Abstract
Multiple sclerosis is a progressive neurological disorder that results in the degradation of myelin sheaths that insulate axons in the central nervous system. Therefore promotion of myelin repair is a major thrust of multiple sclerosis treatment research. Two mouse monoclonal natural autoantibodies, O1 and O4, promote myelin repair in several mouse models of multiple sclerosis. Natural autoantibodies are generally polyreactive and predominantly of the IgM isotype. The prevailing paradigm is that because they are polyreactive, these antibodies bind antigens with low affinities. Despite their wide use in neuroscience and glial cell research, however, the affinities and kinetic constants of O1 and O4 antibodies have not been measured to date. In this work, we developed a membrane biosensing platform based on surface plasmon resonance in gold nanohole arrays with a series of surface modification techniques to form myelin-mimicking lipid bilayer membranes to measure both the association and dissociation rate constants for O1 and O4 antibodies binding to their myelin lipid antigens. The ratio of rate constants shows that O1 and O4 bind to galactocerebroside and sulfated galactocerebroside, respectively, with unusually small apparent dissociation constants (KD ~0.9 nM) for natural autoantibodies. This is approximately one to two orders of magnitude lower than typically observed for the highest affinity natural autoantibodies. We propose that the unusually high affinity of O1 and O4 to their targets in myelin contributes to the mechanism by which they signal oligodendrocytes and induce central nervous system repair.
Keywords: Surface plasmon resonance, supported lipid bilayer, extraordinary optical transmission, nanohole arrays, natural autoantibodies, multiple sclerosis, remyelination, atomic layer deposition
INTRODUCTION
Multiple sclerosis (MS) is a neurologic disorder characterized by the loss of myelin sheaths in the central nervous system. Myelin loss impairs neuronal signal transduction and manifests itself in a number of symptoms including sensory and motor deficits.1 Approximately 1 out of every 1000 Americans has MS,2 but unfortunately there is no cure. Treatment options generally decrease exacerbations, but do not stop the long-term progression of the disease. Therefore, a major thrust in MS treatment research is to develop strategies to promote remyelination and protect vulnerable axons as a means to improve neurologic function. In earlier work, Rodriguez and coworkers showed that the IgM-isotype mouse monoclonal natural autoantibodies O1 and O4 promote remyelination in mouse models of MS.3 Furthermore, O1 and O4 are widely used to define oligodendrocyte lineage and monitor oligodendrocyte differentiation. O4 binds to sulfated galactocerebroside (Sulf) expressed on progenitor glial cells beginning at the time of commitment to the oligodendrocyte lineage through terminal differentiation of the cell and mature myelin membrane formation. O1 recognizes the presence of galactocerebroside (GalC) which is inserted into the oligodendrocyte membrane after the appearance of Sulf signifying later stages of maturation.4 Both O1 and O4 are polyreactive; O4 reacts with Sulf along with seminolipid and cholesterol, while O1 reacts with GalC as well as monogalactosyl-diglyceride and psychosine.5 Asakura et al. showed that both O1 and O4 have very few mutations from germline gene sequences, which indicates that they are natural autoantibodies.6 While these antibodies are useful in evaluating cell lineage and development, they also can initiate cell signaling and differentiation. When treated with O4, both astrocytes and oligodendrocytes display intracellular Ca2+ transients resulting from Ca2+ release from intracellular stores (astrocytes) or influx of extracellular Ca2+ (oligodendrocytes).7 Also, Bansal et al. showed that growing the cells in media containing O4 promoted oligodendrocyte differentiation whereas treating cultures with O1 did not.8
A number of lipid cell-surface antigens can be shown to bind O1 and O4 antibodies, but despite their wide use as detailed above, little is known about the affinity of the interactions or the kinetics by which these antibodies interact with their primary antigens, Sulf and GalC. Indeed, quantifying antibody binding kinetics to lipid bilayers mimicking cellular membranes is a very challenging task that requires label-free biosensors that can incorporate fluid lipid bilayer membranes. Commercial surface plasmon resonance (SPR) instruments such as Biacore have been widely used to measure receptor-ligand binding kinetics using ligands that are covalently attached on functionalized gold sensing surfaces.9,10 However, adapting conventional SPR instruments for sensing with fluid lipid membranes presents a series of challenges. Functionalized gold surfaces used in commercial SPR instruments can incorporate membrane-bound receptors on captured liposomes, however the morphology of the captured lipid membranes is not precisely known and appears to be dominated by intact liposomes rather than a planar lipid bilayer.11,12 Moreover, the capture efficiency of liposomes can be modulated by their charge state,12 making direct comparison between multiple liposome compositions more complex. Alternatively, lipid-antibody binding kinetics can be measured using planar supported lipid bilayer (SLB) membranes, which can be readily integrated with surface-sensitive techniques such as SPR. The majority of conventional SPR sensors utilize gold surfaces modified with self-assembled monolayers, whereas the formation of SLB is much easier on glass or silica surfaces.13 As Dahlin et al. have shown, the formation of SLBs on gold surfaces can be considerably simplified by patterning nanoholes in the gold film to expose the underlying silica surface.14 Thus we have employed the variant of SPR based on nanohole arrays in a gold film as the sensor chip and developed a new surface modification technique to conformally coat the entire sensor surface with an ultra-thin silica (SiO2) shell using atomic layer deposition (ALD) to facilitate the SLB formation without sacrificing the signal-to-noise ratios. This type of silica-based SPR chip functionalization is not available commercially. Furthermore, on the instrumentation side, the nanohole geometry facilitates simple optical measurements of molecular binding kinetics via recording changes in the transmission of light through nanoholes in a gold film, which is governed by a process known as extraordinary optical transmission (EOT).15,16,17 When light strikes a periodic array of nanoholes in a metallic film, surface plasmons, which are surface electromagnetic waves propagating at the metal interface, are launched. In EOT, these surface plasmons funnel through the nanoholes and enhance optical transmission at characteristic wavelengths. The wavelengths at which peaks in the transmission spectrum occur are related to the periodicity of the nanoholes, the dielectric function of the metal film and the refractive index of the overlying media. In our case, the overlying media includes the silica shell and the SLB. Binding of analytes to ligands immobilized in the SLB over the sensing surface increases the local refractive index and shifts the characteristic transmission wavelengths, analogous to the angular shift monitored in conventional SPR instruments. The positions of peaks and minima in the transmission spectrum are measured continually in real-time for label-free measurements of binding kinetics.18 While nanohole SPR sensors have been used for many proof-of-concept experiments,14,19-23 they have not been used for quantitative measurements between therapeutic antibodies and fluid SLBs. In this work, SLBs containing Sulf and GalC were formed on the silica-coated gold nanohole array, and antibody binding kinetics to these lipids was quantified in a label-free manner. Furthermore, the lipid-containing sample is injected through disposable microfluidic channels to avoid clogging, which can be problematic in permanent fluidic systems in commercial SPR instruments. The use of optimized nanohole SPR instruments tailored for SLBs has led to a high signal-to-noise ratio of ~800.
Current dogma states that natural IgM autoantibodies bind to targets with low affinity. The equilibrium-dissociation constants for natural IgM autoantibodies typically range from mM to 10 nM.24-26 In this work, however, we show that O1 and O4 bind to Sulf and GalC with significantly greater affinity; i.e. apparent dissociation constants in the sub-1 nM regime, which is at least an order of magnitude greater affinity than other natural autoantibodies. Furthermore, the nanohole SPR instrument can detect O4 binding to Sulf-containing lipid bilayers at concentrations as low as 310 pM. These high-affinity interactions may partially explain how therapeutic antibodies signal cells in culture and promote remyelination in vivo. In fact, we have shown that a natural IgM autoantibody derived from human serum with character similar to O4 and O1 can promote remyelination with a single 500-ng dose in a mouse model of MS.27 The high affinity of this class of IgMs may facilitate targeting in vivo at very low local tissue concentrations. These observations contrast the observations that, in general, the low affinity and varied pharmacokinetics of IgMs make them poor drug candidates.28
EXPERIMENTAL SECTION
Preparation of antibodies
The mouse monoclonal IgMs O4 and O1 were purified from hybridoma supernatant using PEG precipitation.29 The antibodies were further purified by gel filtration chromatography and the purity was verified by PAGE. Antibody concentration was determined using capture ELISA compared to commercial IgMs.
O1 and O4 binding to myelin by ELISA
Myelin was isolated from SJL/J mouse whole brain according to established procedures.30 Myelin quality was determined by Western blotting for the presence of myelin associated glycoprotein, myelin oligodendrocyte glycoprotein, proteolipid protein, 2′, 3′-cyclic nucleotide 3′-phosphodiesterase, and myelin basic protein, via the binding of well characterized anti-myelin lipid antibodies by direct ELISA. For antibody binding to myelin by ELISA, mouse myelin (10 μg/well diluted in PBS) was first dried overnight onto poly-D-lysine coated 96-well flat bottom plates (Nunc Immunosorp). Wells were washed twice with PBS and blocked for 2 hr with 3 % BSA/PBS, before antibodies diluted as indicated in 1 % BSA/PBS were added and incubated overnight at 4 °C. Following washing with PBS, bound mouse antibodies were detected using a goat anti-mouse μ-chain-specific alkaline phosphatase conjugated secondary antibody (Sigma) diluted in 1 % BSA/PBS for 4 hr followed by Sigma 104 phosphatase substrate (1 mg/mL) in Tris/Glycine buffer. Optical density was read at 405 nm (OD405) using a SpectraMax M2 micro plate reader (Molecular Devices, Sunnyvale, CA). Mean OD405 of triplicate values were calculated.
Immunocytochemistry using O1 and O4 on live oligodendrocytes
Immunocytochemistry surface staining was performed in the cold as previously described on unpermeabilized unfixed cells using 10 μg/mL of primary antibody.31 Briefly, mixed primary glial cells were prepared from Sprague-Dawley neonatal rats. Cells were plated at low density on poly-D-ornithine-coated glass cover slips (Fisher) and cells were grown for 14 days in DMEM 10 % fetal calf serum. After blocking for 10 min with 5 % BSA in HEPES buffered Earle’s balanced salt solution antibodies at 10 μg/mL in 1 % BSA were added for 15 minutes. After washing and fixation for 10 min with 4 % paraformaldehyde, bound primary antibodies were detected using fluorophore-conjugated goat anti-mouse μ-chain specific Fab’2 fragments (Jackson Immunoresearch). Coverslips were mounted in using Vectashield (Vector Labs) and viewed using an epi-fluorescence microscope.
Nanohole array fabrication
Standard microscope slides were cleaned by acetone, methanol, isopropyl alcohol, and water, and dried by a stream of nitrogen gas, then a 200 nm-thick gold film with a 5 nm-thick Cr adhesion layer was deposited by evaporation. Then, a 30 nm-thick Al2O3 masking layer was deposited on the gold-coated slides using ALD at 250 °C. After spin-coating a thermal nanoimprint resist (NXR-1025), a silicon imprint stamp (Lightsmyth) with a two-dimensional array of circular posts with 210 nm in diameter, 350 nm in depth, and 500 nm in periodicity was imprinted onto the resist with a pressure of 300 psi for 2 min at 130 °C. After partially removing residual resist with an oxygen dry etching, the Al2O3 was patterned using a dry etcher. Then the underlying gold film was etched with the patterned Al2O3 layer as a mask using an ion mill and the Al2O3 layer was removed. After making the nanohole array, the gold surface was uniformly covered by an 11 nm-thick SiO2 layer using ALD at 250 °C. The deposition rate was about 12 Å per cycle.
Formation of vesicles and supported lipid bilayers
Vesicles were composed of egg phosphatidylcholine (Avanti Polar Lipids) and GalC and Sulf (Sigma-Aldrich). To render the vesicles fluorescent, 1,2-dimyristoyl-sn-glycero-3-phosphatidylethanolamine-N-(lissamine rhodamine sulfonyl) was included. To form the vesicles, we dissolved the appropriate relative amounts of lipids in chloroform. All lipid compositions were calculated as mole percent. The chloroform was evaporated under vacuum for at least 6 hr to form a dry lipid film in a glass vial. The dry film was then rehydrated with Tris buffer (100 mM NaCl, 10 mM Tris, 1 mM EDTA) to a total lipid concentration of 2 mg/mL and allowed to rest overnight to form a vesicle suspension. Vesicles containing 6 % Sulf and 16 % GalC were rehydrated at 55 °C. The next day, the suspension was vortex mixed then sonicated for 10 min in a bath sonicator at room temperature. The vesicle suspension was then extruded through polycarbonate filters with pore diameter of 200 nm with an Avanti Mini-Extruder. The vesicles were then diluted to 1 mg/mL in Tris buffer containing calcium (100 mM NaCl, 10 mM Tris, 10 mM CaCl2). For FRAP studies, extruded vesicles were exposed to SiO2-coated gold nanohole arrays for 1 hr, then vigorously rinsed with buffer to remove excess unruptured vesicles. The FRAP experiments were carried out with an Olympus FV1000 BX2 confocal microscope equipped with a 60× water immersion objective with NA = 0.90. The diffusion coefficients for lipids in the SLBs were calculated by fitting the normalized fluorescence recovery curves to a single exponential function. The time at which half-maximum recovery occurred (t1/2) was determined and used to determine the diffusion coefficient using the equation D = r2/4t1/2, where r is the radius of the photobleached spot. At least 3 photobleached spots were used to calculate the average D values.
SPR biosensing
For SPR kinetic experiments, a poly(dimethylsiloxane) (PDMS) microfluidic flow-cell was assembled onto the silica-coated nanohole array chip. The microfluidic channels were 300 μm wide and 50 μm high. The assembled sensor chip was mounted on an upright microscope stage and illuminated with a tungsten-halogen light source. The optical transmission spectra were monitored with an Ocean Optics fiber-optic spectrometer and normalized to the spectrum of the light source. The flow of all solutions was controlled with a Harvard Apparatus syringe pump. After flowing a PBS solution through the microfluidic chip for 5 min, an extruded vesicle solution in Tris buffer containing calcium was injected and incubated for 1 hr at a flow rate of 3 μL/min to form a SLB on the silica-coated nanohole sensor surface. The channel was then washed by PBS at 30 μL/min for 30 min to remove excessive vesicles and lipid overlayers. Then the surface was blocked by injecting a 1 mg/mL BSA solution at 30 μL/min for 30 min followed by PBS washing for another 30 min. For kinetic measurement, baseline transmission spectra were measured with PBS at 30 μL/min for 3 min, and then an association curve was measured by flowing an antibody solution at 30 μL/min for 5 min. The injection flow rate was tuned to minimize mass transport limitations. After 5 min, PBS was injected for 15 min to measure a dissociation curve. Between each kinetic measurement, the surface was regenerated by dissociating all antibodies bound on the SLB. This was done by flowing a 2.7 M MgCl2 solution at 30 μL/min for 20 sec followed by PBS washing for 3 min. Instrumental noise was measured by calculating the standard deviation of the peak position when PBS was flowing through the microfluidic chip with a SLB on the sensing surface.
RESULTS AND DISCUSSION
O1 and O4 binding to oligodendrocytes and myelin
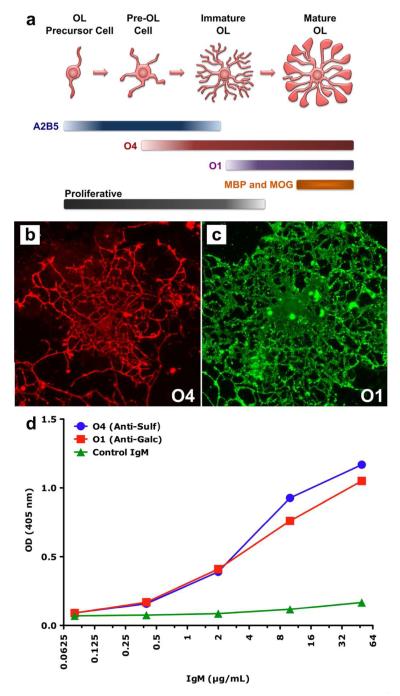
The monoclonal autoantibodies O1 and O4 bind to oligodendrocytes in vitro in a developmentally dependent manner.4 Figure 1a shows a scheme illustrating the stages of oligodendrocyte development as well as which antibodies bind to the cell surface during each developmental stage. When the cells are in immature, proliferative stages O4 will bind, as will A2B5, an antibody that binds several complex gangliosides present on early oligodendrocytes.32 As the oligodendrocytes mature, they insert more Sulf into their plasma membrane and, thus, strongly bind O4. At the preoligodendrocyte stage, the cells do not express GalC or other lipids associated with O1 binding, as such O1 will not bind. When the oligodendrocytes begin the process of myelinogenesis, they express Sulf and GalC, as well as myelin basic protein (MBP) and myelin oligodendrocyte glycoprotein (MOG). Therefore, antibodies O1 and O4 will bind to the cell surface as will antibodies directed toward MBP and MOG. Figure 1b shows an immature oligodendrocyte that strongly binds O4, while Figure 1c shows a mature oligodendrocyte that strongly binds O1. Compared to the cell in Figure 1b, it is clear that the cell in Figure 1c has more fully developed processes and larger soma associated with oligodendrocyte maturity.
Figure 1.
Assessment of oligodendrocyte phenotype by immunocytochemistry. (a) This diagram represents the morphological and antigen expression changes that occur as oligodendrocytes (OL) differentiate from a simple bipolar proliferative precursor cell to a mature complex post-proliferative cell capable of synthesizing myelin membrane. A2B5 is a mouse IgM that recognizes several complex gangliosides and is often used to define oligodendrocyte precursor cells. The binding of O4 to cell surface Sulf indi cates the cell’s commitment to the oligodendrocytes phenotype. The loss of A2B5 binding and the expression of Sulf is associated with the cessation of proliferation. The binding of O1 to surface GalC and the subsequent expression of MBP and MOG are indicative of mature post-mitotic oligodendrocytes. (b and c) An example of O4 binding to the surface of a live immature rat oligodendrocyte (b) and of O1 binding to the surface of a live mature oligodendrocyte (c). (d) ELISA of O1 and O4 binding to purified myelin.
We also examined how O1 and O4 bind to myelin extracted from mouse brain. Figure 1d shows results from ELISA where varying concentrations of O1, O4 and an isotype control IgM (MMEN-OE5 antibacterial lipopolysaccharide IgM κ) were exposed to myelin. Assuming a molecular weight for an IgM pentamer of 900 kDa,33 the mass concentration range shown in Figure 1d corresponds to molar concentrations ranging from 69 pM to 71 nM. At the low end of the concentration scale, there is little difference in optical density between O1, O4 and control samples. However, as the concentrations of antibodies increase, O1 and O4 bind myelin in a concentration-dependent fashion. At ~0.4 μg/mL (0.44 nM), O1 and O4 binding is clearly greater than the control. Furthermore, as the concentrations increase, both O1 and O4 bind myelin with similar intensity.
Myelin is a complex mixture of lipids and proteins; about 70 % and 30 % of the dry weight respectively,34 whereas most other cell membranes are roughly 50 % lipid and 50 % protein. Combined Sulf and GalC make up over 20 % of the lipid weight of myelin.35 Because oligodendrocyte membranes and myelin are complex mixtures of lipids and proteins, we used myelin-mimicking SLBs on a nanohole SPR sensor to quantify the specific interactions between O1/GalC and O4/Sulf.
Supported lipid bilayers on nanohole arrays
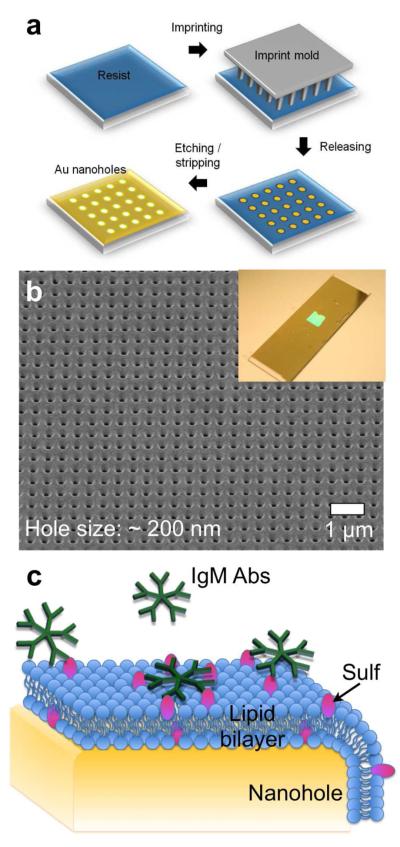
Periodic nanohole arrays with 200 nm hole diameter and 500 nm periodicity were made in a cm2 area of a gold film by nanoimprint lithography. After fabrication, a thin conformal layer (~11 nm) of SiO2 was deposited on the nanohole array by ALD. Figure 2a shows a schematic of the fabrication process, while Figure 2b shows a scanning electron micrograph (SEM) of a nanohole array in a gold film. The size of patterned area is scalable, similar to the nanoimprint stamp. Nanoimprint lithography patterned the nanoholes over a large area (Figure 2b inset) to allow integration with a multi-channel microfluidic flow cell.
Figure 2.
(a) A schematic illustration of the nanohole fabrication process using nanoimprint lithography. (b) A scanning electron micrograph (SEM) of a nanohole array. The hole diameter and periodicity are 200 nm and 500 nm, respectively. The inset to Figure 2(b) shows a photograph of the fabricated nanohole SPR chip. The nanohole array is patterned in a ~cm2 area in the middle of a gold-coated glass slide. (c) Schematic representation of a SLB on a gold nanohole array. The pentameric structures represent antibodies (IgM Abs), such as O1 and O4.
SLBs have been employed widely for studying membrane biochemistry and biophysics.36-38 SLBs were formed by spontaneous rupture of phospholipid vesicles on SiO2, which promotes the rupture of phospholipid vesicles by a well characterized mechanism.39,40 The vesicles used consisted of egg phosphatidylcholine (egg PC) and 2 % GalC or 2 % Sulf. When necessary, 1 % of a fluorescent lipid, 1,2-dimyristoyl-sn-glycero-3-phosphatidylethanolamine-N-(lissamine rhodamine sulfonyl) (Rho-PE), was included for imaging. Figure 2c shows a schematic of a SLB on a nanohole array.
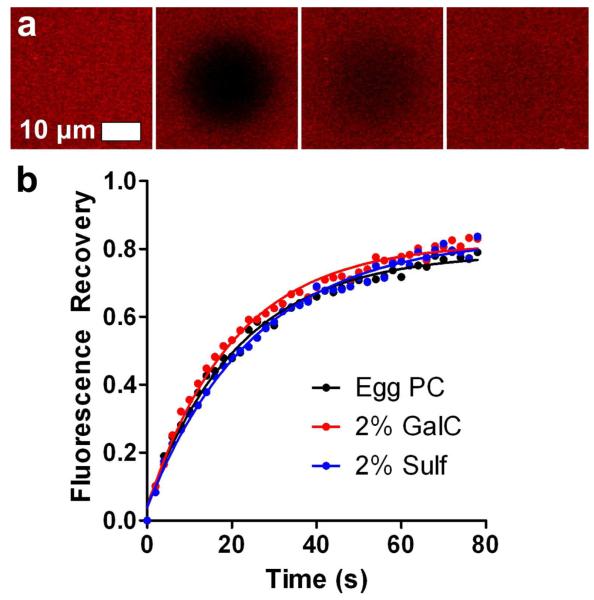
The formation of SLBs on nanohole arrays was confirmed by fluorescence recovery after photobleaching (FRAP).41 For FRAP, small circular areas of the SLBs were photobleached, and the recovery of fluorescence in the photobleached area was monitored as a function of time to generate recovery curves. Figures 3a shows four frames from a photobleached SLB on a nanohole array. Figure 3b shows representative recovery curves obtained from FRAP experiments on SLBs with 2 % GalC, 2 % Sulf and a control SLB free of GalC or Sulf. The recovery curves show that the lipid mixtures do indeed form continuous fluid SLBs on SiO2-coated nanohole arrays. The diffusion coefficients (D) for lipids in the SLBs containing Sulf and GalC SLBs were not significantly different from the Egg PC control, as determined by unpaired t-test. The D for the control was calculated to be 1.84 ± 0.14 μm2/s, while for the 2 % Sulf SLB, D = 1.60 ± 0.14 μm2/s, and for the 2 % GalC SLB, D = 1.95 ± 0.16 μm2/s (mean ± standard deviation). The D values observed here are somewhat higher than those obtained for SLBs on random nanohole arrays in SiO2-coated gold,22 but lower than those observed on nonporous flat SiO2.42 The maximum fluorescence recovery for the SLBs on nanohole arrays is 75 – 80 %, which suggests an immobile lipid fraction and/or unruptured vesicles on the surface. Because the SLBs conform to the nanohole topography,43 there may be pinning of lipids at tightly curved surfaces at the rim of the nanoholes, around the interior of the nanohole surface and around the base of the nanohole which hinders diffusion and leads to an apparent immobile fraction.
Figure 3.
FRAP of SLBs on SiO2-coated gold nanohole arrays. (a) Four frames showing, from left to right, SLB fluorescence before photobleaching, the same SLB immediately after photobleaching, and at 20 and 80 seconds after photobleaching . (b) Fluorescence recovery curves for SLBs composed solely of Egg PC and Egg PC with 2 % GalC or 2 % Sulf.
SPR sensing of O1 and O4 binding to SLBs
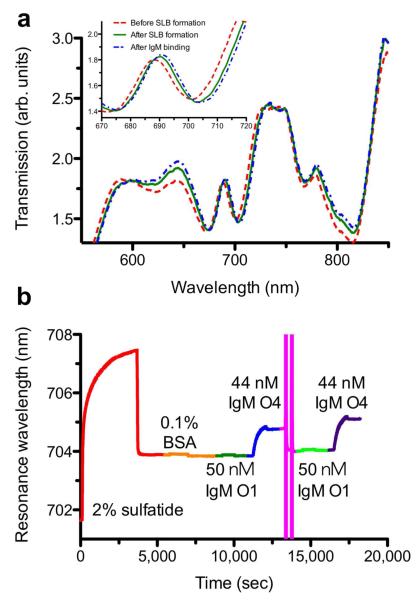
For SPR sensing a poly(dimethylsiloxane) (PDMS) microfluidic chip prepared by soft lithography44 was attached to the nanohole SPR sensor. Figure 4a shows transmission spectra for a SiO2-coated gold nanohole array with hole diameter of 200 nm and periodicity of 500 nm, the same nanohole array after formation of a SLB containing 2 % Sulf and after O4 binds to Sulf in the SLB. When the SLB is formed over the nanohole array the optical transmission spectrum shifts toward longer wavelengths by approximately 2 nm. This shift was consistent between different channels on the chip and from experiment to experiment, which confirmed that the SLBs formed on the nanohole arrays without fluorescence imaging. Yang and Cremer previously showed that SLBs will readily form in microfluidic channels.45 The injection of 44 nM O4 red-shifts the spectrum by 0.8 nm. While the magnitudes of the shifts seem small, the standard deviation of the noise in our instrument is extremely small, typically in the range of 10−3 nm, which means that the limit of detection (defined as a signal to noise (S/N) ratio of 3) is a shift of 0.003 nm and the S/N for a 0.8 nm spectral shift is on the order 800.
Figure 4.
(a) The optical transmission spectra before SLB formation (red), after SLB formation (green) and after IgM antibody binding (blue). (b) A full sensorgram from formation of a SLB containing 2 % Sulf to IgM antibody binding to the SLB surface . The spectral position in minimum transmission around 700 nm was monitored to track the changes on the surface. A 2.7 M MgCl2 solution was used to regenerate for serial kinetic measurements on the same membrane surface.
In nanohole-based SPR sensors, kinetic sensorgrams can be recorded by tracking either the transmission maximum (peak) or minimum (dip) in real time.46 In our system, binding kinetics extracted from the transmission minimum showed a slightly better signal-to-noise ratio, thus we tracked the position of the local minimum in the EOT spectrum around 700 nm (Figure 4a) as a function of time. Figure 4b shows a sensorgram encompassing an entire experiment from SLB formation to multiple antibody injections and surface regeneration. The course of the experiment was as follows. A SLB containing Sulf was formed on the nanohole sensor, resulting in a spectral shift of approximately 2 nm after washing excess vesicles from the sensor. Then the SLB was blocked with BSA to minimize nonspecific adsorption. Following blocking, 50 nM O1 was injected, and no significant shift was observed, indicating that O1 does not bind to a Sulf-containing egg PC membrane, consistent with previous reports.5 After O1, 44 nM O4 was injected and the spectrum appreciably red-shifts to a maximum of about 0.8 nm. After O4 association, the injection solution was switched to antibody-free phosphate buffered saline (PBS) to monitor O4 dissociation. After O4 dissociation, the remaining O4 was removed from the surface with a high ionic strength regeneration solution containing 2.7 M MgCl2.47 The large fluctuations in position seen in the sensorgram during this step are due to the vastly different refractive index of the regeneration solution. After the regeneration wash, the peak position nearly returned to the pre-injection position, indicating that the regeneration wash removed the bound O4, but did not disrupt the SLB on the nanohole sensor. After regenerating the surface, repeated injections of 50 nM O1 and 44 nM O4 confirm that the SLB is intact and that serial measurements could be carried out on our nanohole-SLB platform.
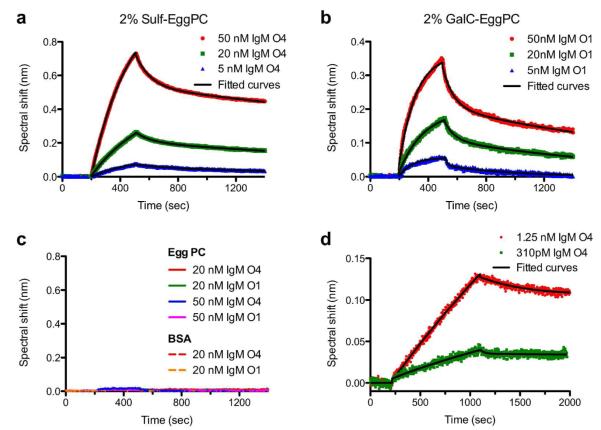
Figures 5a and 5b show binding kinetic curves for multiple concentrations of O4 and O1 binding to SLBs containing 2 % Sulf and 2 % GalC, respectively. The curves in Figures 5a and 5b show that the spectral shift increases with increasing concentrations of antibody injected, as expected. After injection of the antibodies, the injection solution was switched to antibody-free PBS. Upon switching the injection solution, the antibodies dissociate from the SLB surface, and spectral shift decay is observed. The association and dissociation curves were fit to exponential functions to determine the association and dissociation rate constants (kon and koff).48 The kon and koff rate constants for O1 binding to GalC were (6.98 ± 4.92) × 105 M−1 s−1 and (1.10 ± 0.51) × 10−3 s−1 (mean ± standard error of the mean (s.e.m.)), respectively. For O4 binding to Sulf kon and koff were (2.65 ± 1.71) × 105 M−1 s−1 and (3.73 ± 1.15) × 10−4 s −1, respectively. The ratio of the rate constants calculated for each individual experiment were used to determine the apparent dissociation constant (KD) with the equation KD = koff / kon. The KD values from individual experiments (N = 3) were averaged to calculate the mean KD value. The KD values for GalC/O1 and Sulf/O4 were (2.37 ± 0.56) nM and (2.19 ± 0.61) nM, respectively (mean ± s.e.m.). Table 1 shows the KD values obtained from these studies compared to KD values determined for other IgM natural autoantibodies. The KD values for O1/GalC and O4/Sulf interactions are nearly 20-fold smaller than previously reported values, indicating that these antibodies have unexpectedly high affinities for their targets compared to other IgM autoantibodies.
Figure 5.
SPR kinetic curves for IgM autoantibodies binding to SLBs. (a) O4 binding to SLBs containing 2 % Sulf. (b) O1 binding to SLBs with 2 % GalC. (c) Negative controls showing that neither O1 nor O4 bind to SLBs composed solely of egg PC, and that neither O1 nor O4 bind to BSA-blocked, silica-coated nanohole arrays. (d) Kinetic curves for O4 binding to a SLB containing 6 % Sulf and 16 % GalC, which mimics the lipid composition of myelin. The dark lines superimposed upon all of the kinetic curves are exponential fits to the data.
Table 1.
Binding Characteristics of IgM Natural Autoantibodies
| Antibody | Antigen(s) | Method | KD (M) | kon (M−1 s−1)(b) | koff (s−1)(b) | Ref |
|---|---|---|---|---|---|---|
| O1(a) | 2 % GalC in SLB |
SPR | (2.37±0.56)×10−9 | (6.98±4.92)×105 | (1.10±0.51)×10−3 | This study |
| O4(a) | 2 % Sulf in SLB |
SPR | (2.19±0.61)×10−9 | (2.65±1.71)×105 | (3.73±1.15)×10−4 | This study |
| F5-2 | Actin Myosin Tubulin |
SPR | 4.03 × 10−8 2.74 × 10−8 6.80 × 10−8 |
1.85 × 104 2.84 × 104 1.67 × 104 |
7.46 × 10−4 7.78 × 10−4 1.14 × 10−3 |
49 |
| 2E4 | ssDNA β-galactosidase Insulin |
ELISA | 4 × 10−8 7 × 10−7 1 × 10−5 |
N.D. | N.D. | 50 |
| 274.RA.F11 | IgG Fc frag. | ELISA | 4 × 10−5 | N.D. | N.D. | 51 |
| 274.RA.F5 | IgG Fc frag. | ELISA | 3 × 10−7 | N.D. | N.D. | 51 |
| Ab 50 | IgG Fc frag. ssDNA Insulin Tetanus toxoid |
ELISA | 1 × 10−5 5.6 × 10−7 1 × 10−4 6.4 × 10−6 |
N.D. | N.D. | 52 |
| Ab 52 | IgG Fc frag. ssDNA Insulin Tetanus toxoid |
ELISA | 3 × 10−5 1 × 10−6 1 × 10−4 2.5 × 10−6 |
N.D. | N.D. | 52 |
KD, kon and koff values reported as mean ± s.e.m.
N.D. = not determined.
We conducted a number of negative control experiments to confirm that the binding of antibodies was due to specific interactions. (Figure 5c) With both 20 nM and 50 nM O1 and O4, no binding was observed to SLBs composed solely of Egg PC. Also, when a nanohole array without a SLB was exposed to 1 mg/mL BSA, similar to the blocking step mentioned above, and then 20 nM O1 or O4 were injected, no spectrum shifts were observed. This indicates that neither O1 nor O4 nonspecifically adsorb to the egg PC matrix of the SLB, or the BSA used for blocking.
Sulf and GalC make up 6 mole % and 16 mole % of the myelin lipid composition, respectively.53 To better mimic the lipid composition of myelin, a SLB was formed on the nanohole sensor containing 6 % Sulf and 16 % GalC in a matrix of egg PC. With this SLB on the sensor, lower concentrations of O4 were injected to approach the detection limit for this system. We injected 1.25 nM and 310 pM O4 and both concentrations were readily detectable. (Figure 5d) The kinetic rate constants obtained for O4 binding to these myelin-mimicking SLBs were 1.26 × 105 M−1 s−1 (kon) and 1.16 × 10−4 s−1 (koff) and the apparent KD was calculated to be 920 pM. This indicates that O4 binds myelin-like membranes with unusually high affinity for a natural autoantibody.
Because little is known about the affinities of O1 and O4 for GalC and Sulf, it is difficult to compare the results obtained with nanohole SPR sensors to results obtained with commercial SPR instruments or other alternative methods. However, in previous work, our group compared kinetic rate constants and KD values obtained from a nanohole SPR instrument to those obtained from a commercial SPR instrument.16 Antibodies binding to non-lipid antigens with known KD values ranging from 200 pM to 40 nM were examined. For all binding interactions we studied, the rate constants and KD values obtained with nanohole SPR were quite similar to those measured with a commercial Biacore 3000 system. This confirms that the nanohole SPR platform is capable of accurate quantification of binding kinetics.
The KD values obtained in this study were compared to those of other antibodies from previously published studies. (Table 1) The antibodies listed in Table 1 are all IgM natural autoantibodies, and the KD values determined in previous studies range from 100 μM to 40 nM. The smallest KD value is almost 20-fold larger than the values determined in this study. In the previous examples the KD values were determined by SPR or equilibrium assays, such as ELISA. However, because traditional equilibrium assays only offer a static snapshot of the situation at equilibrium, they cannot determine the rate constants kon and koff. Furthermore, the dissociation constant can be calculated from the ratio of the dissociation and association rate constants. Thus, it is possible for two given antibody-antigen interactions to have the same KD value, but drastically different kon and koff values. For any type of potential therapeutic molecule, including remyelinating IgMs, the koff can be a more powerful predictor of potential efficacy than KD.54 Regardless of this fact, in vitro measurements of KD have become acceptable stand-ins for in vivo efficacy in the drug discovery process.55 There are examples, however, where KD and efficacy are well correlated.56 Because drugs are only effective when bound to their receptor, the residence time of a drug at its receptor is a better measure of drug-receptor interactions than the affinity. Residence time is inversely proportional to koff and correlates better with in vivo efficacy and duration of action than does KD.54 Therefore, kinetic-sensing schemes that determine rate constants are preferable when investigating potential therapeutic molecules.
Affinity vs. Avidity
IgM antibodies can bind antigens with high avidity because a single IgM is capable of binding 10 antigens. Therefore it is likely that many of the binding events on our SPR sensors are polyvalent. When an antibody encounters the SLB surface, only one antibody/antigen binding event is required for association and a measured shift in the SPR signal. Subsequent binding events for a single antibody do not contribute to major shifts in the SPR signal because the antibody has already associated with the SLB. On the flip side, for an IgM to completely dissociate from the sensor surface, all of the antibody/antigen interactions must dissociate. Thus, the increased binding avidity of IgMs could contribute to the relatively low apparent KD values observed in this study. We are primarily concerned with comparing O1 and O4 to other IgM natural autoantibodies, which also can display high binding avidity. A particularly apt comparison is to look at kon and koff rate constants for other IgM natural autoantibodies studied by SPR and compare those to the rate constants obtained for O4/Sulf and O1/GalC binding using myelin-mimicking lipid membranes. For example, the F5-2 natural autoantibody studied by Diaw et al. had kon rate constants that were between 9 and 42-times slower (depending on antigen) than those measured for O4/Sulf and O1/GalC.49 (Table 1) The koff rate contants for F5-2 were a maximum of 3-times faster than that observed for O4/Sulf and O1/GalC binding. Because KD is a ratio of these rate constants, the lower KD values observed in our study are largely due to the increased kon of O4 binding to Sulf and O1 binding to GalC. This suggests that the initial recognition process that leads to antibody association, i.e. the affinity between a single antibody binding site and an antigen, is the primary cause of the smaller apparent KD (higher apparent affinity) of O1 and O4 antibodies, rather than increased avidity of O1 and O4 for GalC and Sulf, respectively. An alternative, though unlikely, explanation for the increased kon rates is that the sensor nanohole architecture, to which the SLB with antigens conforms, may artificially increase association rates and/or slow dissociation rates by making lipid antigens more available for initial binding or multivalent binding due to the curvature of the SLB. However, because the nanoholes themselves comprise only 12.5 % of the sensor area, this is unlikely to have a pronounced effect.
CONCLUSION
The present experiments have great significance in defining the mechanism of action by which mouse and human monoclonal antibodies promote remyelination.57 The human antibodies that promote remyelination have also been found to be of the IgM subclass and have the characteristic features of natural autoantibodies. No human IgG antibodies that promote tissue repair in vivo have been identified. Efforts to convert the IgMs to an IgG form, either by spontaneous mutation or by molecular construction, have resulted in the absence of Ca2+ responses in cultured glial cells but, most importantly, an absence of in vivo remyelinating activity.58 Therefore, the high-affinity, high-avidity pentameric structure is essential for the therapeutic response of those antibodies. Traditionally, IgM antibodies, specifically natural autoantibodies, have been considered to have low affinity binding.24-26 The fact that our present experiments show the opposite result, that is, small apparent KD values, may explain why these antibodies have therapeutic efficacy in vivo. In vivo experiments using magnetic resonance imaging show human natural autoantibodies cross the blood brain barrier and indicate that if the antibody has a clear target to bind within the central nervous system (CNS), then the therapeutic antibody can be detected for long as one week in the CNS.59 Similar results have been obtained with 35S-labeled antibodies using EM autoradiography.60 The fact that O1 and O4, despite being IgMs, bind tightly to their antigens is further confirmation of their strong affinity and being IgMs they have high avidity for the myelin target. On the technology side, the combination of nanohole SPR sensor chip with biomimetic SLBs shows great promise for studying these important lipid-antibody reactions in a quantitative manner to reveal not only equilibrium constants but also on-off rates that can help understand the mechanisms of molecular recognition as well as stability of binding. Finally, as human natural autoantibodies begin entering clinical trial, these results may help to determine the dosing schedule for optimum therapeutic efficacy.
ACKNOWLEDGEMENTS
This work was supported by grants to S.H.O. and M.R. from the National Institutes of Health (R01 GM092993) and the Minnesota Partnership Award for Biotechnology and Medical Genomics. S.H.O. also acknowledges support from the Office of Naval Research (ONR) Young Investigator Program and the NSF CAREER Award (DBI 1054191). This work was also supported by grants to M.R. from the National Institutes of Health (NS024180, NS032129, NS048357, R21 NS073684), the National Multiple Sclerosis Society (CA1060A11), the Applebaum Foundation, and the Hilton Foundation.
REFERENCES
- (1).Noseworthy JH, Lucchinetti C, Rodriguez M, Weinshenker BG. New Engl. J. Med. 2000;343:938–952. doi: 10.1056/NEJM200009283431307. [DOI] [PubMed] [Google Scholar]
- (2).Anderson DW, Ellenberg JH, Leventhal CM, Reingold SC, Rodriguez M, Silberberg DH. Ann. Neurol. 1992;31:333–336. doi: 10.1002/ana.410310317. [DOI] [PubMed] [Google Scholar]
- (3).Asakura K, Miller DJ, Pease LR, Rodriguez M. J. Neurosci. 1998:7700–7708. doi: 10.1523/JNEUROSCI.18-19-07700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Sommer I, Schachner M. Dev. Biol. 1981;83:311–327. doi: 10.1016/0012-1606(81)90477-2. [DOI] [PubMed] [Google Scholar]
- (5).Bansal R, Warrington AE, Gard AL, Ranscht B, Pfeiffer SE. J. Neurosci. Res. 1989;24:548–557. doi: 10.1002/jnr.490240413. [DOI] [PubMed] [Google Scholar]
- (6).Asakura K, Miller DJ, Pogulis RJ, Pease LR, Rodriguez M. Mol. Brain Res. 1995;34:283–293. doi: 10.1016/0169-328x(95)00190-4. [DOI] [PubMed] [Google Scholar]
- (7).Paz-Soldan MM, Warrington AE, Bieber AJ, Ciric B, Van Keulen V, Pease LR, Rodriguez M. Mol. Cell. Neurosci. 2003;22:14–24. doi: 10.1016/s1044-7431(02)00018-0. [DOI] [PubMed] [Google Scholar]
- (8).Bansal R, Gard AL, Pfeiffer SE. J. Neurosci. Res. 1988;21:260–267. doi: 10.1002/jnr.490210218. [DOI] [PubMed] [Google Scholar]
- (9).Smith EA, Corn RM. Appl. Spectrosc. 2003;57:320A–332A. doi: 10.1366/000370203322554446. [DOI] [PubMed] [Google Scholar]
- (10).Mrksich M, Sigal GB, Whitesides GM. Langmuir. 1995;11:4383–4385. [Google Scholar]
- (11).Cooper MA, Hansson A, Lofas S, Williams DH. Anal. Biochem. 2000;277:196–205. doi: 10.1006/abio.1999.4389. [DOI] [PubMed] [Google Scholar]
- (12).Anderluh G, Besenicar M, Kladnik A, Lakey JH, Macek P. Anal. Biochem. 2005;344:43–52. doi: 10.1016/j.ab.2005.06.013. [DOI] [PubMed] [Google Scholar]
- (13).Reimhult E, Höök F, Kasemo B. Langmuir. 2003;19:1681–1691. [Google Scholar]
- (14).Dahlin A, Zäch M, Rindzevicius T, Käll M, Sutherland DS, Höök F. J. Am. Chem. Soc. 2005;127:5043–5048. doi: 10.1021/ja043672o. [DOI] [PubMed] [Google Scholar]
- (15).Brolo AG, Gordon R, Leathem B, Kavanagh KL. Langmuir. 2004;20:4813–4815. doi: 10.1021/la0493621. [DOI] [PubMed] [Google Scholar]
- (16).Im H, Sutherland JN, Maynard JA, Oh SH. Anal. Chem. 2012;84:1941–1947. doi: 10.1021/ac300070t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ebbesen TW, Lezec HJ, Ghaemi HF, Thio T, Wolff PA. Nature. 1998;391:667–669. [Google Scholar]
- (18).Gordon R, Sinton D, Kavanagh KL, Brolo AG. Acc. Chem. Res. 2008;41:1049–1057. doi: 10.1021/ar800074d. [DOI] [PubMed] [Google Scholar]
- (19).Stewart ME, Mack NH, Malyarchuk V, Soares J, Lee TW, Gray SK, Nuzzo RG, Rogers JA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17143–17148. doi: 10.1073/pnas.0606216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yang JC, Ji J, Hogle JM, Larson DN. Nano Lett. 2008;8:2718–2724. doi: 10.1021/nl801043t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Im H, Wittenberg NJ, Lesuffleur A, Lindquist NC, Oh SH. Chem. Sci. 2010;1:688–696. doi: 10.1039/C0SC00365D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Jonsson MP, Jonsson P, Dahlin AB, Höök F. Nano Lett. 2007;7:3462–3468. doi: 10.1021/nl072006t. [DOI] [PubMed] [Google Scholar]
- (23).Yang JC, Ji J, Hogle JM, Larson DN. Biosens. Bioelectron. 2009;24:2334–2338. doi: 10.1016/j.bios.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lutz HU, Binder CJ, Kaveri S. Trends Immunol. 2009;30:43–51. doi: 10.1016/j.it.2008.10.002. [DOI] [PubMed] [Google Scholar]
- (25).Notkins AL. Trends Immunol. 2004;25:174–179. doi: 10.1016/j.it.2004.02.004. [DOI] [PubMed] [Google Scholar]
- (26).Zhou Z, Tzioufas AG, Notkins AL. J. Autoimm. 2007;29:219–228. doi: 10.1016/j.jaut.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Warrington AE, Bieber AJ, Ciric B, Pease LR, Van Keulen V, Rodriguez M. J. Neurosci. Res. 2007;85:967–976. doi: 10.1002/jnr.21217. [DOI] [PubMed] [Google Scholar]
- (28).Maynard J, Georgiou G. Annu. Rev. Biomed. Eng. 2000;2:339–379. doi: 10.1146/annurev.bioeng.2.1.339. [DOI] [PubMed] [Google Scholar]
- (29).Cripps AW, Neoh S, Smart IJ. J. Immunol. Meth. 1983;57:197–204. doi: 10.1016/0022-1759(83)90078-9. [DOI] [PubMed] [Google Scholar]
- (30).Norton WT, Poduslo SE. J. Neurochem. 1973;21:749–757. doi: 10.1111/j.1471-4159.1973.tb07519.x. [DOI] [PubMed] [Google Scholar]
- (31).Bieber AJ, Warrington AE, Asakura K, Ciric B, Kaveri SV, Pease LR, Rodriguez M. Glia. 2002;37:241–249. doi: 10.1002/glia.10033. [DOI] [PubMed] [Google Scholar]
- (32).Eisenbarth GS, Walsh FS, Nirenberg M. Proc. Natl. Acad. Sci. U.S.A. 1979;76:4913–4917. doi: 10.1073/pnas.76.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Feinstein A, Munn EA. Nature. 1969;224:1307–1309. doi: 10.1038/2241307a0. [DOI] [PubMed] [Google Scholar]
- (34).Baumann N, Pham-Dinh D. Physiol. Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- (35).Ohler B, Graf K, Bragg R, Lemons T, Coe R, Genain C, Israelachvili J, Husted C. Biochim. Biophys. Acta. 2004;1688:10–17. doi: 10.1016/j.bbadis.2003.10.001. [DOI] [PubMed] [Google Scholar]
- (36).Tamm LK, McConnell HM. Biophys. J. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Castellana ET, Cremer PS. Surf. Sci. Rep. 2006;61:429–444. doi: 10.1016/j.surfrep.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Groves JT, Boxer SG. Acc. Chem. Res. 2002;35:149–157. doi: 10.1021/ar950039m. [DOI] [PubMed] [Google Scholar]
- (39).Keller C, Kasemo B. Biophys. J. 1998;75:1397–1402. doi: 10.1016/S0006-3495(98)74057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Anderson TH, Min Y, Weirich KL, Zeng H, Fygenson D, Israelachvili JN. Langmuir. 2009;25:6997–7005. doi: 10.1021/la900181c. [DOI] [PubMed] [Google Scholar]
- (41).Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Biophy. J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Claesson M, Cho NJ, Frank CW, Andersson M. Langmuir. 2010;26:16630–16633. doi: 10.1021/la102719w. [DOI] [PubMed] [Google Scholar]
- (43).Im H, Lee SH, Wittenberg NJ, Johnson TW, Lindquist NC, Nagpal P, Norris DJ, Oh SH. ACS Nano. 2011;5:6244–6253. doi: 10.1021/nn202013v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Xia YN, Whitesides GM. Annu. Rev. Mater. Sci. 1998;28:153–184. [Google Scholar]
- (45).Yang TL, Jung SY, Mao HB, Cremer PS. Anal. Chem. 2001;73:165–169. doi: 10.1021/ac000997o. [DOI] [PubMed] [Google Scholar]
- (46).Sannomiya T, Scholder O, Jefimovs K, Hafner C, Dahlin AB. Small. 2011;7:1653–1663. doi: 10.1002/smll.201002228. [DOI] [PubMed] [Google Scholar]
- (47).Andersson K, Hamalainen M, Malmqvist M. Anal. Chem. 1999;71:2475–2481. doi: 10.1021/ac981271j. [DOI] [PubMed] [Google Scholar]
- (48).Oshannessy DJ, Brighamburke M, Soneson KK, Hensley P, Brooks I. Anal. Biochem. 1993;212:457–468. doi: 10.1006/abio.1993.1355. [DOI] [PubMed] [Google Scholar]
- (49).Diaw L, Magnac C, Pritsch O, Buckle M, Alzari PM, Dighiero G. J. Immunol. 1997;158:968–976. [PubMed] [Google Scholar]
- (50).Zhou ZH, Zhang YH, Hu YF, Wahl LM, Cisar JO, Notkins AL. Cell Host Microbe. 2007;1:51. doi: 10.1016/j.chom.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Burastero SE, Casali P, Wilder RL, Notkins AL. J. Exp. Med. 1988;168:1979–1992. doi: 10.1084/jem.168.6.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Ueki Y, Goldfarb IS, Harindranath N, Gore M, Koprowski H, Notkins AL, Casali P. J. Exp. Med. 1990;171:19–34. doi: 10.1084/jem.171.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Min Y, Kristiansen K, Boggs JM, Husted C, Zasadzinski JA, Israelachvili J. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3154–3159. doi: 10.1073/pnas.0813110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Copeland RA, Pompliano DL, Meek TD. Nat. Rev. Drug Discov. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- (55).Nunez S, Venhorst J, Kruse CG. Drug Discov. Today. 2012;17:10–22. doi: 10.1016/j.drudis.2011.06.013. [DOI] [PubMed] [Google Scholar]
- (56).Maynard JA, Maassen CBM, Leppla SH, Brasky K, Patterson JL, Iverson BL, Georgiou G. Nat. Biotechnol. 2002;20:597–601. doi: 10.1038/nbt0602-597. [DOI] [PubMed] [Google Scholar]
- (57).Warrington AE, Asakura K, Bieber AJ, Ciric B, Van Keulen V, Kaveri SV, Kyle RA, Pease LR, Rodriguez M. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6820–6825. doi: 10.1073/pnas.97.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Ciric B, Howe CL, Paz-Soldan MM, Warrington AE, Bieber AJ, Van Keulen V, Rodriguez M, Pease LR. Brain Pathol. 2003;13:608–616. doi: 10.1111/j.1750-3639.2003.tb00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Pirko I, Ciric B, Gamez J, Bieber AJ, Warrington AE, Johnson AJ, Hanson DP, Pease LR, Macura SI, Rodriguez M. FASEB J. 2004;18:1577–1579. doi: 10.1096/fj.04-2026fje. [DOI] [PubMed] [Google Scholar]
- (60).Hunter SF, Miller DJ, Rodriguez MJ. Neurol. Sci. 1997;150:103–113. doi: 10.1016/s0022-510x(97)00080-4. [DOI] [PubMed] [Google Scholar]