SUMMARY
Double-strand breaks (DSBs) in heterochromatic repetitive DNAs pose significant threats to genome integrity, but information about how such lesions are processed and repaired is sparse. We observe dramatic expansion and dynamic protrusions of the heterochromatin domain in response to ionizing radiation (IR) in Drosophila cells. We also find that heterochromatic DSBs are repaired by homologous recombination (HR) but with striking differences from euchromatin. Proteins involved in early HR events (resection) are rapidly recruited to DSBs within heterochromatin. In contrast, Rad51, which mediates strand invasion, only associates with DSBs that relocalize outside of the domain. Hetero-chromatin expansion and relocalization of foci require checkpoint and resection proteins. Finally, the Smc5/6 complex is enriched in heterochromatin and is required to exclude Rad51 from the domain and prevent abnormal recombination. We propose that the spatial and temporal control of DSB repair in heterochromatin safeguards genome stability by preventing aberrant exchanges between repeats.
INTRODUCTION
Double-strand breaks (DSBs) are efficiently repaired by two major pathways, HR (homologous recombination) and NHEJ (nonhomologous end-joining). In HR repair, resection of DSBs generates single-stranded DNA (ssDNA) ends that invade homologous sequences, which serve as templates for DNA synthesis and repair. Repair of single-copy sequences through HR is normally error free because a unique homologous sequence is present on the donor sister chromatid or homolog. NHEJ, by contrast, is intrinsically mutagenic because it simply joins broken ends without restoring any missing sequence.
Heterochromatin, a specialized domain enriched for highly repetitive sequences, represents a specific challenge for DSB repair. This cytologically distinct region of the nucleus comprises about 30% of fly and human genomes (Hoskins et al., 2007; Lander et al., 2001). The large number of repeated sequences in heterochromatin and their close proximity within nuclei exacerbate the risk for genome rearrangements in the presence of DSBs, particularly during HR repair. Recombination among repetitive sequences results in loss or duplication of information (Peng and Karpen, 2008). Recombination between identical repeats on nonhomologous chromosomes produces dicentric and acentric chromosomes that are known to contribute to human diseases such as cancer and infertility (Pearson et al., 2005). NHEJ repair of a DSB in repetitive DNA is potentially less problematic because small deletions or mutations do not affect the function of tandem repeats as severely as genes. Thus, two unresolved issues are whether DSBs in heterochromatin are repaired by NHEJ or HR and how repair occurs without threatening the stability of the genome.
Heterochromatin in S. pombe, flies, and mammals is characterized by enrichment of specific histone modifications (e.g., H3K9me2 and H3K9me3) and associated proteins, such as heterochromatin protein 1a (HP1a) and the histone methyltransferase (HMTase) Su(var)3–9 (Grewal and Jia, 2007). Chromatin composition and regulation are integral to many aspects of the DSB response (Downs et al., 2007), and heterochromatin components and structure help to maintain repeat stability (Peng and Karpen, 2008). For example, the absence of Su(var) 3–9 results in the accumulation of DSBs in heterochromatin, formation of extrachromosomal circular repeated DNAs, translocations, and loss of heterozygosity (Peng and Karpen, 2007, 2009).
How heterochromatin components promote genome stability and whether they regulate DSB repair have not been determined, but current evidence suggests three possible mechanisms. First, compaction or chromatin composition could make heterochromatin intrinsically less responsive to DSB formation or processing (Kim et al., 2007). DSBs produced by ionizing radiation (IR) in mammalian cells are detected at lower frequencies and are resolved with slower kinetics in heterochromatin (Cowell et al., 2007; Goodarzi et al., 2008), and delocalization of HP1β and Kap1 (binding partner for HP1) seem to facilitate repair (Ayoub et al., 2008; Goodarzi et al., 2008). Second, heterochromatin components could repress HR and promote less-damaging repair processes, similar to suppression of reciprocal recombination in pericentric heterochromatin during meiosis, which requires heterochromatin components (Westphal and Reuter, 2002). Third, HR repair could occur in heterochromatin, but aberrant recombination is prevented by specific mechanisms, as suggested by studies in S. cerevisiae (Torres-Rosell et al., 2007).
Here, we show that IR-induced DSBs are efficiently formed and processed in Drosophila heterochromatin and that their repair is, surprisingly, dependent on HR. We demonstrate that heterochromatin responds dynamically to IR: proteins involved in early stages of HR are quickly recruited to DSBs and promote expansion of the domain, and repair sites display a dramatic relocalization to outside heterochromatin, where they first recruit a protein (Rad51) required for strand invasion and completion of HR repair. In addition, we identify the Smc5/6 complex as a new heterochromatin component that serves as a key regulator of HR repair in time and space. These results provide new insights into the dynamics of heterochromatic DSB processing and repair and suggest a mechanism for limiting the risk associated with HR repair of repetitive sequences.
RESULTS
DSBs Rapidly Disappear from Heterochromatin and Are Not Associated with Rad51 Foci
In Drosophila, the DAPI-bright heterochromatin is organized as a distinct nuclear domain (Figure S1 available online) that is enriched for the canonical heterochromatin markers histone H3K9me2, H3K9me3, and HP1a (Eissenberg and Reuter, 2009). In contrast, marks associated with gene expression (e.g., H3K4me2 and me3) are excluded from the heterochromatin domain. This striking three-dimensional (3D) separation is maintained throughout the cell cycle (Figures S1D and S1E).
An early marker for DSB formation is a phosphoepitope that appears on histone variant H2Av (H2AX in mammals), which is catalyzed by the ATM and ATR checkpoint kinases and contributes to checkpoint signaling and repair (Downs et al., 2007). Previous studies in mouse cells showed that γH2AX foci are mostly excluded from the heterochromatic chromocenters (DAPI-bright regions) at > 30 min after IR (Cowell et al., 2007), suggesting that DSBs are not formed or processed in hetero-chromatin. However, it is also possible that repair foci form in heterochromatin at earlier time points and are either quickly repaired or moved outside of the domain.
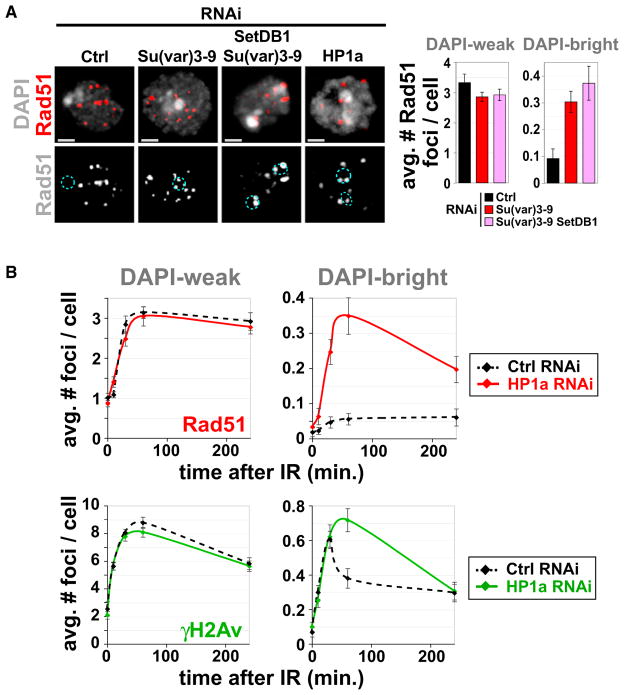
Therefore, we performed a kinetic analysis of the DSB response in heterochromatin, using Kc tissue culture cells fixed at different time points after IR. TUNEL analysis reveals DNA damage in DAPI-bright in most cells at 10 min after IR (Figure S2A), demonstrating that heterochromatin is not refractory to IR-induced damage. However, at 30 min after IR, only 10% of the cells display TUNEL signals in DAPI-bright, when they are still abundant in DAPI-weak regions. Immunofluorescence (IF) analysis and quantitation show that the frequencies of γH2Av and Rad51 (a marker of HR repair) foci peak at 30 min (γH2Av) or 60 min (Rad51) after IR in DAPI-weak euchromatin (Figures 1A and 1B), as observed in mammalian cells (Costes et al., 2009). At later time points, foci frequencies steadily decrease (Figure 1B), which likely reflects ongoing repair. In contrast, the frequencies of γH2Av foci in DAPI-bright hetero-chromatin peak at 10 min after IR and then drop sharply (Figure 1B). Notably, Rad51 foci are rarely observed in DAPI-bright, whereas in DAPI-weak, about half of the γH2Av foci are associated with Rad51 foci (Figure 1B). Similar kinetics are observed in S2 cells (data not shown).
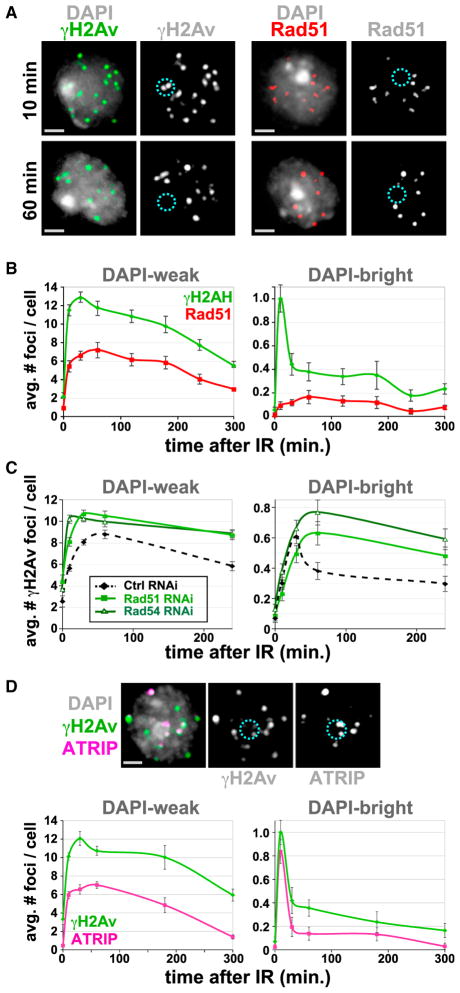
Figure 1. Processing of DSBs in Heterochromatin Occurs with Different Kinetics Than Euchromatin and Requires HR.
(A) γH2Av, but not Rad51, foci form in DAPI-bright (dashed circles) after IR. IF with γH2Av or Rad51 antibodies shows γH2Av foci in DAPI-bright at 10 min and not at 60 min after IR. Rad51 foci are rare in DAPI-bright at both time points (see one example in Figure S2B).
(B) IF was performed at different time points after IR as in (A), and quantitative analysis of foci was performed. In DAPI-weak, frequencies of γH2Av and Rad51 foci peak at 30 and 60 min after IR, respectively, and then show a slow decay. In DAPI-bright, rapid reduction of γH2Av foci is observed between 10 and 30 min after IR (p < 0.001; n > 60). No significant differences in the low numbers of Rad51 foci were observed in DAPI-bright throughout the time course (n > 100).
(C) HR proteins are required for removal of γH2Av foci from heterochromatin. Quantitative kinetic analysis shows persistent γH2Av foci in DAPI-bright and DAPI-weak after Rad51 or Rad54 RNAi at 60 and 240 min after IR (p < 0.01; n > 100). Examples of cells are shown in Figure S2E.
(D) ATRIP foci form in DAPI-bright in response to IR. Cells expressing GFP-ATRIP were fixed 10 min after IR and stained with γH2Av antibodies (top). Quantitation (bottom) reveals rapid reduction of ATRIP and γH2Av foci between 10 and 30 min after IR (p = 0.001; n > 60). In DAPI-weak, the kinetics of ATRIP foci is similar to Rad51 (see B).
Graphs show mean ± SEM. Only Z-stacks including the DAPI-bright are shown as projections in (A) and (D). Scale bars, 1 μm. See also Figure S1 and Figure S2.
We conclude that: (1) DSB formation and processing do occur in heterochromatin soon after damage is induced, (2) DSBs and γH2Av foci rapidly disappear from heterochromatin, suggesting that heterochromatic DSBs are either repaired quickly or relocate elsewhere for repair, and (3) Rad51 foci are rarely observed in heterochromatin throughout the time course.
Heterochromatic DSB Repair Requires the HR Pathway and Is Associated with Early HR Repair Components
Repair by NHEJ would be consistent with the absence of Rad51 foci and the faster kinetics of DSB removal in the heterochromatin domain. To determine which repair pathway is involved, we analyzed the effect of NHEJ or HR inactivation on foci kinetics. Importantly, Kc cells are predominantly in S and G2 phases of the cell cycle (Figures S1A–S1D), when both pathways are functional (Huertas, 2010). In DAPI-weak regions, depleting proteins required for NHEJ (Ku70 and Ku80) or HR (Rad51 or Rad54) results in persistent γH2Av foci after IR (Figure 1C and Figure S2D), confirming the involvement of both pathways in repairing euchromatic DSBs in Kc cells. However, Ku70/80 depletion has no effect on γH2Av or Rad51 foci kinetics in DAPI-bright (Figure S2D), suggesting that NHEJ is not required for DSB repair in heterochromatin. In contrast, depletion of either Rad51 or Rad54 results in persistent γH2Av foci in DAPI-bright (Figure 1C and Figure S2E), reflecting unrepaired DSBs (γH2Av foci overlapping with TUNEL signals) (Figure S2E). We conclude that the fast disappearance of DSBs from Drosophila heterochromatin predominantly depends on the HR pathway.
The requirement for HR is surprising, given the lack of Rad51 foci in heterochromatin. To determine whether HR steps that precede Rad51 loading occur in heterochromatin, we analyzed the kinetics of foci formation of ATRIP, which is recruited to RPA-covered resected DNA (Zou and Elledge, 2003). ATRIP and Rad51 kinetics are similar in DAPI-weak regions (compare Figure 1D to Figure 1B). In sharp contrast, ATRIP foci in DAPI-bright peak at 10 min after IR and then quickly disappear, similar to γH2Av foci (Figure 1D).
We conclude that early steps in DSB processing for HR repair (resection and ATRIP loading) occur within heterochromatin, whereas later steps (assembly of Rad51 foci) are excluded from this domain. This separation of early and late HR events is not observed in euchromatin, in which ATRIP and Rad51 foci display similar kinetics.
Heterochromatin Rapidly Expands and Becomes Dynamic after IR
One reason why HR in heterochromatin is potentially dangerous is because of the close proximity of homologous sequences on different chromosomes. Therefore, we explored whether the organization of this domain changes in response to IR. Time-lapse studies of cells expressing mCherry-tagged HP1a (mCh-HP1a) show that the HP1a domain is compact and contiguous prior to IR treatment. However, soon after IR, this domain expands and becomes fragmented, with rapid extension and retraction of HP1a “fingers” (Figure 2A and Movie S1). HP1a domain volume increases within minutes after IR (Figure S3A) and peaks at 20–40 min (1.5-fold compared to before IR) (Figure 2B and Figure S3A). This is followed by a partial contraction, which is maintained until at least 100 min after IR (Figure 2B and Figure S3B). Fixed cells display a similar increase in HP1a, H3K9me2, and DAPI-bright volumes after IR, with no increase in total nuclear volume (Figures S3C and S3D and data not shown). HP1a expansion was also observed after low dose radiation (1.67 Gy, data not shown; this dose is equivalent to 10cGy treatment of human cells).
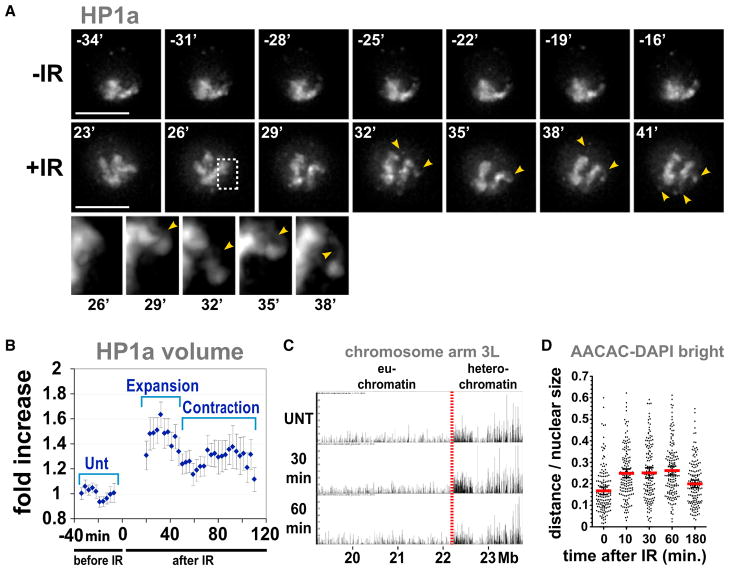
Figure 2. Heterochromatin Expands and Becomes Dynamic in Response to IR.
(A) HP1a expands after IR and forms dynamic protrusions. Stills from Movie S1 are shown, in which a single cell expressing GFP-HP1a was imaged before IR (−IR) and starting 20 min after IR (+IR). Arrows and zoomed detail of the outlined region show HP1a protrusions. Scale bars, 5 μm.
(B) Automated quantitation shows HP1a expansion (p < 0.0001) followed by partial contraction (p = 0.0011) (see examples in Figure S3B). After contraction, the volume of HP1a is still significantly larger than the untreated control (p < 0.0001) (two-tailed unpaired t test; n = 11; graph shows mean ± SEM).
(C) ChIP-array analysis of HP1a localization was performed before (UNT), 30 min, and 60 min after IR. The browser view of the proximal half of chromosome 3L shows the position of the euchromatin-heterochromatin border (red line); x axis = arm coordinates in Mbs. y axis = enrichment p values, normalized to input. No major increases in HP1a enrichments are observed in the euchromatin after IR (other chromosome arms are shown in Figure S3).
(D) IR induces expansion of heterochromatin sequences. Quantitation of the distance between AACAC signals, detected by FISH, and DAPI-bright shows higher values between 10 and 60 min after IR and partial decrease at 180 min (p < 0.001) (n = 150 signals). Red bar, mean; black bars, ± SEM.
The increased volumes of HP1a and H3K9 methylation could result from spreading of heterochromatin into euchromatic regions in response to IR, as observed for chromosome rearrangements that juxtapose euchromatic and heterochromatic sequences (Eissenberg and Reuter, 2009). Alternatively, the heterochromatic domain could physically expand, in which case HP1a would still be restricted to heterochromatic sequences. Genome-wide HP1a ChIP-array analysis shows that HP1a does not become enriched in the euchromatin after IR (Figure 2C and Figure S3E), demonstrating that the volume of the heterochromatin domain expands without spreading into euchromatic sequences.
In addition, we used fluorescence in situ hybridization (FISH) analysis to monitor the locations of the AACAC satellite sequences with respect to DAPI-bright before and after IR. Quantitative analysis demonstrates that these repeats localize predominantly within or close to DAPI-bright in untreated controls (Figure 2D and Figure S1F). However, the average distance between satellite signals and DAPI-bright increases significantly between 10 and 60 min after IR and then decreases by 3 hr (Figure 2D). We conclude that changes in the spatial distribution of heterochromatic repeated sequences occur in concert with the expansion and partial contraction of the HP1a domain.
Heterochromatic Repair Foci Relocate Outside of the Heterochromatin Domain
Relocalization of DSBs to outside of heterochromatin to complete HR repair provides one explanation for the fast disappearance of γH2Av and ATRIP foci, the exclusion of Rad51 foci from heterochromatin, and the requirement for HR repair. To test this hypothesis, we performed live studies of cells expressing fluorescent-tagged versions of HP1a and components of the DSB response. Mu2/Mdc1 recruitment to DSBs relies on direct interactions with γH2Av and is an excellent marker for the early response to DSBs (Dronamraju and Mason, 2009). To detect later steps in HR repair, we tracked proteins recruited to RPA-covered resected DNA (ATRIP and TopBP1) and Rad51, which promotes strand invasion (Su, 2006).
Time-lapse studies and 3D reconstructions of nuclei revealed that Mu2, ATRIP, and TopBP1 foci appear within the HP1a domain by 3 min after IR and then move to the HP1a periphery; by ~30–40 min after IR, most foci are located at the HP1a periphery or outside of the domain (Figures 3A and 3C, Movie S2, Movie S3, Figures S4A–S4E, and Figure S5A). Interestingly, ATRIP and TopBP1 foci are more abundant and much brighter within the HP1a domain at early time points, compared to the euchromatin (Figure 3A and Figure S4B). ATRIP foci dynamically join and split within the HP1a domain (Figure S5B and Movie S4) and often split into multiple foci when relocated outside of the HP1a domain (Figure 3C and Figure S5A). This suggests that the bright ATRIP and TopBP1 foci could result from early clustering of DSBs in heterochromatin, which separate after relocalization. Interestingly, assembly of ATRIP and TopBP1 foci occurs with kinetics similar to Mu2 foci in heterochromatin but is delayed outside of the HP1a domain (Figure 3A and Figures S4A and S4B), consistent with our observations in fixed cells. Thus, DSB recognition (Mu2 and γH2Av foci) occurs with the same kinetics in euchromatin and heterochromatin, whereas subsequent steps (resection or ATRIP/TopBP1 recruitment) are accelerated or enhanced in heterochromatin.
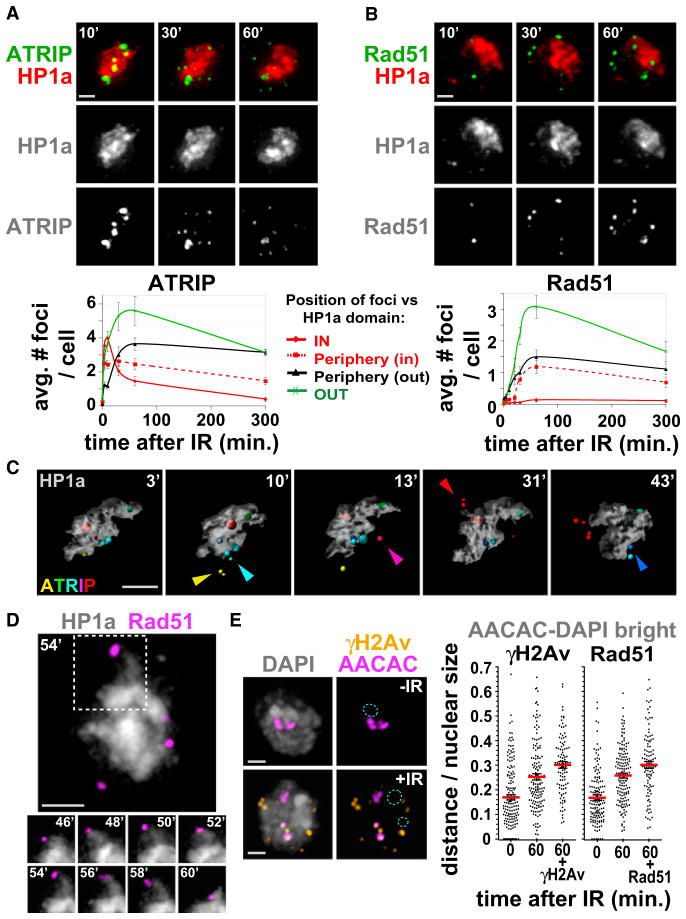
Figure 3. Heterochromatic DSBs Relocalize Outside of the HP1a Domain, Where They Associate with Rad51.
(A) ATRIP foci form within the HP1a domain and relocate outside of the domain. Stills from Movie S2 are shown, in which cells expressing GFP-ATRIP and mCh-HP1a were imaged before and after IR. (Top) Maximum intensity projections of one cell at 10, 30, and 60 min after IR show the relocalization of ATRIP foci to the periphery and outside of the HP1a domain. More time points are shown in Figure S4C. (Bottom) Quantitation shows increased ATRIP foci within the HP1a domain at 4 and 10 min and reductions after 10 min. ATRIP foci accumulate at the periphery of the HP1a domain from 10 to 60 min (p < 0.001; n = 30 cells).
(B) Rad51 foci form at the periphery and outside of the HP1a domain after IR. Cells expressing GFP-Rad51 and mCh-HP1a were analyzed as in (A). (Top) Images of the same cell at 10, 30, and 60 min after IR; more time points are shown in Figure S4E. (Bottom) Quantitation shows that Rad51 foci only appear near the HP1a periphery or outside of the domain (n = 40 cells).
(C) 3D modeling of stills from Movie S3 (Figure S5A), in which cells expressing GFP-ATRIP and mCh-HP1a were imaged before and after IR, shows ATRIP foci formed within the HP1a domain that translocate outside (arrows) and frequently split (see red, cyan, yellow foci).
(D) Cells expressing GFP-Rad51 and mCh-HP1a, imaged before and after IR, show Rad51 foci localized at the “tips” of dynamic HP1a protrusions. Stills from Movie S5 are shown. Representative time points of the region outlined are shown below.
(E) Damaged heterochromatic repeats are more distant from DAPI-bright compared to undamaged repeats. AACAC repeats and γH2Av or Rad51 foci were detected by FISH-IF of cells fixed before (−) and 60 min after (+) IR. Images and quantitation of the distance from DAPI-bright (dashed blue circle) show that AACAC signals that colocalize with γH2Av (60 + γH2Av) or Rad51 (60 + Rad51) are more distant than all AACAC signals at the same time point (60) (p < 0.001 and p < 0.02, respectively, by two-tailed Mann-Whitney test; n > 100 signals). The distances in untreated cells are also shown for each experiment (0). Images are projections of the Z-stacks, including the DAPI-bright.
Graphs show mean ± SEM. Scale bars, 1 μm. See also Figure S4, Figure S5, Movie S2, Movie S3, Movie S4, and Movie S5.
In contrast, Rad51 foci are rarely observed within the HP1a domain at all time points, consistent with the analysis of fixed cells (Figure 3B). Instead, they appear at the periphery of the HP1a domain starting ~30 min after IR (Figure 3B) and move with the ends of the dynamic HP1a fingers (Figure 3D, Figures S4E and S4F, and Movie S5). The assembly of Rad51 foci correlates in space and time with relocalization of ATRIP and TopBP1 foci outside of the HP1a domain, suggesting that relocalization precedes assembly of Rad51 foci at heterochromatic DSBs (compare kinetics in Figure 3A with Figure S4A and images in Figure S4C with Figure S4E). Indeed, colocalization of TopBP1 with Rad51 foci progressively increases at the periphery of HP1a between 10 and 60 min after IR (when TopBP1 foci relocalize) (Figure S4G). The intensity reduction for TopBP1 and ATRIP foci during relocalization and Rad51 foci formation (Figures S4C, S4E, and S4G) likely results from the local removal of RPA during assembly of Rad51 nucleofilaments (Sugiyama and Kowalczykowski, 2002).
We also confirmed that the relocalization of repair foci reflects the behavior of damaged repeated sequences associated with heterochromatic proteins. First, IF for γH2Av or Rad51 combined with DNA FISH (AACAC satellite repeats) shows that satellite sequences associated with γH2Av or Rad51 repair foci are more distant from DAPI-bright compared to all satellite signals at 60 min after IR (Figure 3E). This result directly links repeated heterochromatic sequences to HR repair foci relocalized outside of the heterochromatin domain. Second, coimmunoprecipitation experiments show that ATRIP is strongly associated with HP1a and H3K9me3 at 6 and 40 min after IR (Figure S5C), when ATRIP foci are concentrated in heterochromatin. Reduced association between ATRIP and HP1a at 60 min, after relocalization of foci is complete, could result from the disassembly of ATRIP foci or local heterochromatin changes at the sites of damage. Interestingly, we observe some association of ATRIP with heterochromatin components prior to IR and focus formation.
We conclude that repair foci are highly mobile in Drosophila cultured cells and that early steps in HR repair (e.g., resection) occur within the heterochromatic domain, whereas late HR steps (e.g., strand invasion) do not occur until DSBs relocate to the HP1a periphery.
Heterochromatin Expansion and Relocalization of Foci Require Resection and Checkpoint Proteins
We investigated whether proteins involved in the initial processing of DSBs, specifically checkpoint kinases and resection components, mediate heterochromatin expansion or relocalization of foci. Treatment with caffeine, a potent inhibitor of ATM and ATR kinase activity (Sarkaria et al., 1999), results in defective HP1a expansion after IR (3.6-fold reduction) (Figures 4A and 4B and Movie S6). Caffeine also delays formation of ATRIP foci in euchromatin but prevents formation of ATRIP foci within the HP1a domain (Figures 4A and 4C), suggesting a role for checkpoint kinases in the assembly of ATRIP foci specifically in heterochromatin. We directly addressed the roles of ATM and ATR in heterochromatin expansion by RNAi depletion (Figures S6A and S6B). HP1a expansion is partially defective after ATM RNAi (1.7-fold reduction) and severely impaired after ATR RNAi (3-fold reduction) (Figure 4D and Figure S6E). Simultaneous depletion of ATM and ATR results in defective expansion, but the magnitude of the defect is not significantly different from the removal of ATR alone (Figure 4D and Figure S6E). We conclude that checkpoint kinases are required to promote heterochromatin expansion after IR and that ATR plays a major role.
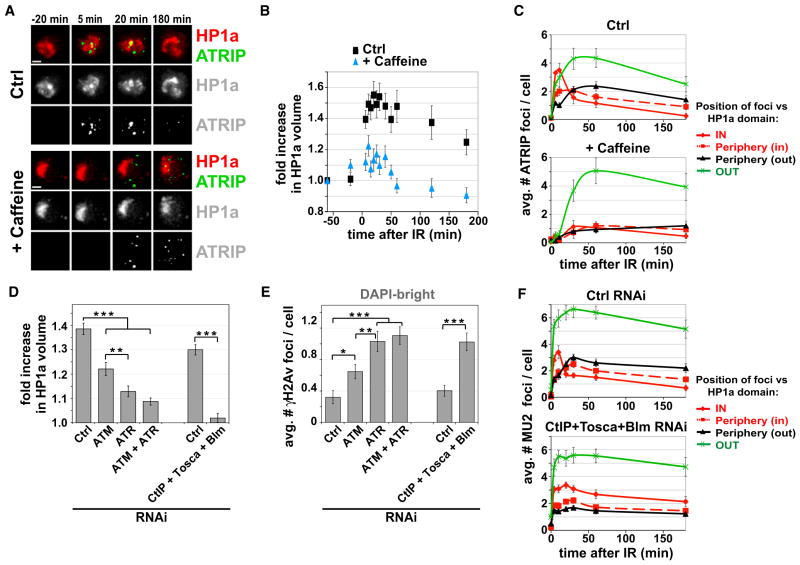
Figure 4. Checkpoint Kinases and Resection Are Required for Heterochromatin Expansion and DSB Relocalization.
(A) Caffeine treatment suppresses HP1a dynamics and prevents ATRIP foci assembly within the HP1a domain. Stills from Movie S6 are shown, in which cells expressing GFP-ATRIP and mCh-HP1a were imaged before and after IR, in the presence (+ caffeine) or absence (Ctrl) of caffeine. Scale bar, 1 μm.
(B) Quantitation of the experiment described in (A) shows defective expansion of the HP1a volume after caffeine treatment compared to untreated cells (p < 0.0001; n > 15).
(C) Quantitation of the experiment described in (A) highlights defective formation of ATRIP foci in the HP1a domain after caffeine treatment (p < 0.001; n > 30). ATM is not required for formation of ATRIP foci in heterochromatin (Figure S6B), suggesting that this defect results from ATR inactivation.
(D) Quantitation of HP1a volume shows defective expansion after depletion of checkpoint or resection proteins. Values = mean fold increase in HP1a volume for time points between 10 and 40 min after IR, compared to control RNAi (***p < 0.0001, **p < 0.01, two-tailed unpaired t test with Welch correction; n > 12). All time points are shown in Figure S6E.
(E) Depletion of checkpoint or resection proteins results in persistent γH2Av foci in DAPI-bright at 60 min after IR (***p < 0.0001, **p < 0.01, *p < 0.05, two-tailed Mann-Whitney test; n > 100). Kinetics are shown in Figure S6F.
(F) RNAi of CtIP+Tosca+Blm results in persistent Mu2 foci within the HP1a domain after IR and defective relocalization to the HP1a periphery (p < 0.0001; n > 20). This analysis could not be performed after ATR depletion because Mu2 foci do not form (Figure S6B). Similarly, ATRIP foci could not be monitored after RNAi of ATR or resection components (Figures S6B–S6D).
Although regulators of resection have not previously been identified in Drosophila, there are homologs of three proteins required for resection in yeast and mammals (CtIP, Exo1 [Tosca in Drosophila], and Blm) (Digilio et al., 1996; Kusano et al., 1999; Uanschou et al., 2007). Simultaneous depletion of all three proteins (Figure S6A) severely impaired the assembly of ATRIP foci and not γH2Av and Mu2 foci (Figures S6C and S6D), indicating that resection is indeed inhibited. Importantly, HP1a expansion is almost completely abolished after CtIP/Tosca/Blm RNAi (15-fold reduction) (Figure 4D and Figure S6E).
Interestingly, loss of resection seems to have a stronger impact than checkpoint inactivation on HP1a expansion (Figure 4D). Moreover, γH2Av and Mu2 foci are weaker and less numerous after ATR RNAi (Figure S6B), but not after depletion of resection components (Figure S6D). Thus, ATR is still partially active after CtIP+Tosca+Blm RNAi, yet expansion is blocked; this suggests that resection could contribute to expansion independent of its role in promoting ATR recruitment.
To address the impact of checkpoint proteins and resection on DSB relocalization, we quantified the frequencies of γH2Av foci within and outside of the DAPI-bright region at 60 min after IR. We observe that γH2Av foci persist in heterochromatin when checkpoint or resection proteins are depleted (Figure 4E and Figure S6F). As for HP1a expansion, ATR, ATR+ATM, and CtIP+ Tosca+Blm depletions display the strongest effects. Time-lapse analysis of GFP-Mu2 confirms that relocalization of heterochromatic DSBs to the periphery and outside of the HP1a domain is defective when resection is inhibited (Figure 4F).
We conclude that IR-induced expansion of the heterochromatin domain and relocalization of repair foci require checkpoint kinases (mainly ATR) and resection.
Heterochromatin Proteins Regulate DSB Repair by Blocking Formation of Rad51 Foci
Careful analysis of fixed and living cells revealed that Rad51 foci assemble in HP1a “holes” or “pockets” at the heterochromatin periphery after IR or where HP1a is not visible after NIR-laser targeting to the heterochromatin (Figure S4F and Figure S7B). This mutual exclusivity suggests that assembly of Rad51 foci could be blocked by heterochromatin proteins.
We therefore tested the effect of heterochromatic protein removal on the distributions of Rad51 foci. Depletion of the Su(var)3–9 HMTase results in severe reduction of H3K9me2 and H3K9me3 levels and disassembly of HP1a, and these effects are exacerbated by simultaneous depletion of the SetDB1 HMTase (Figure S7C). The absence of H3K9 HMTases results in a 3- to 4-fold increased frequency of Rad51 foci after IR, specifically in DAPI-bright (Figure 5A). Similarly, HP1a depletion results in the abnormal presence of Rad51 foci in DAPI-bright (Figure 5A), and the kinetics of γH2Av and Rad51 foci become similar to those observed normally in DAPI-weak euchromatin (Figure 5B, compare to Figure 1). Because H3K9 methylation levels are almost normal after HP1a RNAi (Figure S7D), we conclude that HP1a acts downstream of H3K9 methylation in regulating exclusion of Rad51 foci.
Figure 5. HP1a or H3K9 HMTase Depletion Results in Formation of Rad51 Foci in Heterochromatin.
(A) IF analysis 60 min after IR and quantitation shows that Su(var)3–9, Su(var)3–9 + SetDB1, or HP1a depletion (quantitation is in B) results in increased Rad51 foci in DAPI-bright (dashed blue circles) and not in DAPI-weak (3- to 4-fold increase in DAPI-bright after Su(var)3–9 and SetDB1 RNAi [right]; p < 0.001; n > 100).
(B) Quantitation of γH2Av and Rad51 foci at different time points after IR shows that depletion of HP1a results in increased foci in DAPI-bright (*p < 0.01; n > 100) and not in DAPI-weak.
Graphs show mean ± SEM. Scale bars, 1 μm. See also Figure S7.
Enrichment of the Drosophila Smc5/6 Complex in Heterochromatin Requires HP1a
HP1a could counteract the formation of Rad51 foci directly or by recruiting other proteins that provide antirecombinase functions. One good candidate for this role is the Smc5/6 complex, which prevents the formation of DSB-induced Rad52 foci in the nucleolus and suppresses spontaneous recombination of rDNA repeats in S. cerevisiae (Torres-Rosell et al., 2007). In S. pombe, HU treatment promotes Smc5/6 recruitment to pericentric heterochromatin, suggesting a role in responding to the presence of stalled forks (Pebernard et al., 2008).
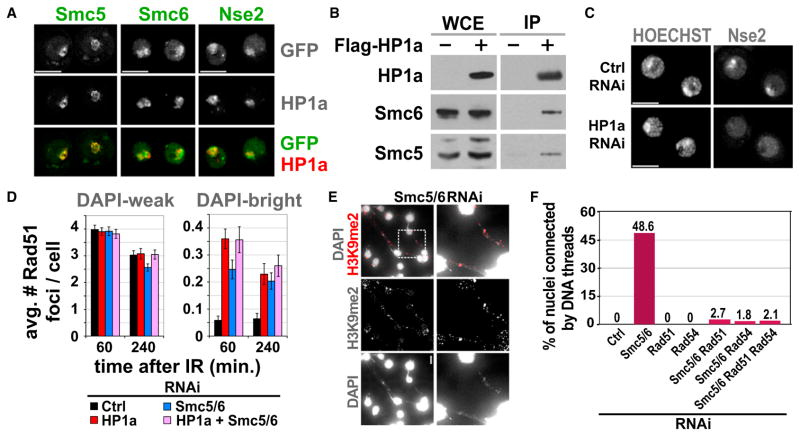
We first determined whether subunits of the Drosophila Smc5/6 complex are localized to heterochromatin. Live imaging shows that GFP-tagged Smc5, Smc6, and Nse2 colocalize with mCh-HP1a through all interphase stages of the cell cycle (Figure 6A, Figure S8A, and data not shown). Similar to HP1a (Kellum et al., 1995), Smc5/6 components disassemble from heterochromatin during mitosis and reassemble in G1 (Figure S8A and data not shown). Finally, Smc5 and Smc6 coimmunoprecipitate with FLAG-HP1a (Figure 6B). These results identify the Smc5/6 complex as a new heterochromatin component in Drosophila that is physically associated with HP1a.
Figure 6. The Smc5/6 Complex Is Required to Prevent IR-Induced Rad51 Foci and Aberrant Recombination in Heterochromatin.
(A) Images of cells expressing GFP-tagged Smc5, Smc6, or Nse2 and mCh-HP1a show that different Smc5/6 subunits colocalize with HP1a.
(B) The Smc5/6 complex interacts with HP1a. FLAG-HP1a (+) expressing S2 cells were used to immunoprecipitate HP1a. Western blot with anti-FLAG (HP1a), Smc5, and Smc6 antibodies shows enrichment of Smc5 and Smc6 compared to control cells (−). Extracts were treated with Benzonase prior to IP, suggesting that Smc5/6-HP1a interaction is not mediated by DNA.
(C) HP1a is required for localization of the Smc5/6 complex to heterochromatin. HP1a RNAi results in delocalization of GFP-Nse2 from heterochromatin (see Figure S8B for quantitation).
(D) Smc5/6 and HP1a are part of the same pathway that excludes Rad51 foci from heterochromatin. Bw (control), HP1a, Smc5/6, or HP1a + Smc5/6 were depleted for 5 days by RNAi, and then cells were fixed after IR. Quantitation shows similar increases in DAPI-bright Rad51 foci after HP1a or Smc5/6 + HP1a RNAi (n > 150). Graphs show mean ± SEM.
(E) Smc5/6 depletion alone (without IR) results in heterochromatic DNA filaments connecting dividing cells. Smc5 and Smc6 were depleted for 6.5 days by RNAi, spun down, mixed, and settled to adhere to the slide. Then, cells were fixed and stained with DAPI and H3K9me2 antibodies. Images are maximum intensity projections. The brightness of the DAPI-only image was uniformly increased to visualize the filaments. Similar results were observed after separate depletion of Smc5 or Smc6 (Figure S8E).
(F) DNA filaments formed in the absence of Smc5/6 are dependent on recombination. Bw (control), Smc5/6, Rad51, and/or Rad54 were depleted and cells were processed as in (E). DNA filaments resulting from Smc5/6 RNAi are suppressed by simultaneous depletion of Rad51 and/or Rad54 (p < 0.0001; n > 500). Scale bars, 5 μm. See also Figure S8.
RNAi depletions were used to determine the interdependency of Smc5/6 and HP1a recruitment to heterochromatin. GFP-Nse2 colocalizes with intense HOECHST staining (which identifies the equivalent of DAPI-bright in living cells) in control cells, but not after HP1a RNAi (Figure 6C). Conversely, depletion of Smc5/6 has no impact on HP1a localization to heterochromatin or on the levels and nuclear distributions of H3K9me2 and H3K9me3 (Figure S8C). Similarly, Smc5/6 RNAi did not increase the DAPI-bright volume, a phenotype observed after depletion of Su(var)3–9, Su(var)3–9 + SetDB1, or HP1a (Figure S8D).
We conclude that the Drosophila Smc5/6 complex is a hetero-chromatin component, and its recruitment to heterochromatin is HP1a dependent.
Smc5 and Smc6 Are Required to Prevent Formation of Rad51 Foci and Aberrant Recombination Events in Heterochromatin
We next determined whether loss of the Smc5/6 complex affects the behavior of DSBs in heterochromatin. RNAi depletion of Smc5, Smc6, or Smc5+Smc6 significantly increased Rad51 foci formation only in DAPI-bright (Figure 6D and Figure S8E), similar to the phenotypes observed after Su(var)3–9 or HP1a depletion (Figure 5). The number of Rad51 foci that were observed in DAPI-bright after simultaneous depletion of Smc5, Smc6, and HP1a by RNAi is comparable to HP1a depletion alone (Figure 6D), suggesting that they control HR repair in heterochromatin through the same pathway. Similar to HP1a RNAi, the kinetics of heterochromatic γH2Av and Rad51 foci become equivalent to euchromatin after Smc5/6 depletion (compare Figures S8F and S8G with Figure 5B). Because Smc5/6 RNAi does not affect levels and distributions of H3K9me2, me3, and HP1a, as well as compaction of the DAPI-bright domain, we conclude that Smc5/6 acts downstream of HP1a in controlling Rad51 exclusion from the heterochromatin domain.
Finally, RNAi depletion of Smc5/6 complex components results in the appearance of extended DNA filaments between ~50% of nuclei (cells do not complete cytokinesis) (Figure 6E and Figure S8E). Similar phenotypes were observed after HP1a or Su(var)3–9 depletion (data not shown). These DNA filaments are enriched in H3K9me2 (Figure 6E and Figure S8E), indicating that they contain heterochromatic DNA. Importantly, the filament phenotype is rescued by blocking HR (simultaneous depletion of Smc5/6, Rad51, and/or Rad54) (Figure 6F), suggesting that they arise from aberrant recombination events.
We conclude that the presence of the Smc5/6 in heterochromatin precludes the assembly of Rad51 foci at early time points after damage. Removal of the Smc5/6 complex or of any of the heterochromatin components responsible for its recruitment results in Rad51 foci formation inside of heterochromatin and failure to properly resolve HR events.
DISCUSSION
Our findings reveal that DSBs are formed in Drosophila heterochromatin after IR and are rapidly recognized and processed for repair by HR. Furthermore, both the heterochromatin domain and heterochromatic repair foci display dramatic, dynamic behaviors in response to IR. Within minutes, γH2Av, Mu2, TopBP1, and ATRIP foci are assembled in heterochromatin, and the HP1a domain globally expands and forms dynamic protrusions. Later, repair foci relocate to the HP1a periphery or outside of the domain. Importantly, we directly show that damaged repeated sequences relocalize, not just DSBs within the HP1a domain. We also demonstrate that resection and checkpoint kinases, predominantly ATR, play crucial roles in both heterochromatin expansion and foci relocalization. Despite the HR requirement for repairing heterochromatin, Rad51 foci only form at the HP1a periphery or outside of the domain, where they associate with the HP1a dynamic protrusions and regions where HP1a is locally absent. These results demonstrate that the initial recognition and resection of heterochromatic DSBs are spatially and temporally separated from Rad51 foci assembly. Heterochromatin components are crucial for this regulation; the Smc5/6 complex, recruited to heterochromatin by HP1a, is the key component that is required to prevent formation of IR-induced Rad51 foci in heterochromatin. Smc5/6 depletion results in recombination-dependent heterochromatic DNA filaments between nuclei, revealing the importance of the Smc5/6 complex in suppressing abnormal HR in heterochromatin.
A Model for How HR Repair Is Controlled in Heterochromatin to Prevent Abnormal Recombination of Repeats
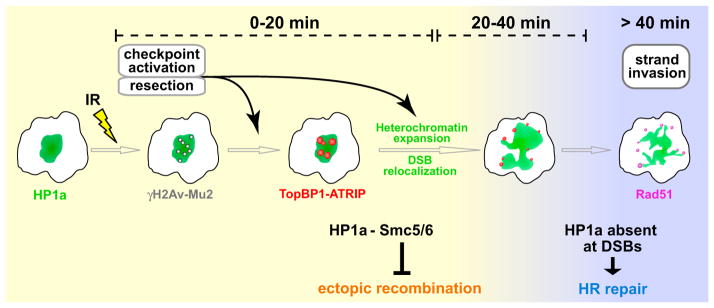
Our results suggest a model for how HR repair of damaged heterochromatic repeats could occur without generating genome instability (Figure 7 and Movie S7). Heterochromatic DSBs are quickly detected by early components of the DNA damage response, resulting in resection and checkpoint signaling, whereas recruitment of Rad51 is initially suppressed by HP1a through its association with the Smc5/6 complex. Expansion of the HP1a domain, triggered by resection and ATR activity, facilitates the relocalization of DSBs to the heterochromatin periphery. Next, the local absence or removal of HP1a at the periphery of the heterochromatin domain allows recruitment of Rad51 and other proteins required for recombination, which may also participate in tethering DSBs outside of the heterochromatin domain until repair is complete.
Figure 7. A Model for the Spatiotemporal Regulation of DSB Repair Events in Heterochromatin.
DSB detection, checkpoint activation, and resection occur quickly in heterochromatin, where ATR and HP1a facilitate the assembly of TopBP1/ATRIP foci. Meanwhile, Rad51 recruitment and strand invasion are suppressed by the Smc5/6 complex, which is recruited to heterochromatin by HP1a (yellow background). Heterochromatin expansion facilitates DSB relocalization to the HP1a periphery; both dynamic behaviors require checkpoint kinases (mainly ATR) and resection proteins. The euchromatic environment or local disassembly of HP1a at DSBs allows Rad51 loading and completion of HR repair (blue background). Once initiated, strand invasion facilitates the retention of DSBs at the HP1a periphery or the euchromatic domain during HP1a contraction. This mechanism prevents ectopic recombination by isolating the damaged site from the undamaged heterochromatic repeats before strand invasion. Homolog or chromatid pairing, maintained during relocalization, likely provide the templates for completing HR repair. Only processes related to DSB repair in heterochromatin are shown. See Movie S7 for animated version.
We propose that the dynamic spatial and temporal regulation of the DSB response in heterochromatin helps to ensure repeat and genome stability. Separation of the repeats with DSBs from the rest of the (undamaged) heterochromatin before strand invasion would preclude the use of sequences on nonhomologous chromosomes as templates for repair, thus preventing chromosome rearrangements. Pairing of sister chromatids and homologous chromosomes is maintained throughout interphase in Drosophila cells (McKee, 2004). Thus, genome stability could be ensured by completing recombination repair using homologous sequences on sister chromatids or homologs that relocalize in concert with the damaged DNA.
Aspects of this model share striking similarities with mechanisms described in S. cerevisiae to preserve rDNA stability, where the Smc5/6 complex plays a role in suppressing formation of Rad51 foci until DSBs relocalize to outside nucleoli (Torres-Rosell et al., 2007). This similarity is surprising because S. cerevisiae lacks H3K9 methylation and HP1 proteins, which in Drosophila are critical components of DSB repair in heterochromatin and are not enriched in nucleoli. In addition, nucleolar rDNAs are the most actively expressed genes in the genome, whereas Drosophila heterochromatin is mostly transcriptionally silent and contains a wider spectrum of repeats (simple, short satellites and transposable elements).
Our work extends the insightful yeast results by demonstrating that the spatiotemporal separation of HR repair events also occurs in the heterochromatin domain and that the response to IR includes physical expansion of HP1a. Moreover, we demonstrate that relocalization is coordinated with heterochromatin expansion and is orchestrated by checkpoint and resection activities, as well as heterochromatin components.
DSB Processing Is Highly Efficient in Heterochromatin
Contrary to previous conclusions (Cowell et al., 2007; Goodarzi et al., 2008; Kim et al., 2007), we show that heterochromatin does not represent an obstacle to DSB recognition and processing. In fact, Mu2 and γH2Av foci are quickly assembled in heterochromatin, with kinetics similar to euchromatin.
Unexpectedly, TopBP1 and ATRIP foci appear even faster in heterochromatin than in euchromatin and are unusually bright compared to euchromatic foci. These observations suggest that heterochromatin features may amplify some aspects of the DSB response, such as resection, or formation or clustering of TopBP1/ATRIP foci. TopBP1/ATRIP foci formation could be enhanced by the interaction that we identified between HP1a and ATRIP, which is increased after IR. Checkpoint kinases could also play a role, as caffeine treatment abrogates ATRIP foci specifically in heterochromatin.
Because resection is required for ATRIP-ATR loading and, together with ATR, for heterochromatin expansion and DSB relocalization, the efficiency of resection and/or ATRIP assembly is likely to play a pivotal role in promoting heterochromatin stability in the presence of DSBs. One advantage of efficient resection in heterochromatin might be to suppress NHEJ and promote HR repair (Huertas, 2010). Because NHEJ can generate translocations from DSBs in close proximity (Soutoglou and Misteli, 2008), safe repair of DSBs in repeats could require both immediate repression of NHEJ plus spatial and temporal separation of early and later steps in HR repair.
What Is the Significance of Heterochromatin Expansion?
We discovered that IR rapidly induces a global change in heterochromatin organization in Drosophila (even at low dose), visualized as dynamic HP1a protrusions, and expansion of both HP1a and DAPI-bright domains. This response may be conserved, as increased size of the HP1β domain was observed after laser microirradiation in mouse cells. However, it is unclear whether the HP1β response reflects heterochromatin relaxation (Ayoub et al., 2008; Kruhlak et al., 2006) or HP1β recruitment to euchromatic DSBs next to heterochromatin (Luijsterburg et al., 2009). In contrast, we never observe HP1a recruitment to γH2Av foci in Drosophila euchromatin, and multiple cell biological and epigenomic analyses demonstrate that heterochromatin volume expands in response to IR in Drosophila cells.
Previous studies suggested that heterochromatin relaxation is required to permit accessibility of repair components (Ayoub et al., 2008; Goodarzi et al., 2008). However, we observe that: (1) the peak of γH2Av, Mu2, ATRIP, and TopBP1 foci formation in heterochromatin occurs before the peak of heterochromatin expansion, (2) depletion of proteins required for expansion (e.g., resection components) does not interfere with the formation of Mu2 and γH2Av foci in heterochromatin, and (3) removal of Smc5/6, which does not interfere with heterochromatin compaction or the presence of heterochromatin marks (H3K9me2/3 and HP1a), permits Rad51 foci formation in heterochromatin. Nevertheless, the fact that heterochromatin expansion requires resection and checkpoint kinases, indicating that it is specifically induced by DSBs, suggests that expansion does play a role in the DSB response. Importantly, the peak of heterochromatin expansion coincides with the timing of DSB relocalization, and conditions that suppress expansion also prevent DSB relocalization. Thus, we propose that heterochromatin expansion may be required for mobilizing DSBs, rather than accessibility of repair components (see below).
What Regulates Relocalization of Foci and the Progression of HR Repair?
One of the most striking observations reported here is the movement of DSBs and repair foci to outside of the heterochromatin domain. Such directional, long-range DSB movements were previously considered a peculiarity of S. cerevisiae (Lisby et al., 2003), in contrast to the positional stability of repair foci in mammalian cells (Kruhlak et al., 2006; Soutoglou et al., 2007). An exception is that unprotected telomeres, which resemble DSBs, become mobilized in mouse cells (Dimitrova et al., 2008). Telomeres are enriched for repeats and HP1, suggesting the intriguing possibility that relocalization of DSBs is conserved in mammals, but only for regions of the genome where proximity of repeated sequences presents a dangerous environment for repair. Relocalization of DSBs to the heterochromatin periphery would also explain the absence of γH2Ax foci in chromocenters of mouse cells at > 30 min after IR (Cowell et al., 2007). Detailed studies of the kinetics of the DSB response and mobility of foci in mammalian heterochromatin are required to determine whether the dynamics that we observe in Drosophila are conserved in vertebrates.
Importantly, we show that relocalization of foci requires the presence of checkpoint and resection components. It is currently unknown whether they affect relocalization directly or indirectly through regulation of expansion. Further investigations are also required to determine the role of interactions between heterochromatin proteins and resection/checkpoint signaling (e.g., HP1a and ATRIP) in promoting DSB relocalization.
In terms of mechanisms, relocalization within nuclei could be an “active” directed process that is mediated by cytoskeletal elements and motors, which regulate other types of chromosome dynamics (Chuang et al., 2006; Sato et al., 2009). The movement of Rad51 foci at the tips of the HP1a protrusions suggest that “pulling” forces could be present that help to isolate DSBs from the main heterochromatin domain. However, inhibition of actin or tubulin polymerization (by latrunculin A or colchicine treatment, respectively) did not affect relocalization of heterochromatic foci (data not shown); additional studies are required to definitively determine whether cytoskeletal elements or motors are required. Alternatively, relocalization of foci could be regulated by “passive” mechanisms. The initial compaction of the domain could limit the motion of DSBs, with expansion allowing increased mobility. One possibility, based on the persistence of DSBs in heterochromatin after Rad51 or Rad54 depletion, is that relocalization is ensured by restricting strand invasion to outside of the heterochromatin domain, where active Rad51 and associated proteins “capture” resected DSBs. Interestingly, the human Ku70/80 complex is required to constrain the local motion of DSBs (Soutoglou et al., 2007). GFP-Ku80 is mostly absent in the HP1a compartment in Drosophila (data not shown), suggesting that suppression of NHEJ repair may contribute to the mobility of DSBs. These models are not mutually exclusive; for example, mobility could be triggered by active processes even if relocation involves a later capture mechanism.
Heterochromatin proteins, and particularly the Smc5/6 complex, could play different roles in relocalization and the progression of HR repair. Studies in yeast demonstrate that the Smc5/6 complex not only suppresses illegitimate HR, but also facilitates sister chromatid exchanges (SCEs) (De Piccoli et al., 2009). Thus, Drosophila Smc5/6 could promote SCEs at heterochromatic DSBs during completion of HR repair, in addition to its earlier role in exclusion of Rad51 foci.
How the Smc5/6 complex directly or indirectly controls HR is unknown. The Smc5/6 complex has an SMC structure, like cohesins and condensins, and the Nse2 subunit is an E3 SUMO-ligase (De Piccoli et al., 2009). An important challenge will be to determine whether the Smc5/6 complex controls the stability of Drosophila heterochromatic sequences by SUMOylating relevant targets of the recombination pathway or by recruiting other proteins, and whether relocalization of DSBs relies on the unique proprieties of SMC complexes, such as the ATP-dependent ability to link DNA molecules and interact with subnuclear structures.
Consequences of Misregulation of HR Repair in Heterochromatin
We previously showed that loss of HP1a or Su(var)3–9 results in excision of repeats and chromosome translocations in flies (Peng and Karpen, 2007, 2009). The results presented here suggest that heterochromatin components ensure genome stability by preventing aberrant recombination at DSBs after IR and that the Smc5/6 complex mediates this function. In addition, Smc5/6 removal without IR results in dramatic accumulation of abnormal, recombination-dependent heterochromatic filaments between dividing nuclei. These connections likely arise from deregulation of repair followed by chromosome segregation in the presence of unresolved recombination intermediates. Thus, heterochromatin components could also help to maintain genome stability during normal replication, when spontaneous damage can result from passage of replication forks through highly repeated DNA (Branzei and Foiani, 2010). Because the Smc5/6 complex is a constitutive heterochromatin component in Drosophila, we suggest that it is a key contributor to ensuring the stability of repeats after both spontaneous and induced damage. We propose that other examples of heterochromatic “threads” in human cells (Baumann et al., 2007) and Drosophila (Hughes et al., 2009; Royou et al., 2010) could arise from failures in control of heterochromatin HR repair or local, directed deregulation of the mechanisms described here.
Despite the challenges that they pose to genome stability, repeats are likely maintained in complex eukaryotes because they encode essential functions such as centromeres, meiotic pairing, and telomeres (Bernard et al., 2001; de Lange, 2005; Dernburg et al., 1996; Karpen et al., 1996). Our results suggest that heterochromatin components play an additional, major role in preventing genome and repeat instability through temporal and spatial regulation of HR repair. It will be important to determine whether the dynamic behaviors and regulatory roles of heterochromatin described here are conserved in vertebrates and whether defects in these processes contribute to human diseases associated with chromosome instability and aberrant heterochromatin, such as cancer (Dialynas et al., 2008) and aging (Shin et al., 2010).
EXPERIMENTAL PROCEDURES
Cell Culture, IR Treatments
Kc167 (Kc) cells were used for all experiments unless indicated otherwise and were maintained as logarithmically growing cultures. In IR experiments, the culture was exposed to 5Gy using a 320 kV x-ray source. In kinetics with fixed cells, time 0 (Unt) corresponds to cells fixed without exposing them to IR. In time-lapse experiments, time 0 (Unt) corresponds to cells imaged 5–10 min before IR treatment, unless otherwise indicated.
Cytological Methods, Time-Lapse Experiments, and Image Processing
IF, FISH, and TUNEL assays were described in Peng and Karpen (2007, 2009). Classification of foci inside or outside of the heterochromatin domain was done by analyzing their positions with respect to DAPI-bright or the HP1a domain in each of the Z stacks and in 3D image reconstructions with soft-WoRx. In time-lapse experiments, the same field of cells was imaged before and after IR, as described in Extended Experimental Procedures. Deltavision movie files were processed with Imaris (Bitplane AG, Zurich, Switzerland) for 3D rendering and compensation of movement, when necessary. Movies were created in Imaris, and time points were added using Photoshop (Adobe). 3D modeling (Figure 3C, Figure S5A, Movie S2, and Movie S3) was done with Imaris.
Quantitation of HP1a Expansion
3D volumes occupied by HP1a were analyzed for consecutive time points in time-lapse images of nuclei. Quantitation was done with custom Matlab scripts, as described in Extended Experimental Procedures. In the graphs, the volume quantified for each cell was normalized to the value (or mean value) of the same cell before IR.
ChIP-Chip
ChIP samples were prepared as described in Extended Experimental Procedures, and data were analyzed using the Affymetrix Tiling Analysis Software. Browser shots were generated with the Affymetrix Integrated Genome Browser.
Statistical Analysis
Unless otherwise indicated, n = number of cells/sample, and p values were calculated using the Kruskal-Wallis test with multiple-comparison Dunnet’s post test. Analyses were done with InStat (Graphpad).
Supplementary Material
Acknowledgments
This work was supported by postdoctoral fellowships from FIRC and Unesco to I.C., by Ruth Kirchstein NIH Postdoctoral Fellowship to S.U.C. (1F32GM086111), and by the Low Dose Radiation Research Program U.S. Department of Energy (DOE) (DE-AC02-05CH11231) to S.V.C. and G.H.K. We are grateful to P.-O. Mari for assistance with the NIR experiments and to S. Elgin, J. Kadonaga, B. Mellone, and D. Rio for reagents. We thank H.V. Le, R. Kunitake, and C. Pham for their help; A. Dernburg, E. Dunleavy, J. Swenson, and W. Zhang for comments on the manuscript; S. Langley for advice on ChIP-array data analysis; C-Y. Chen for software development; and the Karpen lab for sharing reagents and helpful discussions.
Footnotes
Supplemental Information includes Extended Experimental Procedures, eight figures, and seven movies and can be found with this article online at doi:10.1016/j.cell.2011.02.012.
References
- Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- Baumann C, Körner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Chuang C-H, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- Costes SV, Chiolo I, Pluth JM, Barcellos-Hoff MH, Jakob B. Spatiotemporal characterization of ionizing radiation induced DNA damage foci and their relation to chromatin organization. Mutat Res. 2009;704:78–87. doi: 10.1016/j.mrrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS ONE. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- De Piccoli G, Torres-Rosell J, Aragón L. The unnamed complex: what do we know about Smc5-Smc6? Chromosome Res. 2009;17:251–263. doi: 10.1007/s10577-008-9016-8. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- Dialynas GK, Vitalini MW, Wallrath LL. Linking Heterochromatin Protein 1 (HP1) to cancer progression. Mutat Res. 2008;647:13–20. doi: 10.1016/j.mrfmmm.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digilio FA, Pannuti A, Lucchesi JC, Furia M, Polito LC. Tosca: a Drosophila gene encoding a nuclease specifically expressed in the female germline. Dev Biol. 1996;178:90–100. doi: 10.1006/dbio.1996.0200. [DOI] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- Dronamraju R, Mason JM. Recognition of double strand breaks by a mutator protein (MU2) in Drosophila melanogaster. PLoS Genet. 2009;5:e1000473. doi: 10.1371/journal.pgen.1000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Reuter G. Cellular mechanism for targeting heterochromatin formation in Drosophila. Int Rev Cell Mol Biol. 2009;273:1–47. doi: 10.1016/S1937-6448(08)01801-7. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Kennedy C, Acevedo D, Evans-Holm M, Frise E, Wan KH, Park S, Mendez-Lago M, Rossi F, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SE, Gilliland WD, Cotitta JL, Takeo S, Collins KA, Hawley RS. Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes. PLoS Genet. 2009;5:e1000348. doi: 10.1371/journal.pgen.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Le MH, Le H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science. 1996;273:118–122. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- Kellum R, Raff JW, Alberts BM. Heterochromatin protein 1 distribution during development and during the cell cycle in Drosophila embryos. J Cell Sci. 1995;108:1407–1418. doi: 10.1242/jcs.108.4.1407. [DOI] [PubMed] [Google Scholar]
- Kim J-A, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Müller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K, Berres ME, Engels WR. Evolution of the RECQ family of helicases: A drosophila homolog, Dmblm, is similar to the human bloom syndrome gene. Genetics. 1999;151:1027–1039. doi: 10.1093/genetics/151.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee BD. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim Biophys Acta. 2004;1677:165–180. doi: 10.1016/j.bbaexp.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- Pebernard S, Schaffer L, Campbell D, Head SR, Boddy MN. Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J. 2008;27:3011–3023. doi: 10.1038/emboj.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 2009;5:e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A, Gagou ME, Karess R, Sullivan W. BubR1- and Polo-coated DNA tethers facilitate poleward segregation of acentric chromatids. Cell. 2010;140:235–245. doi: 10.1016/j.cell.2009.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907–919. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DM, Kucia M, Ratajczak MZ. Nuclear and chromatin reorganization during cell senescence and aging - a mini-review. Gerontology. 2010;57:76–84. doi: 10.1159/000281882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. On the contribution of spatial genome organization to cancerous chromosome translocations. J Natl Cancer Inst Monogr. 2008;2008:16–19. doi: 10.1093/jncimonographs/lgn017. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragón L, Lisby M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- Uanschou C, Siwiec T, Pedrosa-Harand A, Kerzendorfer C, Sanchez-Moran E, Novatchkova M, Akimcheva S, Woglar A, Klein F, Schlögelhofer P. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007;26:5061–5070. doi: 10.1038/sj.emboj.7601913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal T, Reuter G. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics. 2002;160:609–621. doi: 10.1093/genetics/160.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.