Abstract
Protein misfolding, aggregation and deposition are common disease mechanisms in many neurodegenerative diseases including Parkinson’s disease. Accumulation of damaged or abnormally modified proteins may lead to perturbed cellular function and eventually to cell death. Thus neurons rely on elaborated pathways of protein quality control and removal to maintain intracellular protein homeostasis. Molecular chaperones, the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal pathway (ALP) are critical pathways that mediate the refolding or removal of abnormal proteins.
The successive failure of these protein degradation pathways, as a cause or consequence of early pathological alterations in vulnerable neurons at risk, may present a key step in the pathological cascade that leads to spreading neurodegeneration. A growing number of studies in disease models and patients have implicated dysfunction of the UPS and ALP in the pathogenesis of Parkinson’s disease and related disorders. Deciphering the exact mechanism by which the different proteolytic systems contribute to the elimination of pathogenic proteins, like α-synuclein, is therefore of paramount importance. We herein review the role of protein degradation pathways in Parkinson’s disease and elaborate on the different contributions of the UPS and the ALP to the clearance of altered proteins. We examine the interplay between different degradation pathways and provide a model for the role of the UPS and ALP in the evolution and progression of α-synuclein pathology. With regards to exciting recent studies we also discuss the putative potential of using protein degradation pathways as novel therapeutic targets in Parkinson’s disease.
Keywords: Parkinson’s disease, neurodegeneration, α-synuclein, autophagy, lysosome, ubiquitin-proteasome system, molecular chaperones
INTRODUCTION
Protein misfolding, aggregation and deposition are common disease mechanisms shared by many neurodegenerative diseases that are collectively referred to as “proteinopathies” or “protein conformation disorders”. In Parkinson’s disease (PD) the synaptic protein α-synuclein accumulates to form distinctive aggregates termed Lewy bodies and Lewy neurites. Although the mechanisms by which α-synuclein malforms and causes neurodegeneration are not fully elucidated, strong genetic and molecular evidence implicates a crucial role for this protein in PD. Cells and neurons as post-mitotic cells in particular rely on elaborated pathways of protein quality control and removal to maintain intracellular protein homeostasis. Accumulation of damaged or abnormally modified proteins leads to perturbed cellular function and eventually to cell death. Extensive genetic and molecular data in transgenic disease models suggests that many of the proteins responsible for “proteinopathies” cause neuronal dysfunction and neurodegeneration by a toxic gain of function. Thus, it is of great importance to understand the cellular pathways that regulate levels of toxic protein species.
Functional steady-state levels of any given protein are determined by synthesis, post-translational modification, degradation and release. The first line of defense against misfolded proteins prone to aggregation are molecular chaperones that can stabilize or refold such protein species and that, in addition, are also intimately associated with mechanisms of protein degradation [41,63]. Under pathological conditions with massive protein misfolding and cellular stress, however, the subsequent depletion of chaperones and the rising levels of aggregate prone proteins can exceed the refolding capacity of the chaperone system. In this scenario, proteins that no longer can be refolded may be shuttled to proteolytic systems to be eliminated. This is constantly true for the folding of proteins in the endoplasmic reticulum, where a substantial fraction of polypeptides fails to reach their native state and are thus mislocalized to the cytosol where degradation through the proteasome takes place, a process that is referred to as endoplasmic reticulum associated degradation (ERAD) [30,145].
In neurons, the two major proteolytic systems that participate in normal protein turnover and the removal of altered proteins are the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal pathway (ALP). While the UPS degrades most short-lived soluble proteins, the ALP is a degradation process that refers to the degradation of intracellular components, proteins and organelles, in lysosomes. The following sections will review the role of intracellular protein degradation pathways in PD and the different contributions of the UPS and ALP to the clearance of altered proteins. With regards to exciting findings from recent studies, a discussion of the potential of protein degradation pathways as a novel therapeutic target in PD will conclude this review.
DYSFUNCTION OF THE UBIQUITIN-PROTEASOME-SYSTEM IN PARKINSON’S DISEASE
The UPS is the major pathway that mediates the degradation of soluble intracellular proteins in the cytoplasm, nucleus and endoplasmic reticulum (ERAD) [reviewed in 30,50,145,169]. The clearance of proteins by the UPS involves an elaborated sequence of events: First, a chain of activated ubiquitin monomers, a small and stable globular protein, is covalently linked to lysine residues of the substrate protein in an ATP-dependent manner. This process is initiated and orchestrated by the sequential action of ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2) and ubiquitin ligases (E3). The fact that there is at least one E1 enzyme, but more than ten E2 enzymes and more than a thousand E3 enzymes interacting, underscores the high degree of selectivity of UPS-mediated degradation although it is a vital and constitutively active cell function. The ubiquitin chain serves as a signal for recognition by the proteasome. Proteasomes are small, barrel shapes organelles, that consist of a catalytic core (the 20S proteasome) and regulatory subunits that form two 19S “caps” at either end to regulate the entry and catalytic function of the core. The fully assembled proteasome is referred to as the 26S proteasome. The catalytic core is formed by 28 subunits organized in two outer α-rings and two inner β-rings. The latter comprises the proteolytic chamber that holds three major kinds of protease activity: chymotrypsin-like, trypsin-like and peptidyl-glutamyl-peptide hydrolytic activity. Proteasomal degradation yields short peptide fragments that are then subject to further degradation by peptidases, allowing the recycling of amino acids for the synthesis of novel proteins. The ubiquitin chain is also dissembled by a group of enzymes termed ubiquitin c-terminal hydrolases. It is noteworthy that some proteins do not require ubiquitination to undergo degradation by 26S proteasomes and that other proteins are simply degraded by 20S proteasomes, although the exact relevance of both processes in health and disease remains to be determined.
Following the initial finding that proteins in Lewy bodies are highly ubiquitinated [78,84], numerous components of the UPS, such as proteasomal subunits, ubiquitinating and de-ubiquitinating enzymes and proteasome activators were found to be abundantly present in these aggregates as well [59,79,93,141,183]. Genetic hints for an involvement of the UPS were provided by the discovery of mutations in parkin (PARK2; an E3 ligase that can also modulate 26S proteasome activity) [68,158] and UCH-L1 (PARK5; an ubiquitin hydrolase) [85] that can lead to monogenetic forms of PD.
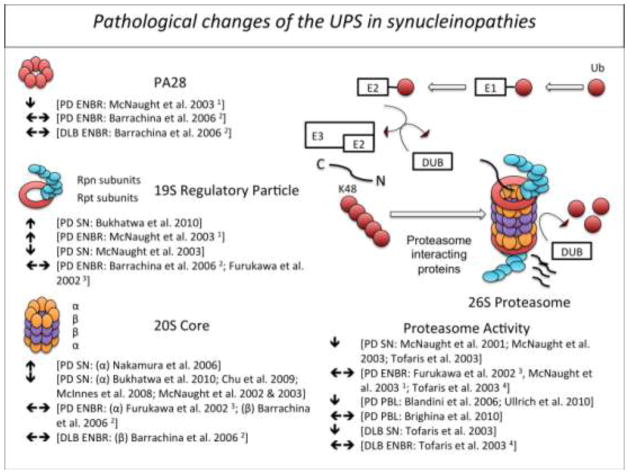
First evidence for malfunction of the UPS in PD was demonstrated by studies that evaluated proteasome activity in post-mortem brain tissue by using enzymatic assays (Table 1A and Figure 1). Importantly, a significant decrease of the chymotrypsin-like and trypsin-like proteasome activity was noticed in the substantia nigra (SN) of PD patients in comparison to age-matched controls [106,108,154]. However, no deficits or even enhanced proteasome activity was found in other brain regions, arguing that a global perturbation of proteasome function is unlikely a cause or consequence of PD pathology [49,154]. It rather seems plausible that the observed reduction of proteasome activity in the SN is a region specific phenomenon that predisposes vulnerable neurons to subsequent pathological changes or vice versa a reduction in proteasome activity could simply represent a secondary phenomenon to the profound neurodegeneration in this region.
Table 1.
UPS and ALP dysfunction in synucleinopathies
| Table 1A. Dysfunction of the UPS in PD & DLB | Reference |
|---|---|
| Reduced proteasome activity in the SN of PD patients | [106,108,154] |
| Reduced expression of proteasomal subunits in the SN of PD patients or cortex of DLB patients | [20,29,95,106,107] |
| Reduced proteasome activity and expression of UPS related proteins in peripheral blood mononuclear cells | [15,157] |
| Altered proteasome function in toxin-based models of PD | [12,21,46,27,163,181] |
| Altered proteasome function in α-synuclein overexpressing rodent models | [24,44,39] |
| Table 1B. Dysfunction of the ALP in PD & DLB | Reference |
| Altered levels of autophagy-related proteins in PD or DLB patients | [1,29,32,35,57,71,86,151,152,179] |
| Altered autophagy function in peripheral blood cells | [123,171] |
| Altered autophagy function in toxin-based models of PD | [86,112,168] |
| Altered autophagy function in α-synuclein overexpressing rodent models | [29,32,179] |
Fig. 1.
Pathological alterations of the ubiquitin-proteasome system in patients with Parkinson’s disease (PD) or Dementia with Lewy bodies (DLB). 1 cerebral cortex, striatum, hippocampus, pons, cerebellum; 2 frontal cortex; 3 putamen, caudate nucleus, frontal cortex, occipital cortex and cerebellar cortex; 4 frontal cortex, cingulate cortex, occipital cortex. DUB (deubiquitinating enzyme); EBNR (extranigral brain region); PBL (peripheral blood lymphocyte); Ub (ubiquitin); SN (substantia nigra)
A similar distribution pattern was observed for levels of proteasomal subunits (Table 1A and Figure 1): Genes for several proteasome subunits were significantly downregulated in the SN of PD patients [53] and expression levels of the 20S proteasome core and α-subunits in particular as well as the 19S regulatory caps were significantly reduced in the SN of PD patients [20,29,106,107] and cortex of Dementia with Lewy bodies (DLB) cases [95], but unchanged or even elevated in other extranigral regions [7,49,106,107]. In addition to studies investigating central proteasome function, a few studies have demonstrated altered proteasome activity in peripheral blood cells of PD patients (Table 1A and Figure 1). Two groups reported reduced 20S proteasome activity in peripheral blood mononuclear cells of PD patients that interestingly seemed to correlate inversely with disease duration and severity suggesting that impaired proteasome function could be an indicator of disease progression [15,157]. Significantly reduced proteasome function was, however, only present in patients treated with both L-dopa and dopamine agonists [15], suggesting an alternative hypothesis by which dopamine levels alter proteasome function, a finding that has been observed under experimental conditions in cultured cells and animal models [66,11,157,177]. A more recent report, however, failed to confirm altered proteasome function in peripheral blood cells of PD patients, arguing that peripheral proteasome function might be a poor disease indicator [19].
A convincing link between proteasome dysfunction and PD has been established by studies in disease models. In α-synuclein-eGFP transfected mouse primary neurons [105], rat ventral mesencephalic primary neurons [109] and cultured PC12 cells [128], application of the proteasome inhibitor lactacystin leads to dose-dependent neurodegeneration and the formation of ubiquitin- and α-synuclein positive inclusions. By systemically applying proteasome inhibitors to rats, McNaught and colleagues seemed to have produced an accurate rat model of PD, both in terms of behavioral or motor deficits and core pathological features [110]. In addition to the progressive and dopamine-responsive motor impairment, treated rats were found to display degeneration of dopaminergic neurons in the SN and decreased levels of dopamine in the striatum. Importantly, surviving neurons were also found to contain α-synuclein and ubiquitin positive inclusion bodies. Hence, this exciting model has received considerable attention, although in the following years several independent laboratories have failed to reproduce most if not all features provided in the initial description [18,75,98,102,140,180]. Only two of five follow-up reports [140,180] replicated dopaminergic cell loss and only one group [180] detected α-synuclein positive aggregates while none could identify a distinct motor phenotype. The extensive variability of findings in this rat model created significant doubt about its utility as an accurate model of PD. Despite this disappointment, related models for proteasome inhibition induced neurodegeneration in rats [45,95,114,161], mice [118,149,172], Medaka fish [103] and cultured-cells [126,139] have been validated and used in subsequent studies.
Besides this, the relation between UPS impairment and sporadic PD has been stressed by studies in toxin-based animal models (Table 1A). In vitro and in vivo studies demonstrated a marked decrease in proteasome activity following exposure to toxins like rotenone or MPTP [12,21,46,27,163,181]. In vivo, toxin induced proteasome inhibition was found to cause neuronal cell loss in the SN, dopamine depletion in the striatum and even a disease phenotype [46]. Surprisingly when repeated in mice lacking α-synuclein, UPS impairment was significantly alleviated suggesting that α-synuclein exacerbates the deleterious effects of MPTP on the UPS [46]. Interestingly, a direct inhibitory effect of α-synuclein on proteasome activity has been reported. A number of studies showed that stably increased levels of α-synuclein can lead to impaired proteasome function, potentially paving the way for subsequent pathological changes [25,44,89,122,146,148,150,182]. The reciprocal interaction between α-synuclein and proteasome function suggests a self-perpetuating process in which permanently raised α-synuclein levels impair the UPS, which can in turn lead to further α-synuclein accumulation.
Apart from the reciprocal interaction with α-synuclein metabolism, a recent study interestingly found that overexpression of LRRK2, a protein in which mutations cause autosomal-dominant forms of PD, in cell culture and zebrafish impairs proteasomal protein clearance without affecting the catalytic activity or expression of proteasome subunits [88]. Another intriguing study found that in the dopamine depleted striatum of MPTP-lesioned monkeys or 6-OHDA-lesioned mice chronic stimulation of D1 receptors via administration of L-Dopa leads to a significantly impaired chymotrypsin-like activity of the proteasome, arguing for a causal role of proteasome dysfunction in L-Dopa induced dyskinesia, a very compelling link between a focal derangement of protein metabolism, neurotransmission and a behavioral phenotype [11].
Finally, an exciting report by Bedford and colleagues has fundamentally underscored the importance of UPS mediated protein homeostasis for neurodegeneration [9]: A mouse model expressing a conditional deletion of the Rpt2/PSMC1 subunit, an ATPase of the 19S regulatory complex, spatially restricted to the forebrain and a second model engineered to lack the Rpt2/PSMC1 subunit in tyrosine hydroxylase (TH) positive neurons were developed and characterized. As the Rpt2/PSMC1 subunit is vital to the assembly and activity of the 26S proteasome this model represents a model for the specific impairment of the 26S proteasome, while the 20S proteasome remains unaffected. Remarkably, α-synuclein and ubiquitin positive inclusions resembling Lewy bodies developed in the brain regions with impaired 26S proteasome function. In the forebrain restricted knockdown model, progressive neurodegeneration and learning deficits were observed while in the knockdown restricted to TH-positive neurons striking neurodegeneration and inclusion body formation in the SN occurred.
DYSFUNCTION OF AUTOPHAGY IN PARKINSON’S DISEASE
The autophagy-lysosomal pathway (ALP) or autophagy (Greek “to eat oneself”) is the general term used to describe pathways that converge into degradation of intracellular proteins or organelles in lysosomes [69,169]. Within the ALP, delivery of targets to the lysosome occurs in three distinct ways that distinguish the respective subtype: macroautophagy, chaperone-mediated-autophagy (CMA) and microautophagy. The final step of all three autophagic processes is the degradation of the substrate protein within lysosomes. Lysosomes are single membrane vesicles that contain a variety of cellular hydrolases including proteases, lipases and glycosidases and that maintain an acidic pH through ATP-dependent proton pumps. Macroautophagy is the “bulk” degradation process that involves the de-novo formation of double membrane bound vesicles that sequester complete regions of the cytosol, including organelles, proteins and lipids. These initial double-membrane vesicles have been termed autophagosomes or autophagic vacuoles. While this compartment lacks proteolytic enzymes, the content can only be degraded by a fusion with lysosomes or late endosomes, which provide the necessary enzymes. The fusion product of autophagosomes and lysosomes is then referred to as an autolysosome.
Interestingly, autophagosomes can be formed anywhere within the cell, independent of the close presence of lysosomes. In neurons for example, autophagosome formation and cargo sequestration can take place in neuronal processes [74]. The long distance from perinuclearly located lysosomes is bridged by cellular transport mechanisms that deliver autophagosomes for the final step of fusion with lysosomes [61]. Without going into much detail, the biogenesis of autophagic vesicles involves 35 autophagy related genes and proteins (Atg) that form two unique ubiquitin-like conjugation systems: The Atg12-Atg5 and the MAP1LC3 (microtubule-associated proteins 1A/1B light chain 3 or LC3 or Atg8)-phosphatidylethanolamine (PE) conjugation [115,117]. Atg12 is linked to Atg5 in a reaction that requires Atg7 and Atg10 (E1- and E2-like enzymes). The Atg12-Atg5 conjugate then associates with Atg16L, which oligomerizes to form a complex. This complex then acts as an E3-like enzyme that determines the sites of LC3 lipidation [48]. Before this step can take place however, LC3 has to be cleaved by Atg4 to generate the cytosolic LC3-I that can subsequently be conjugated to a PE tag by Atg7 and the E2-like enzyme Atg3. The resulting lipidated form of LC3, LC3-II, is attached to both sides of the autophagosome membrane but is removed from the outer membrane before fusion with lysosomes occurs.
Macroautophagy is an evolutionary highly conserved and tightly regulated pathway. Not only a “bulk” degradation process, increasing evidence suggests the existence of very selective subtypes of macroautophagy, governed by specific cargo recognition molecules (e.g. mitophagy or pexophagy – the selective autophagic degradation of mitochondria or peroxisomes respectively [62,70]). Macroautophagy occurs constitutively in cells and is markedly induced under stress conditions, for instance under starvation [77]. This stress response is mediated by a signaling cascade that involves the serine/threonine kinase mTOR (mammalian target of rapamycin), an important target for pharmacological agents that induce or enhance autophagy [116]. More recent studies also demonstrated that macroautophagy can be enhanced in a mTOR-independent manner [138,165], which may provide additional targets for novel autophagy enhancing compounds.
While macroautophagy is a vital high-turnover cellular pathway under stress conditions, e.g. when essential macromolecules are needed under conditions of nutritional scarcity or when altered intracellular components need to be removed to prevent cell death, CMA’s characteristic feature is selectivity. Particular cytosolic proteins that contain a pentapeptide targeting motif (KFERQ) are recognized by molecular chaperones (Hsc70 and co-chaperones) and translocated to the lysosomal surface [5]. Here, chaperones again assist and mediate the binding of the substrate protein to the lysosomal receptor protein LAMP-2a. Assisted by a different set of chaperones located inside the lysosomal compartment, the substrate then crosses the lysosomal membrane in an unfolded state to finally become degraded inside the lysosomal lumen. As it is the case for macroautophagy, CMA is constitutively active in most tissues but becomes recruited under stress conditions. KFERQ-like targeting motifs are estimated to be present in about 30% of all cytosolic proteins, a number that is most likely underrated since post-translational modification can also render proteins prone to recognition by the chaperone-co-chaperone complex.
The third and least well-characterized autophagy subtype, microautophagy, involves the direct sequestration of regions of the cytosol at the lysosomal surface by invagination of the lysosomal membrane. These invaginations then pinch off as vesicles into the lysosomal lumen where rapid degradation takes place [113].
Although each autophagy pathway has individual characteristics, they appear to be interconnected and display compensatory up-regulation upon failure of one of the pathways. Cells respond to a blockage of CMA by activating macroautophagy in a constitutive manner [101]. Although both pathways are not redundant, activation of macroautophagy under basal conditions can preserve protein homeostasis when CMA function is compromised [101]. Vice versa, CMA is enhanced when macroautophagy is dysfunctional [65]. Expanding the crosstalk to non-autophagic pathways, acute UPS inhibition was shown to induce and upregulate autophagic flux in cell culture [38,60,127] and Drosophila melanogaster [120] and recently in human α-synuclein transgenic mice [39]. Chronic blockage of the proteasomal pathway leads to constitutively upregulated macroautophagy that however cannot increase upon exposure to cellular stress, as it would be the case under normal conditions [37]. Interestingly, accumulation of p62, an important autophagic adapter protein, in autophagy deficient cells resulted in impaired clearance of proteins through the UPS, although the catalytic activity of the proteasome remained unaffected [76]. The reduced UPS flux could be relieved upon expression of p97/VCP, a chaperone involved in the acquisition of polyubiquitinated proteins for proteasomal degradation, potentially indicating that the UPS and macroautophagy compete for the clearance of ubiquitinated proteins via ubiquitin-binding proteins and p62 [76]. Crosstalk between the different subtypes of autophagy and coupling to the UPS is of particular interest to neurodegenerative diseases where a primary failure of degradation mechanisms has been proposed [40]. In PD in particular, the accumulation of toxic protein species might be driven by a successive failure of molecular chaperones, the UPS and the ALP [40].
First evidence for an involvement of the ALP in PD was demonstrated by a pathological study that found accumulation of autophagic vacuoles in the SN of PD patients [4]. This accumulation is consistent with either an overproduction (as would be the case following primary UPS failure or as a secondary phenomenon to neurodegenerative changes) or impaired turnover of autophagic vacuoles (as would be the case with primary ALP dysfunction or secondary dysfunction due to neurodegenerative changes). Supportive of both hypotheses, recent studies confirmed the association of autophagic signal molecules and components of CMA [1] and macroautophagy [32,35,151,179] or lysosomes and the ALP in general [29,57,71,86,152] with α-synuclein pathology in both PD and DLB (Table 1B & Figure 2).
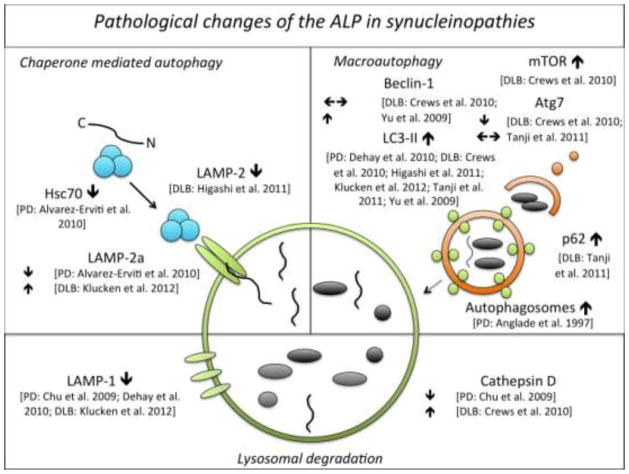
Fig. 2.
Pathological alterations of the autophagy-lysosomal pathway in patients with Parkinson’s disease (PD) or Dementia with Lewy bodies (DLB)
Uniformly, all studies found an increase of the autophagosome marker LC3-II [32,35,57,71,151,179] and a decrease of the lysosomal marker LAMP-1 [29,35,71] in both tissue of PD and DLB patients, suggesting the presence of abundant and dysfunctional autophagosomes and lysosomes in synucleinopathies. Whether the observed increase in LC3 expression directly translates into decreased or increased autophagic processing is currently unknown, however the increase in p62, a selective autophagy substrate that is continuously degraded by macroautophagy [14,73], in DLB as observed by Tanji and colleagues does suggest that autophagic flux is impaired [151]. Furthermore LC3 immunostaining widely co-localized with different types of Lewy pathology arguing for an interaction between autophagy and α-synuclein pathology [1,32,35,57,71,151]. Discrepant findings exist regarding levels of the CMA adapter molecule LAMP-2a and the lysosomal protease cathepsin D: LAMP-2a was significantly reduced in the SN pars compacta and amygdala of PD patients [1], while in the lysosomal enriched fraction of temporal cortex tissue of DLB patients LAMP-2a was significantly increased compared to controls [71]. Likewise, cathepsin D immunoreactivity was significantly reduced in substantia nigra neurons of PD patients with an even greater decrease in α-synuclein inclusion bearing cells [29]. In contrast cortical neurons of DLB patients displaying α-synuclein accumulation were found to show abundant enlarged cathepsin D positive lysosomes [32]. It is currently difficult to interpret these apparent differences. Nevertheless the co-localization of LAMP-2a and cathepsin D with Lewy body pathology implies a pivotal role of these lysosomal proteins in α-synuclein aggregation and/or removal.
In peripheral blood mononuclear cells, two studies identified increased LC3-II levels, suggestive of autophagy activation (Table 1B) [123,171].
Important genetic evidence in support of altered autophagy in PD was provided by studies that linked loss-of function mutation in the lysosomal enzyme ATP13A2, a lysosomal ATPase, to Kufor-Rakeb syndrome, a familial, juvenile onset form of Parkinsonism [125] and by the strong association between Gaucher’s disease, caused by mutations in the lysosomal enzyme glucocerebrosidase, and the incidence of PD [144]. The latter in particular, has recently received considerable attention and exciting new molecular links to α-synuclein pathology are being explored [34,104,134]. Parkin and PINK1, both linked to autosomal-recessive forms of PD, have also been found to be part of the signaling pathway that controls mitophagy, a cargo-specific autophagic process that eliminates damaged mitochondria [178]. The interplay between α-synuclein and CMA or macroautophagy has been explored in recent years. Mutant A53T and A30P α-synuclein was shown to strongly bind the CMA adapter molecule LAMP-2 on the surface of lysosomes, but are unable to become internalized and thus act as inhibitors of CMA, hindering their own processing and the degradation of other CMA substrate proteins [33,173]. Relevant to sporadic PD, dopamine induced conformational changes in wild type α-synuclein revealed the same inhibitory effects [100]. The resulting increase in cytosolic α-synuclein levels could favor pathological misfolding and aggregation. Further supporting the crucial involvement of CMA in PD, the turnover of the neuronal transcription factor MEF2D was shown to dependent on CMA and lysosomal degradation, a process that is disrupted by wild-type and mutant α-synuclein, leading to impaired MEF2D function and thus to neuronal degeneration [176].
In the MPTP mouse model of PD, autophagosome accumulation and dopaminergic cell death were found to be preceded by a disruption of lysosomal integrity and clearance caused by abnormal permeablization of lysosomal membranes through mitochondrial induced oxidative stress [35]. The ectopic release of lysosomal hydrolases into the cytosol may therefore contribute to the dopaminergic cell death observed with these toxins.
On the other hand, mice deficient in cathepsin D, a major lysosomal protease, were found to display α-synuclein accumulation that interestingly formed primarily outside of autophagic vacuoles or lysosomes and that was accompanied by significant proteasome impairment [124]. Conversely, cathepsin D overexpression was protective against α-synuclein overexpression induced cell death in cultured cells and C. elegans [124]. Similarly ALP impairment by pharmacologic inhibition using Bafilomycin A1 was shown to exacerbate α-synuclein induced toxicity but to reduce aggregate formation in both cell culture and transgenic mice, suggesting the production of toxic oligomeric intermediates when autophagic clearance is dysfunctional [71]. Lysosomal dysfunction was also shown to influence the secretion of α-synuclein via exosomes [2], a recently discovered mechanism that might have important implications for the seeding, regional spreading and progression of α-synuclein pathology and neuronal degeneration [43,54].
Another recent study, using cell culture and α-synuclein transgenic mice, demonstrated that excess levels of α-synuclein can inhibit macroautophagy in the very early stages of autophagosome formation via inhibition of the small GTPase Rab1A [167]. Compelling in vivo evidence for this finding was provided by a second study that showed enhanced autophagy in α-synuclein depleted (Snca −/−) mice [31].
Mutations in LRRK2, the most prevalent cause of autosomal-dominant PD, cause alterations in the autophagic pathway as well. LRRK2 not only co-localizes with markers of the endosomal-lysosomal pathway in cases of DLB [58], its knock-out [155,156] or overexpression [51] also significantly alters autophagosome formation.
Integral to the concept that constitutive macroautophagy is essential to neuronal survival, studies in knockout models of essential components of this pathway (Atg5 and Atg7) revealed that genetic inactivation of macroautophagy function leads to profound neurodegeneration and the formation of ubiquitinated inclusion bodies [55,72]. A very recent study also demonstrated that cell specific deletion of Atg7 in midbrain dopamine neurons of knockout mice leads to the formation of ubiquitin und p62 positive inclusions, dendritic and axonal pathology as well as delayed and moderate loss of dopaminergic neurons, accompanied by late-onset locomotor deficits [47]. Intriguingly, whole-brain depletion of Atg7 was found to cause presynaptic accumulation of α-synuclein and LRRK2. While the former was caused by reduced α-synuclein turnover, the latter was shown to be attributable to increased transcription [47].
It should be noted that studies that assessed the effects of genetic depletion of CMA function in mice, did not report a distinct neurological phenotype or neurodegeneration [101], potentially because deleterious effects of CMA dysfunction are masked through a compensatory upregulation of macroautophagy.
Apart from pure dysfunction, aberrant autophagy following exposure to overexpressed A53T mutant α-synuclein [28] neurotoxins [86,112,168] or iron overload [26] importantly demonstrates that prolonged excess levels of autophagy promote cell death. The recent study by Choubey and colleagues in particular demonstrates an interesting example for the concept of “lethal” autophagy in neurodegeneration because it links the overexpression of mutant A53T α-synuclein in primary cortical neurons to excessive autophagy and consequently to mitochondrial loss via mitophagy, bioenergetics deficits and neurodegeneration [28]. Importantly, these findings were recapitulated with the use of rapamycin (with toxicity occurring only at a prolonged exposure of 5 days), a finding that conceivably has broad implication for therapeutic strategies that aim at globally enhancing autophagy (Table 1B).
DEGRADATION OF α-SYNUCLEIN – A CRUCIAL QUESTION FOR PARKINSON’S DISEASE
The simple question how α-synuclein is degraded by neurons has caused much controversy and a body of conflicting results (summarized in Table 2). Deciphering the pathway responsible for degradation of α-synuclein has significant impact for the pathogenesis of α-synuclein aggregation and derived therapeutic approaches in synucleinopathies: α-synuclein may accumulate because of dysfunctional degradation and on the other hand degradation pathways could be targeted pharmacologically to facilitate clearance and to counteract α-synuclein pathology.
Table 2.
Published studies on α-synuclein degradation
| reference | α-synuclein | UPS | ALP | model system |
|---|---|---|---|---|
| Ancolio et al. 2000 | WT & A53T | NO (Lactacystin, PSI) | n.a. | Transiently & stably transfected HEK293 cells or TSM1 neurons |
|
| ||||
| Batelli et al. 2011 | WT | NO (MG132) | Yes (3-MA) | Inducible TetOn(α-syn) PC12 cells |
| A30P | Yes (MG132) | Yes (3-MA) | ||
|
| ||||
| Bennett et al. 19910 | WT & A53T His-tagged | YES (β-lactone) | n.a. | Transiently transfected SH-SY5Y cells |
|
| ||||
| Crews et al. 2010 | WT | n.a | YES (Rapamycin, Atg7 upregulation) | Human WT α-synuclein tg mice, B103 cells |
|
| ||||
| Cuervo et al. 2004 | Rat | NO (Epoxomicin) |
YES (NH4Cl) NO (3-MA) |
Rat ventral midbrain cultures |
| WT | n.a. | YES (NH4Cl, serum removal) | Stably transfected PC12 cells & isolated lysosomes | |
| A53T, A30P | n.a. | NO (CMA) | Isolated lysosomes | |
|
| ||||
| Ebrahimi- Fakhari et al. 2011 | Mouse and human α-synuclein in α-synuclein transgenic mice; α-synuclein-eGFP in a multiphoton imaging based in vivo paradigm | YES (CLBL, MG132) | YES (BafA1, HCQ) | Human α-synuclein transgenic mice, α-synuclein-eGFP transgenic mice and non-transgenic mice |
|
| ||||
| Lee et al. 2004 | WT, A53T, A30P Myc- and His-tagged | YES for aggregates (β-lactone, epoxomicin) |
YES for aggregates (BafA1, E64, cathepsin I) NO for aggregates (3-MA, rapamycin) |
AV-mediated infection of COS-7 cells & SH-SY5Y cells Rotenone induced aggregate formation paradigm |
| n.a. | YES for oligomeric aggregates (BafA1) | |||
| NO for soluble monomer (β-lactone, epoxomicin) | NO for soluble monomer (BafA1, E64) | |||
| Rat | n.a. | YES for oligomeric aggregates (BafA1) | Rat primary cortical neurons | |
|
| ||||
| Liu et al. 2003; 2005 | WT | YES (MG132) | n.a. | Isolated 20S and 26S proteasomes |
|
| ||||
| Machiya et al. 2010 | S129P | YES (Lactacystin, MG132) | YES (3-MA, CQ) | Stably transfected SH- SY5Y cells |
| WT | NO (Lactacystin, MG132) | YES (CQ) | Stably transfected SH- SY5Y cells | |
| S129P | YES (Lactacystin, MG132) | n.a. | Rat primary cortical neurons | |
| Rat | NO (Lactacystin, MG132) | n.a. | Rat primary cortical neurons | |
|
| ||||
| Mak et al. 2010 | Mouse α-synuclein in a paraquat based overexpression model | n.a. | YES | Paraquat treated C57BL/6 mice |
|
| ||||
| Martinez- Vicente et al. 2008 | S129P; S129E; Oligomeric forms induced by nitrating agents; nitrated & dopamine modified forms WT monomers and dimers | n.a. | NO (CMA) | Isolated lysosomes |
| n.a. | YES (CMA) | |||
|
| ||||
| Paxinou et al. 2001 | WT |
YES (MG132) NO (Lactacystin) |
YES (NH4Cl) | Stably transfected HEK293 cells |
|
| ||||
| Rideout et al. 2001 | Rat | NO (Lactacystin, PSI) | n.a. | PC12 cells |
|
| ||||
| Riedel et al. 2010 | A53T | n.a. | YES (17-AAG, NH4Cl, rapamycin) | OLN-93 cells |
|
| ||||
| Rott et al. 2011 | Monoubiquitinated α-synuclein | YES (Lactacystin, MG132) | YES, but only small effect (3-MA, CQ, NH4Cl, Wortmannin) | Transfected SH-SY5Y cells |
| Deubiquitinated α-synuclein | YES, but only small effect (Lactacystin) | YES (3-MA) | ||
|
| ||||
| Sarkar et al. 2007 | WT, A53T, A30P | n.a. |
YES for A53T & A30P (Trehalose) NO for WT (Trehalose) |
Stably transfected PC12 cells |
|
| ||||
| Shin et al. 2005 | SynT (WTSynEGFPΔ 155) | YES (ALLN) | YES (NH4Cl) | Stably and transiently transfected H4 cells |
|
| ||||
| Spencer et al. 2009 | WT, Ser129P | n.a. | YES (Beclin-1 upregulation, 3-MA, BafA1, rapamycin) | B103 cells |
| WT | n.a. | YES (Beclin-1 upregulation) | Human WT α-synuclein tg mice | |
|
| ||||
| Sevlever et al. 2008 | WT | n.a. | Yes (shRNA to cathepsin D) | 3D5 cells |
|
| ||||
| Tofaris et al. 2001 | WT | YES (Lactacystin) | n.a. | Stably transfected SH-SY5Y cells & isolated 20S proteasomes |
|
| ||||
| Tofaris et al. 2011 | Ubiquitinated α-synuclein | YES, but not in Nedd4 overexpressing cells (Bortezomib) | YES (CQ, 3-MA) | SH-SY5Y and HEK293 cells |
|
| ||||
| Vogiatzi et al. 2008 | Rat & overexpressed WT | NO (Epoxomicin) | YES (3-MA, Lamp downregulation, DQWT mutant) | PC12 cells |
| WT | n.a. | YES (DQWT mutant) | SH-SY5Y cells | |
| Rat | NO (Epoxomicin) | YES (3-MA, Lamp2a downregulation) | Rat primary cortical neurons | |
| Mouse α-synuclein | n.a. | YES (Lamp2a downregulation) | Mouse primary cortical neurons | |
| Rat synuclein | n.a. | YES (Lamp2a downregulation, 3- MA) | Primary ventral mid- brain cultures | |
|
| ||||
| Webb et al. 2003 | WT, A53T, A30P HA tagged | YES (Lactacystin, epoxomicin) | YES (BafA1, 3-MA, rapamycin) | Inducible TetOn(α-syn) PC12 cells |
|
| ||||
| Wong et al. 2008 | WT α-synuclein in lactacystin induced aggresomes | n.a. | YES (Starvation, rapamycin, 3-MA) | SH-SY5Y cells |
|
| ||||
| Yamada et al. 2006 | WT, A53T, A30P Myc-His tagged | YES | n.a. | Archaeal 20S proteasomes in HEK293 & Neuro2a cells |
|
| ||||
| Yu et al. 2009 | WT, A53T | n.a | YES for oligomers (Beclin-1 downregulation, Atg5 downregulation; 3MA, rapamycin) | Transfected M17 cells |
Abbreviations: 3-MA (3-Methyladenine); AV (adenovirus); BafA1 (Bafilomycin A1); CLBL (Clasto-lactacystin β-lactone); CMA (chaperone mediated autophagy); CQ (Chloroquine); HCQ (Hydroxychloroquine); PSI (Z-lle-Glu(OtBu)-Ala-Leu-al); WT (wild type)
As a general concept, excess levels of abnormal proteins cause misfolding, aggregation and proteotoxic stress. Clearance of pathological proteins is a vital cell function and essential for cellular integrity. Mechanisms of protein clearance are even more important to neurons, as post mitotic cells with a long life span that renders them vulnerable to protein accumulation over time and proteotoxic stress.
Current literature, nearly exclusively based on in vitro studies and different cell-culture models, provides evidence for the UPS [10,90,91,121,153,164,175] and the ALP [8,32,33,94,100,121,142,147,162,164,177] as the primary mechanism by which unmodified human wild type α-synuclein is degraded, but conflicting findings exist. No role for the UPS was observed in a different set of studies [3,13,82,94,121,129,162,177] and the role of the ALP was challenged by a number of experiments [82,135].
Two groups convincingly demonstrated that unfolded, non-ubiquitinated α-synuclein can undergo degradation via both the 26S and 20S proteasome in vitro [90,91,153]. Degradation through the latter was found to yield truncated α-synuclein species that are known to promote the formation of inclusions [91].
Mutant A53T and A30P α-synuclein is degraded by both the UPS [10,164,175,177] and macroautophagy [130,135,164,177,179], although this was questioned in other studies [3,82]. Importantly, as explained in the previous section, Cuervo and colleagues demonstrated that mutant α-synuclein cannot be degraded by CMA [33] and modified α-synuclein, such as dopamine or nitration induced species, were shown to behave similarly in this respect [100]. A53T mutant α-synuclein was further shown to increase upon inhibition of a specific cytosolic peptidase, the Puromycin-sensitive aminopeptidase, that promotes autophagic clearance by enhancing autophagosome formation [111].
Serine129 phosphorylated α-synuclein was found to be degraded by the UPS [94] and macroautophagy [94,147] but not by CMA [100]. Recent studies also convincingly demonstrated that α-synuclein oligomers are targeted for macroautophagic [179] and lysosomal [82] degradation but are not a substrate for CMA [100], as opposed to the dimeric form of α-synuclein which was found to be translocated and degraded within the lumen of isolated lysosomes [100]. Toxin-induced aggregates and aggregate prone species of α-synuclein were shown to be subject to proteasomal and lysosomal degradation [82,143] although not unequivocally [82]. Alpha-synuclein containing aggresomes were found to be amenable to autophagic clearance induced by starvation or rapamycin treatment in cultured cells [170]. Rat α-synuclein, containing the A53T substitution just like familial cases of PD [80], seems to be degraded by the ALP, macroautophagy [162] and CMA [33,162], although ambiguous findings concerning the latter exist [33]. A definite role for the UPS, however, was negated [33,94,128,162]. Mouse α-synuclein, again sharing the A53T mutation, was shown to be degraded by CMA [96,162] what again contradicts findings for the human A53T mutant protein.
Two recently published in vivo reports using genetic and pharmacologic strategies to upregulate macroautophagy function in human wild type α-synuclein transgenic mice demonstrated enhanced clearance of α-synuclein via this pathway [32,147]. A third in vivo study argued for an increased association of mouse α-synuclein with CMA in a paraquat-based toxin model of α-synuclein overexpression [96].
Our own work addressing the role of UPS and the ALP in PD through a novel in vivo paradigm that employs multiphoton imaging and cranial windows to study α-synuclein turnover in the living mouse brain [159,160], demonstrates clear but distinct roles for both pathways [39,40]. Our results show that α-synuclein turnover is mediated by the UPS both in non-transgenic mice with only endogenous levels of α-synuclein and in transgenic mice with pathologically raised α-synuclein burden. Consistent with previous reports, we found that the UPS becomes increasingly impaired as mice age and importantly that stably raised α-synuclein levels impact proteasome activity in a negative fashion. Turning to autophagy, we found that macroautophagy is not crucially involved in the regular turnover of endogenous levels of α-synuclein but becomes recruited to eliminate excess levels of α-synuclein when the pre-existing burden is raised as it is the case in α-synuclein transgenic mice.
Shedding light on the role of ubiquitination in the degradation process of α-synuclein, another recent study found a pivotal role for the deubiqitinase USP9X in controlling α-synuclein’s fate [131]. Levels of USP9X were found to be significantly reduced in the SN of PD cases and cortex of DLB patients, potentially through sequestration into Lewy bodies, leading to impaired deubiquitinase activity in recovered tissue extracts. Monoubiquitination of α-synuclein by the E3-ubiquitin ligase SIAH [87] and deubiquitination by USP9X might be critical for Lewy body formation as a number of studies have demonstrated that monoubiquitination promotes α-synuclein aggregation and inclusion formation that are toxic to cells [83,87,132]. Important to the degradation of α-synuclein, Rott and colleagues demonstrated that, in the absence of proteolytic impairment, monoubiquitination promotes degradation through the proteasome while deubiquitinated α-synuclein is cleared through autophagy in cultured cells [131]. Another recent study by Tofaris and colleagues showed that Nedd4, a protein of the endosomal-lysosomal pathway, acts as an E3 ligase that mediates α-synuclein ubiquitination at lysine residue 63 and promotes degradation through lysosomes [152].
Taken together, early studies addressing the degradation mechanism of α-synuclein in cell-culture models have produced conflicting results and no clear finding was established for any of the α-synuclein species examined. Variability in the findings might result from the use of different in vitro or cell culture models, the use of non-neuronal cultures, different transfection paradigms, different culture conditions, the use of different pharmacological inhibitors at a different dose and incubation time, different expression levels of α-synuclein in the individual experimental settings, differences in the degradation of different α-synuclein species, e.g. rodent versus human α-synuclein or monomeric, soluble α-synuclein versus aggregated, insoluble α-synuclein species. Recent in vivo studies, including our own work, however, have provided new evidence for distinct roles of the ubiquitination machinery, the UPS and ALP in α-synuclein turnover and pathology (Figure 3).
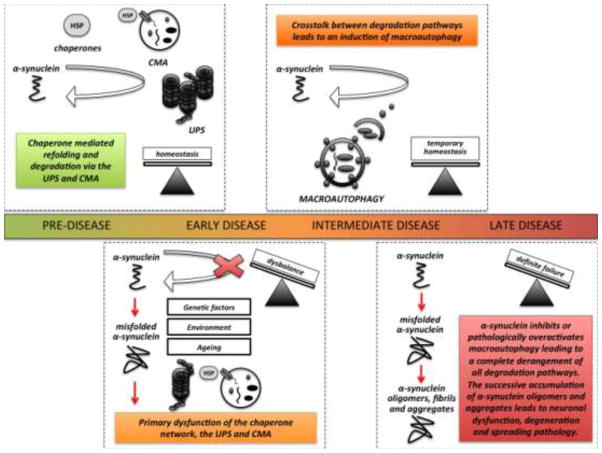
Fig. 3.
The successive failure of protein degradation pathways might provide a key step in the pathological cascade that leads to α-synuclein pathology, neuronal dysfunction and spreading neurodegeneration. The regular turnover of unmodified α-synuclein is largely mediated via both the ubiquitin-proteasome system (UPS) and chaperone-mediated autophagy (CMA). In the early disease stage, primary dysfunction of protein degradation, caused and influenced by genetic, environmental and age-related factors, may lead to excess levels of α-synuclein and the formation of pathologically modified species that overwhelm the capacity of chaperones, the UPS and CMA. Accumulating α-synuclein leads to further and continuous impairment of these pathways therefore eventually creating a vicious cycle. In the next disease stage, crosstalk between degradation pathways drives the induction of macroautophagy and temporary protein homeostasis is achieved before this balance finally tilts. Entering the late disease stage, α-synuclein inhibits or disproportionally overactivates dysfunctional macroautophagy and lysosomal degradation, leading to a complete derangement of all protein degradation pathways. Uncontrolled accumulation of α-synuclein produces oligomeric species that cause neuronal dysfunction and degeneration. At the same time, excess levels of α-synuclein and impaired degradation trigger α-synuclein secretion, thus promoting seeding or protein templating and therefore potentially contributing to a spread of the pathology. CMA (chaperone mediated autophagy); HSP (heat shock proteins); UPS (ubiquitin-proteasome-system)
ENHANCED DEGRADATION OF α-SYNUCLEIN AS A THERAPEUTIC STRATEGY
Advanced understanding of the implications of protein degradation pathways for age-related neurodegenerative diseases has led to an intensive search for small-molecules that can modulate the activity of chaperones, the UPS or ALP. Development of these compounds for clinical use might provide a novel disease modifying strategy for PD and related protein conformation disorders. In the case of molecular chaperones numerous proof-of principle studies in disease models have provided encouraging results [36,41].
As for mechanisms of protein degradation, the UPS has successfully been explored as a drug target for treatment of several forms of cancer, most importantly hematologic malignancies such as multiple myeloma [42]. While proteasome inhibitors like bortezomib are approved and established therapeutics for these diseases, the challenge for neurodegenerative diseases is to develop targeted strategies to facilitate clearance of unwanted proteins. Such “enhancers” of proteasomal degradation could in theory correct or ameliorate a primary UPS dysfunction in PD (Figure 3). Enhanced degradation of α-synuclein and other potentially pathogenic proteins through the proteasome could possibly slow down or even prevent protein misfolding and accumulation. A recent and exciting study for example explored the proteasome-associated deubiquitinating enzyme USP14 as a target for proteasome enhancement [81]. USP14 is known to inhibit proteasomal degradation by trimming of ubiquitin chains on target proteins that would otherwise determine their clearance. By blocking USP14, proteasomal degradation of substrates, including the proteins tau and TDP-43, both linked to neurodegenerative diseases, can be facilitated [81]. Studies of proteasome enhancement in models of PD or α-synucleinopathies respectively are awaited eagerly.
Autophagy enhancement has emerged as a very promising route to clear unwanted protein species and to prevent neurodegeneration [56,99,133]. Autophagy is constitutively active at a low basal level but is induced in response to various forms of cell stress. It therefore represents an essential mechanism by which neurons can adapt to acute changes in intracellular protein homeostasis (Figure 3). Small molecule enhancers of autophagosome formation are being explored for neurodegenerative diseases and can be classified into two groups: mTOR-dependent and mTOR-independent enhancers of autophagy.
Rapamycin (Sirolimus) inhibits the activity of mTOR, a protein-kinase that has a key role in controlling autophagy [184], and thus rapamycin stimulates autophagosome formation and autophagic protein turnover. As a therapeutic, rapamycin is an FDA- and EMA-approved immunosuppressant used to prevent graft rejection after transplantation of organs or coronary stents and has shown promising results in studies for various forms of cancer [17]. Important to neurodegenerative diseases associated with aging, rapamycin strongly ameliorates the disease phenotype in models of Alzheimer’s disease, Huntington’s disease, amyotrophic lateral sclerosis and PD [17] and has been shown to exhibit strong anti-aging effects in several species [184]. In toxin based mouse models of PD, the beneficial effects of rapamycin [92,118,119] has been explained by two distinct mechanisms: The first study by Malagelada et al. reported that mTOR inhibition using rapamycin effectively blocks translation of the pro-cell death signaling molecule RTP180 and thus prevents neurodegeneration [97]. In a second study by Dehay et al. the neuroprotective effect was attributed to an induction of autophagy and a restoration of lysosomal deficiency by a boost in lysosomal biogenesis [35]. As discussed above rapamycin and related small molecule enhancers of rapamycin attenuate α-synuclein accumulation in neuronal cell bodies and synaptic terminals through autophagy enhancement [32,130,138,147,164,179].
While mTOR inhibitors clearly show potential as future therapeutics their use might be limited because of their significant impact on other pathways that are regulated in a mTOR-dependent manner. mTOR-independent inducers of autophagy comprise a heterogeneous group of compounds of which trehalose is probably studied best. Trehalose is a stable disaccharide with unique physicochemical properties: It directly acts as a chemical chaperone that can stabilize misfolded proteins but it also facilitates the clearance of aggregate-prone proteins including α-synuclein via the autophagic pathway [22,135]. Interestingly, simultaneous treatment with trehalose and rapamycin had an additive effect on the clearance of mutant A30P α-synuclein [135]. L-type calcium channel blockers such as verapamil were identified as autophagy enhancers in a small-molecule screen and were found to effectively promote degradation of A53T mutant α-synuclein [165]. Lithium, a mood-stabilizing drug used in bipolar disorder, is an autophagy inducer through its inhibitory effect on inositol monophosphatases therefore depleting the intracellular signaling molecule IP3 [136]. Although lithium enhances autophagic flux independent of mTOR signaling, it has the opposite effect through its inhibition of GSK3β, which leads to mTOR activation and therefore alleviates autophagy [137]. Nevertheless enhanced clearance of mutant α-synuclein [136] and neuroprotective effects [67,174] through lithium were demonstrated in cell culture models and mice, underscoring the potential of lithium and related molecules for further evaluation.
Two important caveats to the putative potential of enhanced protein degradation have to be kept in mind: 1) Enhanced protein degradation and enhanced autophagy in particular is a double-edged sword, since both impaired and unregulated, excessive protein degradation can be detrimental to neuronal integrity. Hence, for example simply upregulating autophagic flux by applying autophagy-enhancing drugs may not be a feasible strategy and should be assessed with great caution. Determining the primary molecular mechanism responsible for dysregulation of protein degradation pathways in PD will allow the development of tailored therapeutic interventions that restore deficits. 2) The exact role of mature protein aggregates or Lewy bodies respectively for neurodegeneration remains unclear, therefore the consequences of preventing, dissolving or removing mature aggregates are uncertain. Notably the presence of soluble oligomers as opposed to mature aggregates correlates best with neurotoxicity [52,64,166]. In fact neurons may employ aggregate formation as a protective mechanism to prolong survival [6,23]. Supporting this theory, promotion of aggregate formation, which may act as scavengers for toxic intermediate oligomeric species, might be a promising neuroprotective approach [16].
CONCLUDING REMARKS AND FUTURE CHALLENGES
Protein degradation pathways play an integral role in the development of neurodegenerative diseases. Their successive failure, as a cause or consequence of early pathological alterations in vulnerable neurons at risk, might provide a key step in the pathological cascade that leads to spreading neurodegeneration (Figure 3). Exploring the crosstalk between the UPS and the different subtypes of autophagy will reveal crucial insights into progression of dysfunctional protein degradation and/or the prevention thereof.
Keeping the balance in protein homeostasis is a primary goal for interventions that aim at mechanisms for protein removal. The targeted development of small molecules that specifically induce the ubiquitination and proteasomal or lysosomal degradation of aberrant and potentially toxic proteins will provide a novel generation of therapeutics for PD and related diseases. Deciphering the exact mechanism by which the different proteolytic systems contribute to the elimination of pathogenic proteins, like α-synuclein, is therefore of paramount importance. The short review of the existing studies on α-synuclein degradation highlights the difficulties faced in the attempt to answer this question. The few in vivo studies addressing α-synuclein’s degradation in a more relevant and accurate experimental paradigm have provided new and much needed insights into the roles of the UPS and ALP in PD. It is now on future research to translate mechanistic insights into therapeutic strategies that are amenable to further testing and evaluation in disease models and patients. Therapeutic use of degradation pathways in PD to overcome a dysfunction in protein homeostasis may turn a “curse” into a “blessing”.
Acknowledgments
We thank the peer reviewers for their valuable and thoughtful comments, which have led to a substantial improvement of the manuscript. This work was supported by NIH NS063963 and NS073740 (P.J.M.), the German National Academic Foundation (Studienstiftung des deutschen Volkes to D.E.-F. & L.W.), the Hamburg Foundation for International Research and Studies (D.E.-F.) and the Parkinson’s Disease Foundation (D.E-F.).
Contributor Information
Darius Ebrahimi-Fakhari, Division: Institute of Anatomy and Cell Biology; Organization: Ruprecht-Karls University Heidelberg; Address: INF 307, 69120 Heidelberg, Germany; Division: MassGeneral Institute for Neurodegenerative Disease; Organization: Department of Neurology, Massachusetts General Hospital, Harvard Medical School; Address: 114 16th Street, Charlestown, MA 02129, USA.
Lara Wahlster, Division: Institute of Physiology and Pathophysiology; Organization: Ruprecht-Karls University Heidelberg; Address: INF 326, 69120 Heidelberg, Germany; Division: MassGeneral Institute for Neurodegenerative Disease; Organization: Department of Neurology, Massachusetts General Hospital, Harvard Medical School; Address: 114 16th Street, Charlestown, MA 02129, USA.
Pamela J. McLean, Division: Department of Neuroscience; Organization: Mayo Clinic Florida; Address: 4500 San Pablo Rd, Jacksonville, FL 32224, USA Division: MassGeneral Institute for Neurodegenerative Disease; Organization: Department of Neurology, Massachusetts General Hospital, Harvard Medical School; Address: 114 16th Street, Charlestown, MA 02129, USA.
References
- 1.Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper JM, Caballero C, Ferrer I, Obeso JA, Schapira AH. Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol. 2010;67 (12):1464–1472. doi: 10.1001/archneurol.2010.198. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Erviti L, Seow Y, Schapira AH, Gardiner C, Sargent IL, Wood MJ, Cooper JM. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol Dis. 2011 doi: 10.1016/j.nbd.2011.01.029. S0969–9961(11)00050-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ancolio K, Alves da Costa C, Ueda K, Checler F. Alpha-synuclein and the Parkinson’s disease-related mutant Ala53Thr-alpha-synuclein do not undergo proteasomal degradation in HEK293 and neuronal cells. Neurosci Lett. 2000;285(2):79–82. doi: 10.1016/s0304-3940(00)01049-1. S0304-3940(00)01049-1 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J, Mouatt-Prigent A, Ruberg M, Hirsch EC, Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol Histopathol. 1997;12 (1):25–31. [PubMed] [Google Scholar]
- 5.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23(2):184–9. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431 (7010):805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 7.Barrachina M, Castano E, Dalfo E, Maes T, Buesa C, Ferrer I. Reduced ubiquitin C-terminal hydrolase-1 expression levels in dementia with Lewy bodies. Neurobiol Dis. 2006;22 (2):265–273. doi: 10.1016/j.nbd.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Batelli S, Peverelli E, Rodilossi S, Forloni G, Albani D. Macroautophagy and the proteasome are differently involved in the degradation of alpha-synuclein wild type and mutated A30P in an in vitro inducible model (PC12/TetOn) Neuroscience. 2011;195:128–37. doi: 10.1016/j.neuroscience.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedford L, Hay D, Devoy A, Paine S, Powe DG, Seth R, Gray T, Topham I, Fone K, Rezvani N, Mee M, Soane T, Layfield R, Sheppard PW, Ebendal T, Usoskin D, Lowe J, Mayer RJ. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci. 2008;28 (33):8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. 28/33/8189 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett MC, Bishop JF, Leng Y, Chock PB, Chase TN, Mouradian MM. Degradation of alpha-synuclein by proteasome. J Biol Chem. 1999;274 (48):33855–33858. doi: 10.1074/jbc.274.48.33855. [DOI] [PubMed] [Google Scholar]
- 11.Berthet A, Bezard E, Porras G, Fasano S, Barroso-Chinea P, Dehay B, et al. L-DOPA Impairs Proteasome Activity in Parkinsonism through D1 Dopamine Receptor. J Neurosci. 2012;32(2):681–91. doi: 10.1523/JNEUROSCI.1541-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betarbet R, Canet-Aviles RM, Sherer TB, Mastroberardino PG, McLendon C, Kim JH, Lund S, Na HM, Taylor G, Bence NF, Kopito R, Seo BB, Yagi T, Yagi A, Klinefelter G, Cookson MR, Greenamyre JT. Intersecting pathways to neurodegeneration in Parkinson’s disease: effects of the pesticide rotenone on DJ-1, alpha-synuclein, and the ubiquitin-proteasome system. Neurobiol Dis. 2006;22 (2):404–420. doi: 10.1016/j.nbd.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Biasini E, Fioriti L, Ceglia I, Invernizzi R, Bertoli A, Chiesa R, Forloni G. Proteasome inhibition and aggregation in Parkinson’s disease: a comparative study in untransfected and transfected cells. J Neurochem. 2004;88 (3):545–553. doi: 10.1046/j.1471-4159.2003.02152.x. [DOI] [PubMed] [Google Scholar]
- 14.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171 (4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blandini F, Sinforiani E, Pacchetti C, Samuele A, Bazzini E, Zangaglia R, Nappi G, Martignoni E. Peripheral proteasome and caspase activity in Parkinson disease and Alzheimer disease. Neurology. 2006;66 (4):529–534. doi: 10.1212/01.wnl.0000198511.09968.b3. [DOI] [PubMed] [Google Scholar]
- 16.Bodner RA, Outeiro TF, Altmann S, Maxwell MM, Cho SH, Hyman BT, McLean PJ, Young AB, Housman DE, Kazantsev AG. Pharmacological promotion of inclusion formation: a therapeutic approach for Huntington’s and Parkinson’s diseases. Proc Natl Acad Sci U S A. 2006;103 (11):4246–4251. doi: 10.1073/pnas.0511256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bove J, Martinez-Vicente M, Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat Rev Neurosci. 2011;12 (8):437–452. doi: 10.1038/nrn3068. [DOI] [PubMed] [Google Scholar]
- 18.Bove J, Zhou C, Jackson-Lewis V, Taylor J, Chu Y, Rideout HJ, Wu DC, Kordower JH, Petrucelli L, Przedborski S. Proteasome inhibition and Parkinson’s disease modeling. Ann Neurol. 2006;60 (2):260–264. doi: 10.1002/ana.20937. [DOI] [PubMed] [Google Scholar]
- 19.Brighina L, Prigione A, Begni B, Galbussera A, Andreoni S, Piolti R, Ferrarese C. Lymphomonocyte alpha-synuclein levels in aging and in Parkinson disease. Neurobiol Aging. 2010;31 (5):884–885. doi: 10.1016/j.neurobiolaging.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Bukhatwa S, Zeng BY, Rose S, Jenner P. A comparison of changes in proteasomal subunit expression in the substantia nigra in Parkinson’s disease, multiple system atrophy and progressive supranuclear palsy. Brain Res. 2010;1326:174–183. doi: 10.1016/j.brainres.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 21.Caneda-Ferron B, De Girolamo LA, Costa T, Beck KE, Layfield R, Billett EE. Assessment of the direct and indirect effects of MPP+ and dopamine on the human proteasome: implications for Parkinson’s disease aetiology. J Neurochem. 2008;105 (1):225–238. doi: 10.1111/j.1471-4159.2007.05130.x. [DOI] [PubMed] [Google Scholar]
- 22.Casarejos MJ, Solano RM, Gomez A, Perucho J, de Yebenes JG, Mena MA. The accumulation of neurotoxic proteins, induced by proteasome inhibition, is reverted by trehalose, an enhancer of autophagy, in human neuroblastoma cells. Neurochem Int. 2011;58 (4):512–520. doi: 10.1016/j.neuint.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Chen L, Feany MB. Alpha-synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat Neurosci. 2005;8 (5):657–663. doi: 10.1038/nn1443. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Thiruchelvam MJ, Madura K, Richfield EK. Proteasome dysfunction in aged human alpha-synuclein transgenic mice. Neurobiol Dis. 2006;23(1):120–126. doi: 10.1016/j.nbd.2006.02.004. S0969–9961(06)00038-6 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Chen Q, Thorpe J, Keller JN. Alpha-synuclein alters proteasome function, protein synthesis, and stationary phase viability. J Biol Chem. 2005;280(34):30009–30017. doi: 10.1074/jbc.M501308200. M501308200 [pii] [DOI] [PubMed] [Google Scholar]
- 26.Chew KC, Ang ET, Tai YK, Tsang F, Lo SQ, Ong E, Ong WY, Shen HM, Lim KL, Dawson VL, Dawson TM, Soong TW. Enhanced autophagy from chronic toxicity of iron and mutant A53T alpha-synuclein: implications for neuronal cell death in Parkinson disease. J Biol Chem. 2011;286 (38):33380–33389. doi: 10.1074/jbc.M111.268409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou AP, Li S, Fitzmaurice AG, Bronstein JM. Mechanisms of rotenone-induced proteasome inhibition. Neurotoxicology. 2010;31(4):367–72. doi: 10.1016/j.neuro.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choubey V, Safiulina D, Vaarmann A, Cagalinec M, Wareski P, Kuum M, Zharkovsky A, Kaasik A. Mutant A53T alpha-synuclein induces neuronal death by increasing mitochondrial autophagy. J Biol Chem. 2011;286 (12):10814–10824. doi: 10.1074/jbc.M110.132514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu Y, Dodiya H, Aebischer P, Olanow CW, Kordower JH. Alterations in lysosomal and proteasomal markers in Parkinson’s disease: relationship to alpha-synuclein inclusions. Neurobiol Dis. 2009;35 (3):385–398. doi: 10.1016/j.nbd.2009.05.023. [DOI] [PubMed] [Google Scholar]
- 30.Claessen JH, Kundrat L, Ploegh HL. Protein quality control in the ER: balancing the ubiquitin checkbook. Trends Cell Biol. 2012;22 (1):22–32. doi: 10.1016/j.tcb.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corrochano S, Renna M, Carter S, Chrobot N, Kent R, Stewart M, Cooper J, Brown SD, Rubinsztein DC, Acevedo-Arozena A. alpha-Synuclein levels modulate Huntington’s disease in mice. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crews L, Spencer B, Desplats P, Patrick C, Paulino A, Rockenstein E, Hansen L, Adame A, Galasko D, Masliah E. Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of alpha-synucleinopathy. PLoS One. 2010;5(2):e9313. doi: 10.1371/journal.pone.0009313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305 (5688):1292–1295. doi: 10.1126/science.1101738. 305/5688/1292 [pii] [DOI] [PubMed] [Google Scholar]
- 34.Cullen V, Sardi SP, Ng J, Xu YH, Sun Y, Tomlinson JJ, Kolodziej P, Kahn I, Saftig P, Woulfe J, Rochet JC, Glicksman MA, Cheng SH, Grabowski GA, Shihabuddin LS, Schlossmacher MG. Acid beta-glucosidase mutants linked to Gaucher disease, Parkinson disease, and Lewy body dementia alter alpha-synuclein processing. Ann Neurol. 2011;69 (6):940–953. doi: 10.1002/ana.22400. [DOI] [PubMed] [Google Scholar]
- 35.Dehay B, Bove J, Rodriguez-Muela N, Perier C, Recasens A, Boya P, Vila M. Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci. 2010;30 (37):12535–12544. doi: 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dimant H, Ebrahimi-Fakhari D, McLean PJ. Molecular Chaperones and Co-Chaperones in Parkinson Disease. Neuroscientist. 2012 doi: 10.1177/1073858412441372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding Q, Dimayuga E, Martin S, Bruce-Keller AJ, Nukala V, Cuervo AM, Keller JN. Characterization of chronic low-level proteasome inhibition on neural homeostasis. J Neurochem. 2003;86 (2):489–497. doi: 10.1046/j.1471-4159.2003.01885.x. [DOI] [PubMed] [Google Scholar]
- 38.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171(2):513–524. doi: 10.2353/ajpath.2007.070188. ajpath.2007.070188 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ebrahimi-Fakhari D, Cantuti-Castelvetri I, Fan Z, Rockenstein E, Masliah E, Hyman BT, McLean PJ, Unni VK. Distinct Roles In Vivo for the Ubiquitin-Proteasome System and the Autophagy-Lysosomal Pathway in the Degradation of {alpha}-Synuclein. J Neurosci. 2011;31 (41):14508–14520. doi: 10.1523/JNEUROSCI.1560-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebrahimi-Fakhari D, McLean PJ, Unni VK. Alpha-synuclein’s degradation in vivo: opening a new (cranial) window on the roles of degradation pathways in Parkinson disease. Autophagy. 2012;8 (2):281–283. doi: 10.4161/auto.8.2.18938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ebrahimi-Fakhari D, Wahlster L, McLean PJ. Molecular Chaperones in Parkinson’s Disease - Present and Future. J Parkinsons Dis. 2011;1 (4):299–320. [PMC free article] [PubMed] [Google Scholar]
- 42.Edelmann MJ, Nicholson B, Kessler BM. Pharmacological targets in the ubiquitin system offer new ways of treating cancer, neurodegenerative disorders and infectious diseases. Expert Rev Mol Med. 2011;13:e35. doi: 10.1017/S1462399411002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L, Vekrellis K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci. 2010;30 (20):6838–6851. doi: 10.1523/JNEUROSCI.5699-09.2010. 30/20/6838 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emmanouilidou E, Stefanis L, Vekrellis K. Cell-produced alpha-synuclein oligomers are targeted to, and impair, the 26S proteasome. Neurobiol Aging. 2010;31(6):953–968. doi: 10.1016/j.neurobiolaging.2008.07.008. S0197-4580(08)00252-2 [pii] [DOI] [PubMed] [Google Scholar]
- 45.Fornai F, Lenzi P, Gesi M, Ferrucci M, Lazzeri G, Busceti CL, Ruffoli R, Soldani P, Ruggieri S, Alessandri MG, Paparelli A. Fine structure and biochemical mechanisms underlying nigrostriatal inclusions and cell death after proteasome inhibition. J Neurosci. 2003;23 (26):8955–8966. doi: 10.1523/JNEUROSCI.23-26-08955.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fornai F, Schluter OM, Lenzi P, Gesi M, Ruffoli R, Ferrucci M, Lazzeri G, Busceti CL, Pontarelli F, Battaglia G, Pellegrini A, Nicoletti F, Ruggieri S, Paparelli A, Sudhof TC. Parkinson-like syndrome induced by continuous MPTP infusion: convergent roles of the ubiquitin-proteasome system and alpha-synuclein. Proc Natl Acad Sci U S A. 2005;102 (9):3413–3418. doi: 10.1073/pnas.0409713102. 0409713102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, Komatsu M, Holstein GR, Yue Z. Disrupted Autophagy Leads to Dopaminergic Axon and Dendrite Degeneration and Promotes Presynaptic Accumulation of alpha-Synuclein and LRRK2 in the Brain. J Neurosci. 2012;32 (22):7585–7593. doi: 10.1523/JNEUROSCI.5809-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19 (5):2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furukawa Y, Vigouroux S, Wong H, Guttman M, Rajput AH, Ang L, Briand M, Kish SJ, Briand Y. Brain proteasomal function in sporadic Parkinson’s disease and related disorders. Ann Neurol. 2002;51 (6):779–782. doi: 10.1002/ana.10207. [DOI] [PubMed] [Google Scholar]
- 50.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426 (6968):895–899. doi: 10.1038/nature02263. nature02263 [pii] [DOI] [PubMed] [Google Scholar]
- 51.Gomez-Suaga P, Luzon-Toro B, Churamani D, Zhang L, Bloor-Young D, Patel S, Woodman PG, Churchill GC, Hilfiker S. Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gosavi N, Lee HJ, Lee JS, Patel S, Lee SJ. Golgi fragmentation occurs in the cells with prefibrillar alpha-synuclein aggregates and precedes the formation of fibrillar inclusion. J Biol Chem. 2002;277 (50):48984–48992. doi: 10.1074/jbc.M208194200. [DOI] [PubMed] [Google Scholar]
- 53.Grunblatt E, Mandel S, Jacob-Hirsch J, Zeligson S, Amariglo N, Rechavi G, Li J, Ravid R, Roggendorf W, Riederer P, Youdim MB. Gene expression profiling of parkinsonian substantia nigra pars compacta; alterations in ubiquitin-proteasome, heat shock protein, iron and oxidative stress regulated proteins, cell adhesion/cellular matrix and vesicle trafficking genes. J Neural Transm. 2004;111 (12):1543–1573. doi: 10.1007/s00702-004-0212-1. [DOI] [PubMed] [Google Scholar]
- 54.Hansen C, Angot E, Bergstrom AL, Steiner JA, Pieri L, Paul G, Outeiro TF, Melki R, Kallunki P, Fog K, Li JY, Brundin P. alpha-Synuclein propagates from mouse brain to grafted dopaminergic neurons and seeds aggregation in cultured human cells. J Clin Invest. 2011;121 (2):715–725. doi: 10.1172/JCI43366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. nature04724 [pii] [DOI] [PubMed] [Google Scholar]
- 56.Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol. 2011 doi: 10.1038/nrneurol.2011.200. [DOI] [PubMed] [Google Scholar]
- 57.Higashi S, Moore DJ, Minegishi M, Kasanuki K, Fujishiro H, Kabuta T, Togo T, Katsuse O, Uchikado H, Furukawa Y, Hino H, Kosaka K, Sato K, Arai H, Wada K, Iseki E. Localization of MAP1-LC3 in vulnerable neurons and Lewy bodies in brains of patients with dementia with Lewy bodies. J Neuropathol Exp Neurol. 2011;70 (4):264–280. doi: 10.1097/NEN.0b013e318211c86a. [DOI] [PubMed] [Google Scholar]
- 58.Higashi S, Moore DJ, Yamamoto R, Minegishi M, Sato K, Togo T, Katsuse O, Uchikado H, Furukawa Y, Hino H, Kosaka K, Emson PC, Wada K, Dawson VL, Dawson TM, Arai H, Iseki E. Abnormal localization of leucine-rich repeat kinase 2 to the endosomal-lysosomal compartment in lewy body disease. J Neuropathol Exp Neurol. 2009;68 (9):994–1005. doi: 10.1097/NEN.0b013e3181b44ed8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ii K, Ito H, Tanaka K, Hirano A. Immunocytochemical co-localization of the proteasome in ubiquitinated structures in neurodegenerative diseases and the elderly. J Neuropathol Exp Neurol. 1997;56 (2):125–131. doi: 10.1097/00005072-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 60.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280(48):40282–40292. doi: 10.1074/jbc.M508786200. M508786200 [pii] [DOI] [PubMed] [Google Scholar]
- 61.Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9(4):574–587. doi: 10.1111/j.1600-0854.2008.00701.x. TRA701 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7 (3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalia SK, Kalia LV, McLean PJ. Molecular chaperones as rational drug targets for Parkinson’s disease therapeutics. CNS Neurol Disord Drug Targets. 2010;9 (6):741–753. doi: 10.2174/187152710793237386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karpinar DP, Balija MB, Kugler S, Opazo F, Rezaei-Ghaleh N, Wender N, Kim HY, Taschenberger G, Falkenburger BH, Heise H, Kumar A, Riedel D, Fichtner L, Voigt A, Braus GH, Giller K, Becker S, Herzig A, Baldus M, Jackle H, Eimer S, Schulz JB, Griesinger C, Zweckstetter M. Pre-fibrillar alpha-synuclein variants with impaired beta-structure increase neurotoxicity in Parkinson’s disease models. Embo J. 2009;28 (20):3256–3268. doi: 10.1038/emboj.2009.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19 (5):2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keller JN, Huang FF, Dimayuga ER, Maragos WF. Dopamine induces proteasome inhibition in neural PC12 cell line. Free Radic Biol Med. 2000;29 (10):1037–1042. doi: 10.1016/s0891-5849(00)00412-3. [DOI] [PubMed] [Google Scholar]
- 67.Kim YH, Rane A, Lussier S, Andersen JK. Lithium protects against oxidative stress-mediated cell death in alpha-synuclein-overexpressing in vitro and in vivo models of Parkinson’s disease. J Neurosci Res. 2011;89 (10):1666–1675. doi: 10.1002/jnr.22700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392 (6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 69.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8 (11):931–937. doi: 10.1038/nrm2245. nrm2245 [pii] [DOI] [PubMed] [Google Scholar]
- 70.Klionsky DJ, Cuervo AM, Dunn WA, Jr, Levine B, van der Klei I, Seglen PO. How shall I eat thee? Autophagy. 2007;3 (5):413–416. doi: 10.4161/auto.4377. [DOI] [PubMed] [Google Scholar]
- 71.Klucken J, Poehler AM, Ebrahimi-Fakhari D, Schneider J, Nuber S, Rockenstein E, Schlotzer-Schrehardt U, Hyman BT, McLean PJ, Masliah E, Winkler J. Alpha-synuclein aggregation involves a bafilomycin A 1-sensitive autophagy pathway. Autophagy. 2012;8(5) doi: 10.4161/auto.19371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441 (7095):880–884. doi: 10.1038/nature04723. nature04723 [pii] [DOI] [PubMed] [Google Scholar]
- 73.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131 (6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 74.Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104 (36):14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kordower JH, Kanaan NM, Chu Y, Suresh Babu R, Stansell J, 3rd, Terpstra BT, Sortwell CE, Steece-Collier K, Collier TJ. Failure of proteasome inhibitor administration to provide a model of Parkinson’s disease in rats and monkeys. Ann Neurol. 2006;60 (2):264–268. doi: 10.1002/ana.20935. [DOI] [PubMed] [Google Scholar]
- 76.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33 (4):517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432 (7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 78.Kuzuhara S, Mori H, Izumiyama N, Yoshimura M, Ihara Y. Lewy bodies are ubiquitinated. A light and electron microscopic immunocytochemical study. Acta Neuropathol. 1988;75 (4):345–353. doi: 10.1007/BF00687787. [DOI] [PubMed] [Google Scholar]
- 79.Kwak S, Masaki T, Ishiura S, Sugita H. Multicatalytic proteinase is present in Lewy bodies and neurofibrillary tangles in diffuse Lewy body disease brains. Neurosci Lett. 1991;128 (1):21–24. doi: 10.1016/0304-3940(91)90751-e. 0304-3940(91)90751-E [pii] [DOI] [PubMed] [Google Scholar]
- 80.Lavedan C. The synuclein family. Genome Res. 1998;8 (9):871–880. doi: 10.1101/gr.8.9.871. [DOI] [PubMed] [Google Scholar]
- 81.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467 (7312):179–184. doi: 10.1038/nature09299. nature09299 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee HJ, Khoshaghideh F, Patel S, Lee SJ. Clearance of alpha-synuclein oligomeric intermediates via the lysosomal degradation pathway. J Neurosci. 2004;24 (8):1888–1896. doi: 10.1523/JNEUROSCI.3809-03.2004. 24/8/1888 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee JT, Wheeler TC, Li L, Chin LS. Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Hum Mol Genet. 2008;17 (6):906–917. doi: 10.1093/hmg/ddm363. [DOI] [PubMed] [Google Scholar]
- 84.Lennox G, Lowe J, Morrell K, Landon M, Mayer RJ. Anti-ubiquitin immunocytochemistry is more sensitive than conventional techniques in the detection of diffuse Lewy body disease. J Neurol Neurosurg Psychiatry. 1989;52 (1):67–71. doi: 10.1136/jnnp.52.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach PJ, Wilkinson KD, Polymeropoulos MH. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395 (6701):451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 86.Li L, Wang X, Fei X, Xia L, Qin Z, Liang Z. Parkinson’s disease involves autophagy and abnormal distribution of cathepsin L. Neurosci Lett. 2011;489 (1):62–67. doi: 10.1016/j.neulet.2010.11.068. [DOI] [PubMed] [Google Scholar]
- 87.Liani E, Eyal A, Avraham E, Shemer R, Szargel R, Berg D, Bornemann A, Riess O, Ross CA, Rott R, Engelender S. Ubiquitylation of synphilin-1 and alpha-synuclein by SIAH and its presence in cellular inclusions and Lewy bodies imply a role in Parkinson’s disease. Proc Natl Acad Sci U S A. 2004;101 (15):5500–5505. doi: 10.1073/pnas.0401081101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lichtenberg M, Mansilla A, Zecchini VR, Fleming A, Rubinsztein DC. The Parkinson’s disease protein LRRK2 impairs proteasome substrate clearance without affecting proteasome catalytic activity. Cell Death Dis. 2011;2:e196. doi: 10.1038/cddis.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lindersson E, Beedholm R, Hojrup P, Moos T, Gai W, Hendil KB, Jensen PH. Proteasomal inhibition by alpha-synuclein filaments and oligomers. J Biol Chem. 2004;279 (13):12924–12934. doi: 10.1074/jbc.M306390200. M306390200 [pii] [DOI] [PubMed] [Google Scholar]
- 90.Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299 (5605):408–411. doi: 10.1126/science.1079293. 1079293 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu CW, Giasson BI, Lewis KA, Lee VM, Demartino GN, Thomas PJ. A precipitating role for truncated alpha-synuclein and the proteasome in alpha-synuclein aggregation: implications for pathogenesis of Parkinson disease. J Biol Chem. 2005;280 (24):22670–22678. doi: 10.1074/jbc.M501508200. [DOI] [PubMed] [Google Scholar]
- 92.Liu K, Liu C, Shen L, Shi J, Zhang T, Zhou Y, Zhou L, Sun X. Therapeutic effects of rapamycin on MPTP-induced Parkinsonism in mice. Neurochem Int. 2011 doi: 10.1016/j.neuint.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 93.Lowe J, McDermott H, Landon M, Mayer RJ, Wilkinson KD. Ubiquitin carboxyl-terminal hydrolase (PGP 9.5) is selectively present in ubiquitinated inclusion bodies characteristic of human neurodegenerative diseases. J Pathol. 1990;161 (2):153–160. doi: 10.1002/path.1711610210. [DOI] [PubMed] [Google Scholar]
- 94.Machiya Y, Hara S, Arawaka S, Fukushima S, Sato H, Sakamoto M, Koyama S, Kato T. Phosphorylated alpha-synuclein at Ser-129 is targeted to the proteasome pathway in a ubiquitin-independent manner. J Biol Chem. 2010;285 (52):40732–40744. doi: 10.1074/jbc.M110.141952. M110.141952 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.MacInnes N, Iravani MM, Perry E, Piggott M, Perry R, Jenner P, Ballard C. Proteasomal abnormalities in cortical Lewy body disease and the impact of proteasomal inhibition within cortical and cholinergic systems. J Neural Transm. 2008;115 (6):869–878. doi: 10.1007/s00702-008-0027-6. [DOI] [PubMed] [Google Scholar]
- 96.Mak SK, McCormack AL, Manning-Bog AB, Cuervo AM, DiMonte DA. Lysosomal degradation of alpha-synuclein in vivo. J Biol Chem. 2010;285 (18):13621–13629. doi: 10.1074/jbc.M109.074617. M109.074617 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Malagelada C, Jin ZH, Jackson-Lewis V, Przedborski S, Greene LA. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson’s disease. J Neurosci. 2010;30 (3):1166–1175. doi: 10.1523/JNEUROSCI.3944-09.2010. 30/3/1166 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Manning-Bog AB, Reaney SH, Chou VP, Johnston LC, McCormack AL, Johnston J, Langston JW, DiMonte DA. Lack of nigrostriatal pathology in a rat model of proteasome inhibition. Ann Neurol. 2006;60 (2):256–260. doi: 10.1002/ana.20938. [DOI] [PubMed] [Google Scholar]
- 99.Marino G, Madeo F, Kroemer G. Autophagy for tissue homeostasis and neuroprotection. Curr Opin Cell Biol. 2011;23 (2):198–206. doi: 10.1016/j.ceb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 100.Martinez-Vicente M, Talloczy Z, Kaushik S, Massey AC, Mazzulli J, Mosharov EV, Hodara R, Fredenburg R, Wu DC, Follenzi A, Dauer W, Przedborski S, Ischiropoulos H, Lansbury PT, Sulzer D, Cuervo AM. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118 (2):777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2006;103 (15):5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mathur BN, Neely MD, Dyllick-Brenzinger M, Tandon A, Deutch AY. Systemic administration of a proteasome inhibitor does not cause nigrostriatal dopamine degeneration. Brain Res. 2007;1168:83–89. doi: 10.1016/j.brainres.2007.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matsui H, Ito H, Taniguchi Y, Inoue H, Takeda S, Takahashi R. Proteasome inhibition in medaka brain induces the features of Parkinson’s disease. J Neurochem. 2010;115 (1):178–187. doi: 10.1111/j.1471-4159.2010.06918.x. [DOI] [PubMed] [Google Scholar]
- 104.Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, Sidransky E, Grabowski GA, Krainc D. Gaucher Disease Glucocerebrosidase and alpha-Synuclein Form a Bidirectional Pathogenic Loop in Synucleinopathies. Cell. 2011;146 (1):37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McLean PJ, Kawamata H, Hyman BT. Alpha-synuclein-enhanced green fluorescent protein fusion proteins form proteasome sensitive inclusions in primary neurons. Neuroscience. 2001;104 (3):901–912. doi: 10.1016/s0306-4522(01)00113-0. S0306-4522(01)00113-0 [pii] [DOI] [PubMed] [Google Scholar]
- 106.McNaught KS, Belizaire R, Isacson O, Jenner P, Olanow CW. Altered proteasomal function in sporadic Parkinson’s disease. Exp Neurol. 2003;179 (1):38–46. doi: 10.1006/exnr.2002.8050. [DOI] [PubMed] [Google Scholar]
- 107.McNaught KS, Belizaire R, Jenner P, Olanow CW, Isacson O. Selective loss of 20S proteasome alpha-subunits in the substantia nigra pars compacta in Parkinson’s disease. Neurosci Lett. 2002;326 (3):155–158. doi: 10.1016/s0304-3940(02)00296-3. [DOI] [PubMed] [Google Scholar]
- 108.McNaught KS, Jenner P. Proteasomal function is impaired in substantia nigra in Parkinson’s disease. Neurosci Lett. 2001;297 (3):191–194. doi: 10.1016/s0304-3940(00)01701-8. [DOI] [PubMed] [Google Scholar]
- 109.McNaught KS, Mytilineou C, Jnobaptiste R, Yabut J, Shashidharan P, Jennert P, Olanow CW. Impairment of the ubiquitin-proteasome system causes dopaminergic cell death and inclusion body formation in ventral mesencephalic cultures. J Neurochem. 2002;81 (2):301–306. doi: 10.1046/j.1471-4159.2002.00821.x. [DOI] [PubMed] [Google Scholar]
- 110.McNaught KS, Olanow CW. Proteasome inhibitor-induced model of Parkinson’s disease. Ann Neurol. 2006;60 (2):243–247. doi: 10.1002/ana.20936. [DOI] [PubMed] [Google Scholar]
- 111.Menzies FM, Hourez R, Imarisio S, Raspe M, Sadiq O, Chandraratna D, O’Kane C, Rock KL, Reits E, Goldberg AL, Rubinsztein DC. Puromycin-sensitive aminopeptidase protects against aggregation-prone proteins via autophagy. Hum Mol Genet. 2010;19 (23):4573–4586. doi: 10.1093/hmg/ddq385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meredith GE, Totterdell S, Petroske E, Santa Cruz K, Callison RC, Jr, Lau YS. Lysosomal malfunction accompanies alpha-synuclein aggregation in a progressive mouse model of Parkinson’s disease. Brain Res. 2002;956 (1):156–165. doi: 10.1016/s0006-8993(02)03514-x. [DOI] [PubMed] [Google Scholar]
- 113.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7 (7):673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 114.Miwa H, Kubo T, Suzuki A, Nishi K, Kondo T. Retrograde dopaminergic neuron degeneration following intrastriatal proteasome inhibition. Neurosci Lett. 2005;380 (1–2):93–98. doi: 10.1016/j.neulet.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 115.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395 (6700):395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 116.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273 (7):3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 117.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2 (3):211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 118.Pan T, Kondo S, Zhu W, Xie W, Jankovic J, Le W. Neuroprotection of rapamycin in lactacystin-induced neurodegeneration via autophagy enhancement. Neurobiol Dis. 2008;32 (1):16–25. doi: 10.1016/j.nbd.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 119.Pan T, Rawal P, Wu Y, Xie W, Jankovic J, Le W. Rapamycin protects against rotenone-induced apoptosis through autophagy induction. Neuroscience. 2009;164 (2):541–551. doi: 10.1016/j.neuroscience.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 120.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447 (7146):859–863. doi: 10.1038/nature05853. nature05853 [pii] [DOI] [PubMed] [Google Scholar]
- 121.Paxinou E, Chen Q, Weisse M, Giasson BI, Norris EH, Rueter SM, Trojanowski JQ, Lee VM, Ischiropoulos H. Induction of alpha-synuclein aggregation by intracellular nitrative insult. J Neurosci. 2001;21 (20):8053–8061. doi: 10.1523/JNEUROSCI.21-20-08053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Petrucelli L, O’Farrell C, Lockhart PJ, Baptista M, Kehoe K, Vink L, Choi P, Wolozin B, Farrer M, Hardy J, Cookson MR. Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron. 2002;36 (6):1007–1019. doi: 10.1016/s0896-6273(02)01125-x. S089662730201125X [pii] [DOI] [PubMed] [Google Scholar]
- 123.Prigione A, Piazza F, Brighina L, Begni B, Galbussera A, Difrancesco JC, Andreoni S, Piolti R, Ferrarese C. Alpha-synuclein nitration and autophagy response are induced in peripheral blood cells from patients with Parkinson disease. Neurosci Lett. 2010;477 (1):6–10. doi: 10.1016/j.neulet.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 124.Qiao L, Hamamichi S, Caldwell KA, Caldwell GA, Yacoubian TA, Wilson S, Xie ZL, Speake LD, Parks R, Crabtree D, Liang Q, Crimmins S, Schneider L, Uchiyama Y, Iwatsubo T, Zhou Y, Peng L, Lu Y, Standaert DG, Walls KC, Shacka JJ, Roth KA, Zhang J. Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Mol Brain. 2008;1:17. doi: 10.1186/1756-6606-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ramirez A, Heimbach A, Grundemann J, Stiller B, Hampshire D, Cid LP, Goebel I, Mubaidin AF, Wriekat AL, Roeper J, Al-Din A, Hillmer AM, Karsak M, Liss B, Woods CG, Behrens MI, Kubisch C. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006;38 (10):1184–1191. doi: 10.1038/ng1884. ng1884 [pii] [DOI] [PubMed] [Google Scholar]
- 126.Rideout HJ, Dietrich P, Wang Q, Dauer WT, Stefanis L. alpha-synuclein is required for the fibrillar nature of ubiquitinated inclusions induced by proteasomal inhibition in primary neurons. J Biol Chem. 2004;279 (45):46915–46920. doi: 10.1074/jbc.M405146200. [DOI] [PubMed] [Google Scholar]
- 127.Rideout HJ, Lang-Rollin I, Stefanis L. Involvement of macroautophagy in the dissolution of neuronal inclusions. Int J Biochem Cell Biol. 2004;36 (12):2551–2562. doi: 10.1016/j.biocel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 128.Rideout HJ, Larsen KE, Sulzer D, Stefanis L. Proteasomal inhibition leads to formation of ubiquitin/alpha-synuclein-immunoreactive inclusions in PC12 cells. J Neurochem. 2001;78 (4):899–908. doi: 10.1046/j.1471-4159.2001.00474.x. [DOI] [PubMed] [Google Scholar]
- 129.Rideout HJ, Stefanis L. Proteasomal inhibition-induced inclusion formation and death in cortical neurons require transcription and ubiquitination. Mol Cell Neurosci. 2002;21 (2):223–238. doi: 10.1006/mcne.2002.1173. [DOI] [PubMed] [Google Scholar]
- 130.Riedel M, Goldbaum O, Schwarz L, Schmitt S, Richter-Landsberg C. 17-AAG induces cytoplasmic alpha-synuclein aggregate clearance by induction of autophagy. PLoS One. 2010;5(1):e8753. doi: 10.1371/journal.pone.0008753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rott R, Szargel R, Haskin J, Bandopadhyay R, Lees AJ, Shani V, Engelender S. alpha-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc Natl Acad Sci U S A. 2011;108 (46):18666–18671. doi: 10.1073/pnas.1105725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rott R, Szargel R, Haskin J, Shani V, Shainskaya A, Manov I, Liani E, Avraham E, Engelender S. Monoubiquitylation of alpha-synuclein by seven in absentia homolog (SIAH) promotes its aggregation in dopaminergic cells. J Biol Chem. 2008;283 (6):3316–3328. doi: 10.1074/jbc.M704809200. [DOI] [PubMed] [Google Scholar]
- 133.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6 (4):304–312. doi: 10.1038/nrd2272. nrd2272 [pii] [DOI] [PubMed] [Google Scholar]
- 134.Sardi SP, Clarke J, Kinnecom C, Tamsett TJ, Li L, Stanek LM, Passini MA, Grabowski GA, Schlossmacher MG, Sidman RL, Cheng SH, Shihabuddin LS. CNS expression of glucocerebrosidase corrects {alpha}-synuclein pathology and memory in a mouse model of Gaucher-related synucleinopathy. Proc Natl Acad Sci U S A. 2011;108 (29):12101–12106. doi: 10.1073/pnas.1108197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282 (8):5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 136.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170 (7):1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sarkar S, Krishna G, Imarisio S, Saiki S, O’Kane CJ, Rubinsztein DC. A rational mechanism for combination treatment of Huntington’s disease using lithium and rapamycin. Hum Mol Genet. 2008;17 (2):170–178. doi: 10.1093/hmg/ddm294. [DOI] [PubMed] [Google Scholar]
- 138.Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O’Kane CJ, Schreiber SL, Rubinsztein DC. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol. 2007;3 (6):331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sawada H, Kohno R, Kihara T, Izumi Y, Sakka N, Ibi M, Nakanishi M, Nakamizo T, Yamakawa K, Shibasaki H, Yamamoto N, Akaike A, Inden M, Kitamura Y, Taniguchi T, Shimohama S. Proteasome mediates dopaminergic neuronal degeneration, and its inhibition causes alpha-synuclein inclusions. J Biol Chem. 2004;279 (11):10710–10719. doi: 10.1074/jbc.M308434200. [DOI] [PubMed] [Google Scholar]
- 140.Schapira AH, Cleeter MW, Muddle JR, Workman JM, Cooper JM, King RH. Proteasomal inhibition causes loss of nigral tyrosine hydroxylase neurons. Ann Neurol. 2006;60 (2):253–255. doi: 10.1002/ana.20934. [DOI] [PubMed] [Google Scholar]
- 141.Schlossmacher MG, Frosch MP, Gai WP, Medina M, Sharma N, Forno L, Ochiishi T, Shimura H, Sharon R, Hattori N, Langston JW, Mizuno Y, Hyman BT, Selkoe DJ, Kosik KS. Parkin localizes to the Lewy bodies of Parkinson disease and dementia with Lewy bodies. Am J Pathol. 2002;160 (5):1655–1667. doi: 10.1016/S0002-9440(10)61113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sevlever D, Jiang P, Yen SH. Cathepsin D is the main lysosomal enzyme involved in the degradation of alpha-synuclein and generation of its carboxy-terminally truncated species. Biochemistry. 2008;47 (36):9678–9687. doi: 10.1021/bi800699v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280 (25):23727–23734. doi: 10.1074/jbc.M503326200. M503326200 [pii] [DOI] [PubMed] [Google Scholar]
- 144.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, Chen CM, Clark LN, Condroyer C, De Marco EV, Durr A, Eblan MJ, Fahn S, Farrer MJ, Fung HC, Gan-Or Z, Gasser T, Gershoni-Baruch R, Giladi N, Griffith A, Gurevich T, Januario C, Kropp P, Lang AE, Lee-Chen GJ, Lesage S, Marder K, Mata IF, Mirelman A, Mitsui J, Mizuta I, Nicoletti G, Oliveira C, Ottman R, Orr-Urtreger A, Pereira LV, Quattrone A, Rogaeva E, Rolfs A, Rosenbaum H, Rozenberg R, Samii A, Samaddar T, Schulte C, Sharma M, Singleton A, Spitz M, Tan EK, Tayebi N, Toda T, Troiano AR, Tsuji S, Wittstock M, Wolfsberg TG, Wu YR, Zabetian CP, Zhao Y, Ziegler SG. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009;361 (17):1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334 (6059):1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Snyder H, Mensah K, Theisler C, Lee J, Matouschek A, Wolozin B. Aggregated and monomeric alpha-synuclein bind to the S6′ proteasomal protein and inhibit proteasomal function. J Biol Chem. 2003;278 (14):11753–11759. doi: 10.1074/jbc.M208641200. M208641200 [pii] [DOI] [PubMed] [Google Scholar]
- 147.Spencer B, Potkar R, Trejo M, Rockenstein E, Patrick C, Gindi R, Adame A, Wyss-Coray T, Masliah E. Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in alpha-synuclein models of Parkinson’s and Lewy body diseases. J Neurosci. 2009;29 (43):13578–13588. doi: 10.1523/JNEUROSCI.4390-09.2009. 29/43/13578 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stefanis L, Larsen KE, Rideout HJ, Sulzer D, Greene LA. Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J Neurosci. 2001;21 (24):9549–9560. doi: 10.1523/JNEUROSCI.21-24-09549.2001. 21/24/9549 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun F, Anantharam V, Zhang D, Latchoumycandane C, Kanthasamy A, Kanthasamy AG. Proteasome inhibitor MG-132 induces dopaminergic degeneration in cell culture and animal models. Neurotoxicology. 2006;27 (5):807–815. doi: 10.1016/j.neuro.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 150.Tanaka Y, Engelender S, Igarashi S, Rao RK, Wanner T, Tanzi RE, Sawa A, VLD, Dawson TM, Ross CA. Inducible expression of mutant alpha-synuclein decreases proteasome activity and increases sensitivity to mitochondria-dependent apoptosis. Hum Mol Genet. 2001;10 (9):919–926. doi: 10.1093/hmg/10.9.919. [DOI] [PubMed] [Google Scholar]
- 151.Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K. Alteration of autophagosomal proteins (LC3, GABARAP and GATE-16) in Lewy body disease. Neurobiol Dis. 2011;43 (3):690–697. doi: 10.1016/j.nbd.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 152.Tofaris GK, Kim HT, Hourez R, Jung JW, Kim KP, Goldberg AL. Ubiquitin ligase Nedd4 promotes alpha-synuclein degradation by the endosomal-lysosomal pathway. Proc Natl Acad Sci U S A. 2011;108 (41):17004–17009. doi: 10.1073/pnas.1109356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tofaris GK, Layfield R, Spillantini MG. alpha-synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett. 2001;509 (1):22–26. doi: 10.1016/s0014-5793(01)03115-5. S0014-5793(01)03115-5 [pii] [DOI] [PubMed] [Google Scholar]
- 154.Tofaris GK, Razzaq A, Ghetti B, Lilley KS, Spillantini MG. Ubiquitination of alpha-synuclein in Lewy bodies is a pathological event not associated with impairment of proteasome function. J Biol Chem. 2003;278 (45):44405–44411. doi: 10.1074/jbc.M308041200. [DOI] [PubMed] [Google Scholar]
- 155.Tong Y, Giaime E, Yamaguchi H, Ichimura T, Liu Y, Si H, Cai H, Bonventre JV, Shen J. Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway. Mol Neurodegener. 2012;7(1):2. doi: 10.1186/1750-1326-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tong Y, Yamaguchi H, Giaime E, Boyle S, Kopan R, Kelleher RJ, 3rd, Shen J. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice. Proc Natl Acad Sci U S A. 2010;107 (21):9879–9884. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ullrich C, Mlekusch R, Kuschnig A, Marksteiner J, Humpel C. Ubiquitin enzymes, ubiquitin and proteasome activity in blood mononuclear cells of MCI, Alzheimer and Parkinson patients. Curr Alzheimer Res. 2010;7 (6):549–555. doi: 10.2174/156720510792231766. [DOI] [PubMed] [Google Scholar]
- 158.Um JW, Im E, Lee HJ, Min B, Yoo L, Yoo J, Lubbert H, Stichel-Gunkel C, Cho HS, Yoon JB, Chung KC. Parkin directly modulates 26S proteasome activity. J Neurosci. 2010;30 (35):11805–11814. doi: 10.1523/JNEUROSCI.2862-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Unni VK, Ebrahimi-Fakhari D, Vanderburg CR, McLean PJ, Hyman BT. Studying protein degradation pathways in vivo using a cranial window-based approach. Methods. 2011;53 (3):194–200. doi: 10.1016/j.ymeth.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Unni VK, Weissman TA, Rockenstein E, Masliah E, McLean PJ, Hyman BT. In vivo imaging of alpha-synuclein in mouse cortex demonstrates stable expression and differential subcellular compartment mobility. PLoS One. 2010;5(5):e10589. doi: 10.1371/journal.pone.0010589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vernon AC, Johansson SM, Modo MM. Non-invasive evaluation of nigrostriatal neuropathology in a proteasome inhibitor rodent model of Parkinson’s disease. BMC Neurosci. 2010;11:1. doi: 10.1186/1471-2202-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vogiatzi T, Xilouri M, Vekrellis K, Stefanis L. Wild type alpha-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J Biol Chem. 2008;283 (35):23542–23556. doi: 10.1074/jbc.M801992200. M801992200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang XF, Li S, Chou AP, Bronstein JM. Inhibitory effects of pesticides on proteasome activity: implication in Parkinson’s disease. Neurobiol Dis. 2006;23 (1):198–205. doi: 10.1016/j.nbd.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 164.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278 (27):25009–25013. doi: 10.1074/jbc.M300227200. M300227200 [pii] [DOI] [PubMed] [Google Scholar]
- 165.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, O’Kane CJ, Floto RA, Rubinsztein DC. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4 (5):295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Winner B, Jappelli R, Maji SK, Desplats PA, Boyer L, Aigner S, Hetzer C, Loher T, Vilar M, Campioni S, Tzitzilonis C, Soragni A, Jessberger S, Mira H, Consiglio A, Pham E, Masliah E, Gage FH, Riek R. In vivo demonstration that alpha-synuclein oligomers are toxic. Proc Natl Acad Sci U S A. 2011;108 (10):4194–4199. doi: 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM, Ravikumar B, Imarisio S, Brown S, O’Kane CJ, Rubinsztein DC. alpha-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol. 2010;190 (6):1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wong AS, Lee RH, Cheung AY, Yeung PK, Chung SK, Cheung ZH, Ip NY. Cdk5-mediated phosphorylation of endophilin B1 is required for induced autophagy in models of Parkinson’s disease. Nat Cell Biol. 2011;13 (5):568–579. doi: 10.1038/ncb2217. [DOI] [PubMed] [Google Scholar]
- 169.Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2(12):a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wong ES, Tan JM, Soong WE, Hussein K, Nukina N, Dawson VL, Dawson TM, Cuervo AM, Lim KL. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet. 2008;17 (16):2570–2582. doi: 10.1093/hmg/ddn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu G, Wang X, Feng X, Zhang A, Li J, Gu K, Huang J, Pang S, Dong H, Gao H, Yan B. Altered expression of autophagic genes in the peripheral leukocytes of patients with sporadic Parkinson’s disease. Brain Res. 2011;1394:105–111. doi: 10.1016/j.brainres.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 172.Xie W, Li X, Li C, Zhu W, Jankovic J, Le W. Proteasome inhibition modeling nigral neuron degeneration in Parkinson’s disease. J Neurochem. 2010;115 (1):188–199. doi: 10.1111/j.1471-4159.2010.06914.x. [DOI] [PubMed] [Google Scholar]
- 173.Xilouri M, Vogiatzi T, Vekrellis K, Park D, Stefanis L. Abberant alpha-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS One. 2009;4(5):e5515. doi: 10.1371/journal.pone.0005515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Xiong N, Jia M, Chen C, Xiong J, Zhang Z, Huang J, Hou L, Yang H, Cao X, Liang Z, Sun S, Lin Z, Wang T. Potential autophagy enhancers attenuate rotenone-induced toxicity in SH-SY5Y. Neuroscience. 2011;199:292–302. doi: 10.1016/j.neuroscience.2011.10.031. [DOI] [PubMed] [Google Scholar]
- 175.Yamada S, Niwa J, Ishigaki S, Takahashi M, Ito T, Sone J, Doyu M, Sobue G. Archaeal proteasomes effectively degrade aggregation-prone proteins and reduce cellular toxicities in mammalian cells. J Biol Chem. 2006;281 (33):23842–23851. doi: 10.1074/jbc.M601274200. [DOI] [PubMed] [Google Scholar]
- 176.Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, Mao Z. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323 (5910):124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yoshimoto Y, Nakaso K, Nakashima K. L-dopa and dopamine enhance the formation of aggregates under proteasome inhibition in PC12 cells. FEBS Lett. 2005;579 (5):1197–1202. doi: 10.1016/j.febslet.2004.12.091. [DOI] [PubMed] [Google Scholar]
- 178.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12 (1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yu WH, Dorado B, Figueroa HY, Wang L, Planel E, Cookson MR, Clark LN, Duff KE. Metabolic activity determines efficacy of macroautophagic clearance of pathological oligomeric alpha-synuclein. Am J Pathol. 2009;175 (2):736–747. doi: 10.2353/ajpath.2009.080928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zeng BY, Bukhatwa S, Hikima A, Rose S, Jenner P. Reproducible nigral cell loss after systemic proteasomal inhibitor administration to rats. Ann Neurol. 2006;60 (2):248–252. doi: 10.1002/ana.20932. [DOI] [PubMed] [Google Scholar]
- 181.Zeng BY, Iravani MM, Lin ST, Irifune M, Kuoppamaki M, Al-Barghouthy G, Smith L, Jackson MJ, Rose S, Medhurst AD, Jenner P. MPTP treatment of common marmosets impairs proteasomal enzyme activity and decreases expression of structural and regulatory elements of the 26S proteasome. Eur J Neurosci. 2006;23 (7):1766–1774. doi: 10.1111/j.1460-9568.2006.04718.x. [DOI] [PubMed] [Google Scholar]
- 182.Zhang NY, Tang Z, Liu CW. alpha-Synuclein protofibrils inhibit 26 S proteasome-mediated protein degradation: understanding the cytotoxicity of protein protofibrils in neurodegenerative disease pathogenesis. J Biol Chem. 2008;283 (29):20288–20298. doi: 10.1074/jbc.M710560200. M710560200 [pii] [DOI] [PubMed] [Google Scholar]
- 183.Zhou Y, Gu G, Goodlett DR, Zhang T, Pan C, Montine TJ, Montine KS, Aebersold RH, Zhang J. Analysis of alpha-synuclein-associated proteins by quantitative proteomics. J Biol Chem. 2004;279 (37):39155–39164. doi: 10.1074/jbc.M405456200. [DOI] [PubMed] [Google Scholar]
- 184.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12 (1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]