This commentary is about the article by Feng, Cho, and Doolittle (1), a paper that addresses genealogical relationships between the three domains of organisms. First, however, I would like to be as forthcoming as possible on issues of nepotism and reveal another genealogy—that relating this paper’s senior author and me. Russell F. Doolittle and I recently have ascertained that we descend from a remote common ancestral couple—Ebeneezer and Hannah (nee Hall) Doolittle–via eight intermediate nodes on Russell’s side and seven, including two more Ebeneezers, on mine. We share Y chromosomes, if there have been no adoptions or other irregularities in either lineage, but eight generations should surely be enough to attenuate bias!
In the deeper genealogy Feng, Cho and Doolittle (1) examine in this issue of the Proceedings of the National Academy of Sciences, there are serious irregularities, some even akin to adoption in their confusing effects. Difficulty in recognizing them led to an unexpected date for the last common ancestor of bacteria and eukaryotes, reported by these same authors (plus S. Tsang and E. Little) in a much-discussed recent article in Science (2). That date was slightly more than 2 billion years ago. In this commentary, I discuss how it was obtained, why it was unexpected, how others explained the “error,” and how the current analysis adroitly fixes the problem, only to point the way to further issues that the community of cell evolutionists must now address.
For the Science paper (2), Doolittle and colleagues assembled from the databases 531 amino acid sequences, representing 57 different enzymes from the broadest phylogenetic spectrum (animals, fungi, plants, protists, Archaea, and Bacteria). Evolutionary distances [amino acid differences weighted by use of accepted point mutation and BLOSUM (BLocks SUbstitution Matrix) scales] were calculated for pairwise comparisons of homologous sequences within and between groups. Distance between chordate species with divergence times known from the fossil record were used to calibrate the distance “clock,” and divergence times for animals vs. fungi, animal vs. plants, all eukaryotes vs. Archaea, and all eukaryotes vs. Bacteria were estimated by extrapolation as 0.96, 1.0, 1.9, and 2.2 billion years, respectively.
The protein sequences used in these analyses were on average 37% identical between Bacteria and eukaryotes. If all amino acid positions are equally changeable (if the number of changes follows a Poisson distribution), this degree of divergence remains within the safe range, Doolittle et al. (2) argued. Most positions will have undergone only single substitutions, and the amino acid distance vs. time curves would not have begun to “plateau out” before 2 billion years because of undetected multiple changes. Of course, amino acid positions in proteins are not equally changeable, but even corrections these authors believed to be extreme, like the assumption that 15% of a typical protein’s residues could not vary at all, would not push the Bacteria/eukaryote divergence beyond 2.5 billion years.
Problems
Two and one-half billion years, however, is not enough! The molecular evolutionary community demanded 3.5 billion years, pursuing what seems an unassailable four-step chain of argument (3, 4).
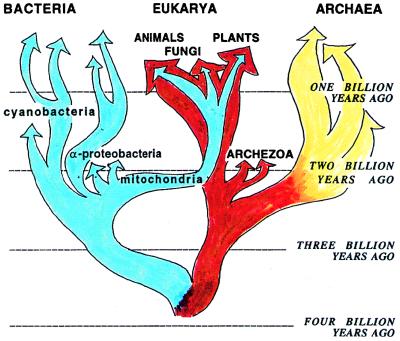
First, we had reached a rough consensus (5) about the structure of a “universal tree of Life” with the key features shown in Fig. 1. We believe: (i) that the deepest branching separates Bacteria from the line leading to Archaea and Eukarya; (ii) that mitochondrial genomes are reduced versions of the genomes of α-proteobacteria that invaded (as endosymbionts) some early eukaryotic “host” cells; (iii) that these host cells arose from an Archaea-like ancestor, so eukaryotic nuclear genomes are primarily retooled archaeal genomes and should be found in pure form in those eukaryotes (“Archezoa”) that diverged from the main stock before the invasion of the α-proteobacteria; and (iv) that after that invasion, a few α-proteobacterial genes whose products still function in mitochondria were transferred to the nucleus.
Second (the second step in the chain of argument), phylogenetic trees based on several large, robust, and independent RNA and protein data sets showed cyanobacteria to be only a moderately deep branch within Bacteria, consistent with the “common sense” view that these complex (often multicellular and often differentiated) oxygen-evolving photosynthesizers are not primitive.
But third, the earliest fossils, well documented by William Schopf (6) and his collaborators and rivals, are 3.5 billion years old at least, and they look very much like cyanobacteria. Even if they are not cyanobacteria, they are complex differentiated multicellular prokaryotes, and independent geochemical evidence does say that biological oxygen evolution began before this time.
Thus, fourth (the last step), if all of this is true, several branchings within the Bacterial side of the tree predate 3.5 billion years ago, and the deepest branch on the whole tree, that separating Bacteria from Archaea/Eukarya, must be pushing hard against the upper limit of Earth’s habitability [3.8 billion years, after major asteroid and comet impacts stopped vaporizing the oceans (7)]; 2.5 billion won’t do it.
Figure 1.
A consensus view of cellular evolution. Dates approximately as in Feng et al. (1).
Resolution
So how could Doolittle et al. (2) have missed the mark by more than 1 billion years? Helpful suggestions came from all quarters. Hasegawa and Fitch (8) argued that, even though Doolittle et al. (2) found that 95% of amino acid positions varied at least once in the whole data set, this does not mean that all positions are variable at all times in evolution. Fitch’s “covarion” model envisions that different constellations of residues are functionally constrained at different periods in a protein’s history, so “many of the varied sites may nevertheless have spent a considerable portion of their time in the invariable category” (8). And even were the covarion model invalid, some positions will surely vary more than others, and the distributions of mutations across sites might be better matched by a γ distribution, not the Poisson. Using differently parameterized γ distributions and assumptions they considered reasonable for absolutely invariant sites, Miyamoto and Fitch (9) and Gogarten and coworkers (10) could push the bacterial/eukaryote divergence back to 3.5 billion, in fact even back to 6 billion, years ago!
The latter critics, together with W. F. Martin (4), suggested as well a second source of error. Doolittle et al. (2) had overlooked the possibility that many eukaryotic proteins used in their analyses might actually derive, like mitochondrial proteins, from more recently acquired eubacterial genes—by hitherto largely unsuspected events of “horizontal gene transfer.”
As it turns out, it was some of both but mostly the latter. Although Doolittle et al. (2) had in fact used a modified Poisson distribution that corrects for some of the faults attributed to it by their critics (11), Feng et al. (1) have abandoned this method for determining evolutionary distance and now calculate distances according to a formula of Grishin (12), which corrects for nature of amino acid interchange and site-to-site variations in rate. They also have expanded the data analyzed to 64 enzyme sets, in particular taking advantage of completed genome sequences to beef up the archaeal contribution. Archaea were represented in only 9 of the original 57 enzyme sets; now they are included in 34.
As a result, it is possible to see that the “consensus” tree of Fig. 1 does not represent most of the data after all. Of the enzyme sets that include at least one archaeal sequence, only eight show strongest sequence similarity between archaeal and eukaryotic enzymes (and would thus give distance trees that are mid-point-rooted between Bacteria and Archaea/Eukarya as in Fig. 1). Twice as many (17) show a stronger sequence similarity between Bacteria and eukaryotes, traditionally the hallmark of nuclear genes “transferred from the mitochondrion” during the conversion of α-proteobacterial symbiont to specialized organelle. And the remaining 13 show (some) archaeal and (some) bacterial sequences as nearest relatives, in most cases looking like instances of horizontal gene transfer from Bacteria to Archaea.
So, much of the data that gave the much-maligned 2 billion-year result of Doolittle et al. (2) do not in fact measure the full length of the tree (from eukaryotes down to the root and back up to Bacteria). Instead, they trace a path like, if not identical to, mitochondrial and mitochondrion-derived sequences—from eukaryotes down to the horizontal transfer event and then back up to modern Bacteria, bypassing the extra billion or so years down to the root. This shorter path could well correspond to a date of ≈2 billion years ago! That is about as far back as the least conservative micropaleontologists would put the origin of eukaryotic cells (13). Furthermore, there is ample support in theory [not only the classical endosymbiont hypothesis as in Fig. 1 but various modern notions of genomic chimerism (14)] for the creation or at least the radical transformation of eukaryotic cells by the coming together of archaeal and bacterial genes.
Nice resolution, but the surprise now is how many bacterial genes eukaryotic nuclear genomes seem to harbor, genes such as that for triose phosphate isomerase (15), which have little specifically to do with the maintenance of mitochondria and which we would have assumed (Fig. 1) to be part of the archaeal heritage of the eukaryotic nuclear genome. I would wager that most of us thought that the bacterial “contamination” of the eukaryotic nuclear genome was limited to (i) those relatively few genes “transferred from the mitochondrion” but still devoted specifically to maintaining the structure and metabolic activities of this organelle and (ii) scattered individual instances of prokaryote-to-eukaryote gene transfer [whose quantitative importance in the larger scheme of things had been earlier scrutinized critically by Doolittle himself (16)].
Admittedly, this is not a surprise for which we were unprepared altogether. Recent work from several labs had shown that even Archezoa, those anaerobic amitochondriate protists that branch near the bottom of rRNA trees and that we had been thinking of as direct descendants of the first eukaryotes (as “host” cells that had never welcomed α-proteobacterial symbionts), have been tarred with the α-proteobacterial brush. Trichomonas (a parabasalan) and Giardia (a metamonad) bear α-proteobacterial genes for chaperonins in their nuclear genomes (refs. 17–19 and A. Roger and M. Sogin, personal communication). They also surely harbor other bacterial genes involved in cytosolic biochemistry. We can account for this by assuming (i) that many genes for many kinds of functions were transferred from symbiont to nuclear genome during the reduction to organellar status or (ii) that there were many independent events of lateral transfer from α-proteo and other Bacteria (that early eukaryotes probably ate for a living, after all) or (iii) that some other form of coming together of bacterial and archaeal genes, some other sort of relationship or symbiosis, possibly involving forms of metabolic interaction other than currently observed between mitochondria and cytosol, explains the structural and genomic complexity of eukaryotes.
But surely some genes should trace out the full tree, from eukaryotes down through an Archaea-like intermediate branch point to the root and back up to Bacteria. Feng et al. (1) show that, indeed, for the 25 enzyme sets with both archaeal and bacterial sequences and no evidence for horizontal transfer between them, average sequence identity (33%) corresponds (on their freshly calibrated distance vs. time plot) to something between 3.1 and 3.8 billion years. They also estimate the divergence of Archaea and Eukarya (using those enzyme sets in which the eukaryotic versions are not of apparent bacterial origin) as occurring 2.3 billion years ago—reasonable indeed on the assumption that eukaryotes (cells with nuclei and cytoskeletons) arose from Archaea-like ancestors before, but not too long before, the mitochondrial invasion.
New problems
So the paper of Feng et al. (1) goes some long distance toward restoring the respectability of attempts to hang dates on the deepest branches of trees and fits in nicely with what else is going on in the community of cellular evolutionists. There are still serious difficulties with the fossil record, however. Feng et al. (1) calculate that divergences within the Bacteria (including that which gave rise to cyanobacteria) go back only 2.1–2.5 billion years. Schopf’s fossils can then only be still be cyanobacteria if we swing the root of the tree in Fig. 1 a bit to the left so that deep bacterial lineages such as Thermotoga become part of the Archaeal/Eukaryal clade. Indeed, Cavalier-Smith (20) has suggested just such a maneuver on the basis of cell biological arguments, but we would do considerable damage to the careful sequence analyses that led to the current general acceptance of the “Iwabe-Gogarten” root (5).
There are also politically charged taxonomic issues. If most of the eukaryotic nuclear genome comes from Bacteria, should we still insist on Archaeal/Eukaryal sisterhood because we think the genes that show this (most of those of the replication, transcription, and translation machinery) are less subject to horizontal gene transfer? Are they really? To what extent is our desire to look at early evolution in terms of cellular lineages preventing us from seeing that it is about genes and their promiscuous spread across taxonomic boundaries, which then have no permanent significance?
Probably the situation is not that dire, and a new consensus view and language that retains cellular lineages but more adequately copes with horizontal gene transfer will emerge. We will probably never be as certain about when the universal common ancestor lived as we are about R. F. Doolittle’s and my common ancestor Ebeneezer (who lived from 1672 to 1711), but progress has been made, interesting and serious hypothesis are being tested, and genealogical research is proving to be great fun.
References
- 1.Feng D-F, Cho G, Doolittle R F. Proc Natl Acad Sci USA. 1997;94:13028–13033. doi: 10.1073/pnas.94.24.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doolittle R F, Feng D-F, Tsang S, Cho G, Little E. Science. 1996;271:470–477. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- 3.Mooers A O, Redfield R. Nature (London) 1996;379:587–588. doi: 10.1038/379587a0. [DOI] [PubMed] [Google Scholar]
- 4.Martin W F. BioEssays. 1996;18:523–527. [Google Scholar]
- 5.Brown J R, Doolittle W F. Proc Natl Acad Sci USA. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schopf J W. Science. 1993;260:640–646. doi: 10.1126/science.260.5108.640. [DOI] [PubMed] [Google Scholar]
- 7.Chyba C F. Nature (London) 1990;343:129–133. [Google Scholar]
- 8.Hasegawa M, Fitch W M. Science. 1996;274:1750. [PubMed] [Google Scholar]
- 9.Miyamoto M M, Fitch W M. Syst Biol. 1996;45:566–573. doi: 10.1093/sysbio/45.4.568. [DOI] [PubMed] [Google Scholar]
- 10.Gogarten J P, Olendzenski L, Hilario E, Simon C, Holsinger K E. Science. 1996;274:1751. doi: 10.1126/science.274.5293.1750b. [DOI] [PubMed] [Google Scholar]
- 11.Doolittle R F, Feng D-F, Tsang S, Cho G, Little E. Science. 1996;274:1751–1753. doi: 10.1126/science.274.5293.1751. [DOI] [PubMed] [Google Scholar]
- 12.Grishin N V. J Mol Evol. 1995;41:675–679. doi: 10.1007/BF00175826. [DOI] [PubMed] [Google Scholar]
- 13.Han T H, Runnegar B. Science. 1992;257:232–235. doi: 10.1126/science.1631544. [DOI] [PubMed] [Google Scholar]
- 14.Golding G B, Gupta R S. Mol Biol Evol. 1995;12:1–6. doi: 10.1093/oxfordjournals.molbev.a040178. [DOI] [PubMed] [Google Scholar]
- 15.Keeling P J, Doolittle W F. Proc Natl Acad Sci USA. 1996;94:1270–1275. doi: 10.1073/pnas.94.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith M W, Feng D-F, Doolittle R F. Trends Biochem Sci. 1992;17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- 17.Bui E T N, Bradley P J, Johnson P J. Proc Natl Acad Sci USA. 1996;93:9651–9656. doi: 10.1073/pnas.93.18.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roger A J, Clark C G, Doolittle W F. Proc Natl Acad Sci USA. 1996;93:7749–7754. doi: 10.1073/pnas.93.15.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horner D S, Hirt R P, Kilvington S, Lloyd D, Embley T M. Proc R Soc Biol Sci. 1996;263:1053–1059. doi: 10.1098/rspb.1996.0155. [DOI] [PubMed] [Google Scholar]
- 20.Cavalier-Smith T. Cold Spring Harbor Symp Quant Biol. 1987;52:805–824. doi: 10.1101/sqb.1987.052.01.089. [DOI] [PubMed] [Google Scholar]