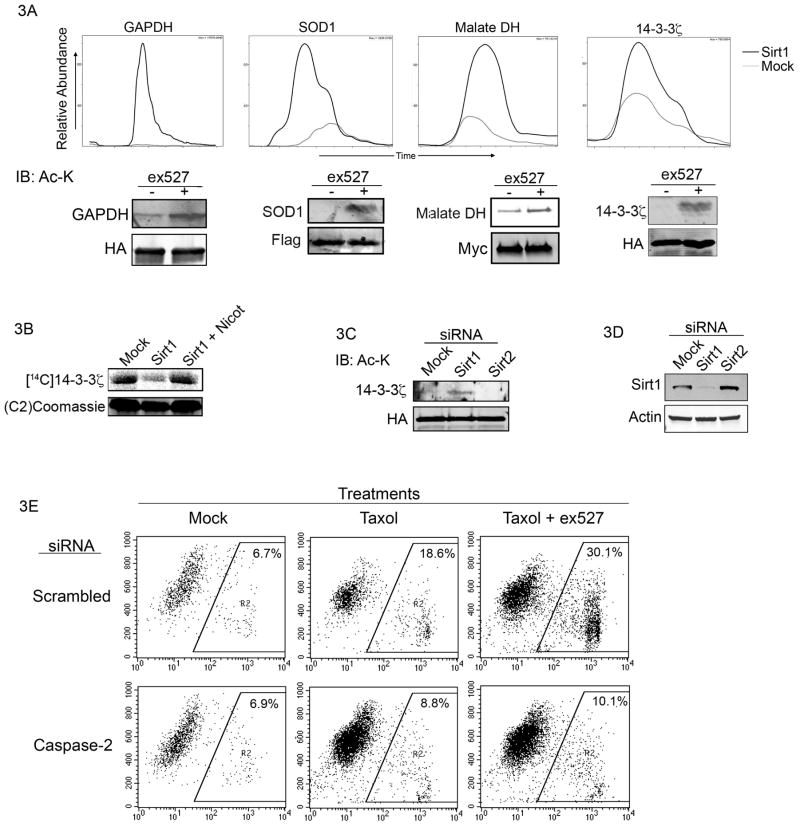
Using the acetyl-bst method, we identified several putative Sirt1 substrates in purified egg cytosol, as shown in Table 1. These substrates primarily group into the areas of cytoskeletal rearrangement, metabolism, and apoptosis/stress, all areas in which Sirt1 has been previously implicated. To validate a subset of substrates in mammalian cells, we expressed epitope-tagged superoxide dismutase-1 (SOD-1), glyceraldehyde-3-phosphate dehydrogenase (GAPHD), malate dehydrogenase (MDH), and 14-3-3ζ in 293T cells treated with ex527 or vehicle control. Individual tagged proteins were then immunoprecipitated and blotted with anti-acetyl lysine antibody. While baseline acetylation levels varied between proteins, all showed an increase in acetylation in the presence of Sirt1 inhibitor (figure 3A). These results indicate that Sirt1 mediates deacetylation of these substrates in intact cells.
Table 1.
Putative Sirt1 substrates identified by acetyl-bst proteomics approach.
| NCBI accession number | Putative Sirt1 substrate | Biological process/pathway | Putative Sirt1-targeted residue |
|---|---|---|---|
| 148224415 | L-lactate dehydrogenase | Glycolysis | K82 |
| 27882192 | Glyceraldehyde 3- phosphate dehydrogenase | Glycolysis, apoptosis | K92, K249, K252 |
| 147903225 | Glutamic-oxaloacetic transaminase | Amino acid metabolism | K274, K288 |
| 148236351 | Triosephosphate isomerase | Glycolysis, pentose shunt, fatty acid biosynthesis, gluconeogenesis | K140, K151, K165 |
| 1655706 | Nucleoside diphosphate kinase | Citric acid cycle | K101 |
| 148223868 | Malate dehydrogenase | TCA cycle | K236 |
| 6468 | α–enolase | Glycolysis, transcription | K28 |
| 1360640 | 14-3-3ζ | Apoptosis | K49 |
| 147906753 | Superoxide dismutase [Cu, Zn] | Oxidative stress | K95 |
| 148233020 | Suppressor of tumorigenicity 13 (HIP) | HSP90, HSP70 signaling | K184 |
| 147905238 | Park7 (DJ-1) | Oxidative stress, apoptosis | K158 |
| 148231458 | Stathmin | Microtubule dynamics | K119, K126, K128, K129 |
| 148232082 | Cofilin-1-A | Actin regulation, cell cycle | K73, K92 |
| 147902214 | Protein LBH | Transcription, MAPK pathway | K37 |
| 147906019 | 14-3-3theta | Transcription, cell cycle | K49 |
| 148223435 | RAN binding protein 1 | RAN signaling, cell cycle | K68 |
| 148236373 | Calmodulin | Ca2+ signaling | K95 |
| 112419222 | Superoxide dismutase [Cu, Zn]-like protein | Oxidative stress | K126 |
| 49118079 | Similar to fatty acid binding protein-4 | Fatty acid transport | K12 |
| 32766481 | Polyubiquitin c | DNA damage, cell cycle, apoptosis | K63 |
Figure 3. Validation of a subset of putative Sirt1 substrates, and C2 involvement in the effect of Sirt1 inhibition on taxol-induced death.
(A) GAPDH, SOD-1, 14-3-3ζ, and malate dehydrogenase were expressed in 293T cells, immunoprecipitated and immunoblotted with anti-acetylated lysine antibody. The chromatographs show LC-MS/MS quantitation of the relative abundance of each protein captured by acetyl-bst in the presence or absence of Sirt1. (B) His-14-3-3ζ was acetylated (14C acetyl-CoA) then retrieved, washed, and incubated with Sirt1 in the presence or absence of nicotinamide. Radiolabled 14-3-3ζ was resolved by SDS-PAGE/phosphorimager. (C) MDA-MB-231 cells were transfected with HA-tagged 14-3-3ζfollowed by transfection with siRNA (20 uM) 24 h later. 48 h after transfection, HA-14-3-3ζ was immunoprecipated and immunoblotted with anti-acetylated lysine antibody. (D) MCF-7 cells were transfected with 20 uM scrambled or C2-targeted smartpool siRNA then treated with ex527/taxol and analyzed for cell death as in panel A.
Validation of 14-3-3ζ as a substrate of Sirt1 implicates C2 in the apoptotic effect of Sirt1 inhibition
To further validate 14-3-3ζ as a direct target of Sirt1, we acetylated 14-3-3ζ with the recombinant catalytic domain of the acetylase p300 in the presence of acetyl-1-14C acetyl-CoA (though it is not known if p300 is the bona fide acetylase for 14-3-3, we achieved robust acetylation in vitro), and incubated the acetylated 14-3-3ζ with recombinant Sirt1, with or without nicotinamide. As shown in figure 3B, recombinant Sirt1 deacetylates 14-3-3ζ in vitro. To measure the effect of Sirt1 inhibition on 14-3-3ζ acetylation in breast cancer cells, in which we had previously observed cytoplasmic Sirt1 and a Sirt1-dependent response to taxol, MDA-MB-231 cells were co-transfected with epitope-tagged 14-3-3ζ and siRNA to Sirt1, Sirt2 or a scrambled control. As shown in figure 3C, RNAi to Sirt1, but not Sirt2, led to an increase in 14-3-3ζ acetylation. Knockdown efficiency was assesed by Western blot (figure 3D). Given these data and our previous observation that 14-3-3ζ regulates C2 activation, we decided to revisit our initial observation (figure 1A) to see if the enhancement of apoptosis by Sirt1 inhibition was dependent on C2. As shown in figure 3E, knockdown of C2 by siRNA in MCF-7 cells markedly reduced cell death induced by taxol, and abrogated the additional effect of Sirt1 inhibition on cell death.
Acetylation of 14-3-3ζ is metabolically regulated and modulates its binding to C2 in Xenopus egg extract and human tissue culture cells
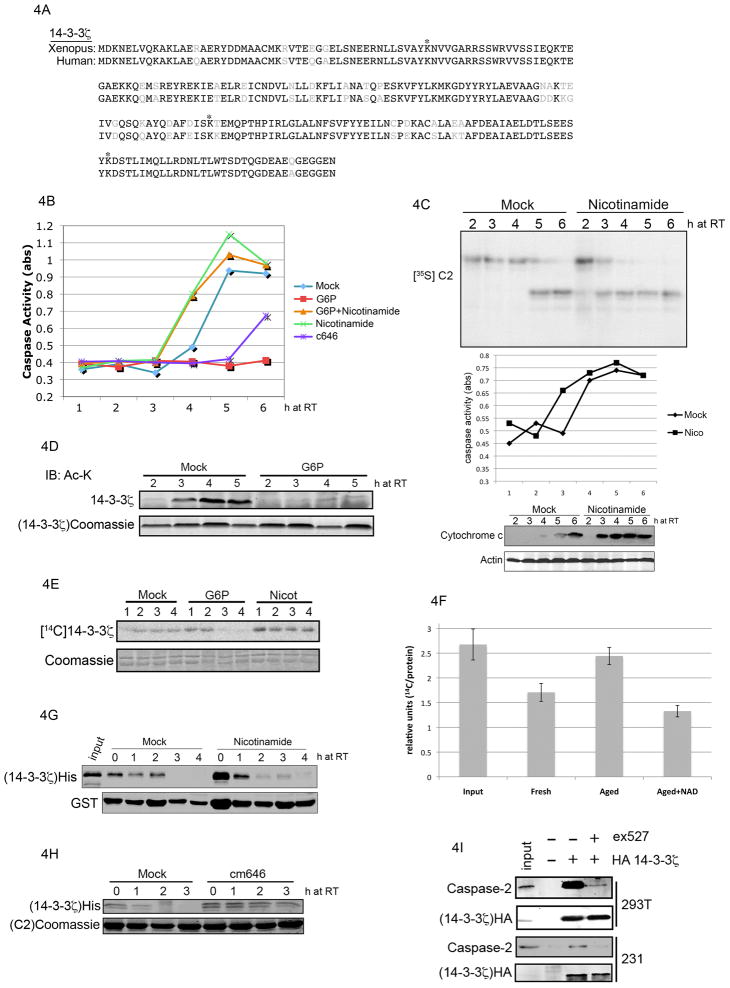
We had previously observed in Xenopus oocytes and egg extracts that declining nutrient levels trigger apoptosis by inducing release of 14-3-3ζ from C2, which leads to C2 activation and downstream apoptotic events (Nutt et al., 2009). However, the molecular events linking a drop in nutrient abundance to 14-3-3ζ release from C2 in extract were unclear. The validation of 14-3-3ζ as a target of Sirt1 suggested that Sirt1 and C2 might be linked via 14-3-3ζ. As shown in figure 4A, Xenopus and human 14-3-3ζ display a high degree of sequence conservation. As a first step to determine the plausibility of 14-3-3ζ acetylation as a regulatory event in the Xenopus system, we performed tandem mass spectrometry with recombinant 14-3-3ζ that had been incubated in control or G6P-supplemented (i.e., nutrient replete) extract. As denoted by asterisks in figure 4A, MS sequencing revealed acetylation at lysine 49, 157 and 212. A quantitative comparison between 14-3-3ζ from the mock- and G6P-treated extract showed lower levels of 14-3-3ζ acetylation in the presence of excess G6P.
Figure 4. Metabolic regulation of 14-3-3 and C2 via sirtuin deacetylase activity.
(A) Sequence alignment of human and Xenopus 14-3-3ζ showing acetylated lysines (asterisks) identified by mass spectrometry in Xenopus egg extract. (B) Egg extract was incubated with combinations of nicotinamide (20 mM), c646 (500 uM), and G6P (5mM). Caspase activity was determined by a colorimetric measurement of DEVD-pNA cleavage. (C) 35S-labeled C2 was incubated in mock- or nicotinamide-treated egg extract, and samples were resolved by SDS-PAGE/phosphorimager (top panel). Caspase activity was measured (middle panel) as in panel B. In parallel, aliquots of extract were fractionated to produce mitochondria-free cytosol, and immunoblotted for cytochrome c and actin (lower panel). (D) His-tagged recombinant 14-3-3ζon resin was incubated in extract and retrieved for immunoblotting with anti-acetylated lysine antibodies. Lower panel shows caspase activity in parallel. (E) His-tagged recombinant 14-3-3ζ on resin was acetylated with 14C-labeled acetyl CoA and incubated in mock-, G6P- or nicotinamide-treated egg extract. 14-3-3ζ was retrieved and resolved by SDS-PAGE/phosphorimager. (F) 14C-acetylated 14-3-3ζ was incubated in fresh, or aged (3 hr) extract supplemented with 2mM NAD or buffer control. Deacetylation was carried out for 60 min and quantified by phosphorimager and normalized to protein levels as determined by coomassie blue staining and Li-Cor imaging. Results of 3 experiments are summarized (error bars represent standard error). (G) Resin-bound GST-C2 was phosphorylated by CaMKII in buffer then allowed to bind His-14-3-3ζ in vitro Bound complex was incubated in mock- or 20 mM nicotinamide-treated extract. GST-C2 was retrieved and immunoblotted for 14-3-3ζ (His). (H) The experiment was performed as in panel F, but in the presence or absence of the acetylase inhibitor c646. (I) 239T and MDA-MB-231 cells were transfected with HA-tagged 14-3-3ζ followed by ex527 (100 uM) treatment for 6 h. HA-14-3-3ζ immunoprecipitates were immunblotted for endogenous C2.
To step back and more broadly test whether sirtuin inhibition might regulate caspase activation in Xenopus egg extracts, we treated extract with the sirtuin inhibitor nicotinamide and measured caspase activity over time. Since we had previously shown that G6P-mediated stimulation of the PPP blocked C2 activation by promoting 14-3-3ζ binding, we were most interested in determining whether sirtuin inhibition would override the anti-apoptotic effect of G6P. Indeed, treatment with nicotinamide accelerated caspase activation and overrode the effects of G6P (figure 4B), while inhibitors of class I and II HDACs had no effect (data not shown). Conversely, treatment of extract with c646, an acetylase inhibitor (Bowers et al., 2010), partially suppressed caspase activation (figure 4B). Moreover, the acceleration of caspase-3 activation was due to the modulation of upstream apoptotic signaling event(s), as mitochondrial cytochrome c release and C2 processing were also accelerated (figure 4C).
We had previously reported that mutation of lysine 49 (one of the residues targeted by Sirt1 based on our proteomics results) of 14-3-3ζ to the negatively charged glutamic acid, abrogated 14-3-3ζ/C2 binding (Nutt et al., 2009). We postulated that the addition of an acetyl group would have a similar effect. This idea is supported by recent work from Choudhary et al. (2009). We reasoned that if acetylation of 14-3-3ζ were promoting its dissociation from C2, its acetylation should correlate with caspase activation and be suppressed by G6P. As shown in figure 4D, we observed 14-3-3ζ acetylation prior to detecting caspase-3 activity in the extract, and supplementation with G6P suppressed both 14-3-3ζ acetylation and caspase-3 activity (corroborated by proteomics results in panel A). Taken together, these data suggest that G6P either suppresses acetylase activity or enhances deacetylase activity. Our observation that nicotinamide accelerated caspase activation suggested that G6P might exert its antiapoptotic effect by enhancing/sustaining deacetylase activity. To test this, we acetylated resin-bound 14-3-3ζ with 14C radiolabel (as in figure 2B) and incubated it in extract treated with G6P, nicotinamide or control. As shown in figure 4E, supplementation of extract with G6P accelerated deacetylation of 14-3-3ζ, while nicotinamide suppressed deacetylation. Moreover, we observed a drop in 14-3-3ζ-directed deacetylase activity in extracts aged on the bench to deplete nutrients; importantly, supplementation of aged extract with 2 mM NAD recovered deactylation of 14-3-3ζ, (figure 4F).
Given the data above and our previous observation that nicotinamide accelerated C2 processing in extract, we were interested in determining whether nicotinamide treatment might accelerate 14-3-3ζ release from C2. To test this, we allowed His-14-3-3ζ and resin-bound GST-C2 to bind in buffer then incubated the bound complex in extract treated with nicotinamide or vehicle control. At specified timepoints, we retrieved the C2 resin, and immunoblotted for His-14-3-3ζ. As shown in figure 4G, nicotinamide accelerated the release of 14-3-3ζ from C2. Conversely, treatment of extract with acetylase inhibitor delayed release (figure 4H). Taken together, our results suggest that nutrient levels modulate the binding of 14-3-3ζ to C2 by regulating its acetylation. Based on this model, assuming conservation between amphibian and mammalian systems, one prediction would be that human cultured cells grown in abundant glucose should show 14-3-3ζ/C2 binding. Moreover, inhibition of Sirt1 should override the effects of glucose and trigger 14-3-3ζ release from C2. To test this, we immunoprecipitated overexpressed HA-tagged 14-3-3ζ from HEK 293T and MDA-MB-231 cells grown in abundant glucose and incubated with or without Sirt1 inhibitor, followed by immunoblotting for C2. In support of our previous observations in extract, we detected binding between 14-3-3ζ and C2, while inhibition of Sirt1 disrupted the interaction between 14-3-3ζ and C2 (figure 4I). Taken together, these data suggest that nutrient-regulated deacetylation of 14-3-3ζ by Sirt1 controls 14-3-3ζ/C2 interactions.
Discussion
Despite the high prevelance of acetylation among metabolic enzymes and many other cytoplasmic proteins, relatively few Sirtuin substrates have been identified, at least partly due to the fact that linking substrates to particular deacetylases has been done largely by co-immunoprecipitation or testing specific deacetylases based on prior knowledge of a given target’s acetylation. The acetyl-bst approach provides a means to isolate deacetylase targets in a high throughput and unbiased manner, providing that the deacetylase can be made recombinantly. The acetyl-bst approach might be enhanced by using lysates from tissue culture cells in which endogenous deacetylases are knocked down by RNAi. In addition, this approach is amenable to identifying substrates within a specific subcellular compartment (Sirt3 substrates in mitochondria, for example) by using enriched biochemical fractions as the starting material.
Our data raise the question of how G6P upregulates the activity of Sirtuins. One possibile explanation is that PPP-mediated production of the ribose-5-phosphate backbone may lead to increased NAD+ levels via de novo nucleotide synthesis. Indeed, addition of NAD+ to nutrient-depleted extracts was able to restore 14-3-3ζ deacetylation (Fig. 4F). In addition, while there is thought to be low glycolytic activity in the oocytes/eggs, the addition of G6P may yield metabolic intermediates (pyruvate, for example) that feed into the TCA cycle to maintain a high NAD/NADH ratio. NADPH produced by the PPP might also function as an allosteric inhibitor of the NAD+ kinase required for conversion of NAD+ to NADP; thus NADPH might decrease NAD+ utilization, leading to its build-up (Zerez et al., 1987). It is also plausible that NADPH is converted directly to NAD+, but there is no known eukaryotic NADPH phosphatase (though there is an archae inositolmonphosphatase with reportedly high NADPH phosphatase activity; Fukuda et al., 2007).
Interestingly, perturbation of nutrient levels has been previously shown to affect 14-3-3 binding. In both arabidopsis and human tissue culture cells, glucose withdrawal/serum starvation induces the release of 14-3-3 from various binding partners (Cotelle et al., 2000; Pozuelo-Rubio, 2011). Moreover, exposure of cells to survival factors induces binding between 14-3-3 and the pro-apoptotic protein Bad, resulting in Bad suppression (Datta et al., 2000). In light of these studies and the possibility that acetylation of 14-3-3 might regulate its binding to substrates in these scenarios, we were intrigued by the fact that, in our hands, 14-3-3ζ acetylation induced by Sirt1 inhibition (in the absence of an apoptotic stimulus like taxol) did not result in dramatic cell death despite release of C2. One possible explanation is that release of 14-3-3ζ from C2 simply primes a cell for death, and a second stimulus (e.g., Pidd/RAIDD induction or activation of BH3 only proteins) is needed to drive a cell into apoptosis.
In addition to 14-3-3ζ, we identified a number of other putative substrates of Sirt1, a subset of which were validated in tissue culture cells. SOD-1 and DJ-1 stand out as candidates that may play a role in Sirt1-mediated inhibition of oxidative stress. Consistent with the emerging notion that acetylation is a central PTM in metabolic pathways (Wang et al., 2010; Zhao et al., 2010), the largest subset of putative Sirt1 targets identified here includes metabolic enzymes, suggesting that cytoplasmic Sirt1, in addition to regulating the transcription of metabolic genes via PGC-1α in the nucleus, may directly modulate these pathways when expressed in the cytosol. Interestingly, nearly all of the proteins identified in our screen here have been reported as acetylated proteins previously (Choudhary et al., 2009). Like kinase/phosphatase systems, acetylases/deacetylases are likely to regulate a diverse array of cellular processes. The development of methods to identify such substrates, such as the one presented here, will allow broad isolation of deacetylase substrates, which will be illuminating in much the same way that identification of phosphatase targets has provided insight into phospho-dependent signaling pathways.
Materials and Methods
Acetyl-biotin switch and mass spectrometry analyses
See supplemental methods for detailed protocols
Apoptosis and biochemical assays
35S-labeled proteins were made as described previously (Andersen et al., 2009). For anti-acetyl-lysine Western blots, combinations of the following antibodies were used: Immunechem (ICP0380-100), Cell Signaling Technology (9441), and Upstate (06-933). Caspase assays, cytochrome c fractionation and propidium iodide staining were performed as described previously (Andersen et al., 2009). In vitro CaMKII assay was done as described previously (Nutt et al., 2009). For 14-3-3ζ release experiments, resin-bound GST-C2 prodomain was incubated with His-14-3-3ζ in PBS for 30 min at room temperature, followed by washing with PBS and 300 mM NaCl. C2/14-3-3 complex was then dipped in extract as described in the text. For deacetylation assays, recombinant resin-bound His-14-3-3ζ was acetylated with p300 catalytic domain (Enzo Life Sciences) and radiolabeled acetyl CoA [acetyl-1-14C] (PerkinElmer) in buffer (50 mM HEPES pH 8, 10% glycerol) for 30 min at room temperature. 14-3-3ζ was then washed and dipped in Xenopus extract or in buffer (25 mM Tris pH 7.5, 100 mM NaCl, 0.5 mM NAD, and 1 mM DTT) with recombinant Sirt1 (Enzo Life Sciences).
Supplementary Material
Highlights.
Inhibition of cytoplasmic Sirt1 in breast tumor cell lines sensitizes to apoptosis
A proteomics approach identifies cytoplasmic substrates of Sirt1
Sirt1 deaceytlates 14-3-3ζ
Acetylation of 14-3-3ζ modulates its binding to caspase-2
Acknowledgments
We thank Dr. Philip Cole for providing acetylase inhibitor and all members of the Kornbluth lab for scientific input. This work was supported by a National Institutes of Health R01 grant GM080333 to SK and an American Cancer Society New England Division-SpinOdyssey Postdoctoral Fellowship PF-08-263-01-CCG to JLA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen JL, Johnson CE, Freel CD, Parrish AB, Day JL, Buchakjian MR, Nutt LK, Thompson JW, Moseley MA, Kornbluth S. Restraint of apoptosis during mitosis through interdomain phosphorylation of caspase-2. EMBO J. 2009;28:3216–3227. doi: 10.1038/emboj.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Byles V, Chimelewski LK, Wang J, Zhu L, Forman LW, Faller DV, Dai Y. Aberrant Cytoplasm Localization and Protein Stability of SIRT1 is Regulated by PI3K/IGF-1R Signaling in Human Cancer Cells. Int J Biol Sci. 2010;6:599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Chu F, Chou PM, Zheng X, Mirkin BL, Rebbaa A. Control of multidrug resistance gene mdr1 and cancer resistance to chemotherapy by the longevity gene sirt1. Cancer Res. 2005;65:10183–10187. doi: 10.1158/0008-5472.CAN-05-2002. [DOI] [PubMed] [Google Scholar]
- Cotelle V, Meek SE, Provan F, Milne FC, Morrice N, MacKintosh C. 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J. 2000;19:2869–2876. doi: 10.1093/emboj/19.12.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, Greenberg ME. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda C, Kawai S, Murata K. NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3′-phosphoadenosine 5′-phosphate phosphatase. Appl Environ Microbiol. 2007;73:5447–5452. doi: 10.1128/AEM.02703-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi M, Meth JL, von Klitzing C, Wei W, Esposito D, Rodgers L, Walsh T, Welcsh P, King MC, Wigler MH. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc Natl Acad Sci U S A. 2002;99:13647–13652. doi: 10.1073/pnas.212516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- Kim JE, Lou Z, Chen J. Interactions between DBC1 and SIRT 1 are deregulated in breast cancer cells. Cell Cycle. 2009;8:3784–3785. doi: 10.4161/cc.8.22.10055. [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69:1702–1705. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Norris KL, Lee JY, Yao TP. Acetylation goes global: the emergence of acetylation biology. Sci Signal. 2009;2:pe76. doi: 10.1126/scisignal.297pe76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Nutt LK, Buchakjian MR, Gan E, Darbandi R, Yoon SY, Wu JQ, Miyamoto YJ, Gibbons JA, Andersen JL, Freel CD, et al. Metabolic control of oocyte apoptosis mediated by 14-3-3zeta-regulated dephosphorylation of caspase-2. Dev Cell. 2009;16:856–866. doi: 10.1016/j.devcel.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt LK, Margolis SS, Jensen M, Herman CE, Dunphy WG, Rathmell JC, Kornbluth S. Metabolic regulation of oocyte cell death through the CaMKII-mediated phosphorylation of caspase-2. Cell. 2005;123:89–103. doi: 10.1016/j.cell.2005.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozuelo-Rubio M. Regulation of autophagic activity by 14-3-3zeta proteins associated with class III phosphatidylinositol-3-kinase. Cell Death Differ. 2011;18:479–492. doi: 10.1038/cdd.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerez CR, Moul DE, Gomez EG, Lopez VM, Andreoli AJ. Negative modulation of Escherichia coli NAD kinase by NADPH and NADH. J Bacteriol. 1987;169:184–188. doi: 10.1128/jb.169.1.184-188.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yamashita H, Toyama T, Sugiura H, Omoto Y, Ando Y, Mita K, Hamaguchi M, Hayashi S, Iwase H. HDAC6 expression is correlated with better survival in breast cancer. Clin Cancer Res. 2004;10:6962–6968. doi: 10.1158/1078-0432.CCR-04-0455. [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.