Abstract
RATIONALE
Neoglycoconjugate vaccines synthesized by the squaric acid spacer method allow single point attachment of the carbohydrate antigen to the protein carrier. However, the localization of the carbohydrate antigen sites of conjugation on the protein carrier has been an elusive task difficult to achieve.
METHOD
Covalent attachment of the lactose antigen to the bovine serum albumin (BSA) was prepared by the squaric acid method using a hapten:BSA ratio of 20:1. Different reaction times were used during the conjugation reaction and two different lactose-BSA glycoconjugate vaccines were obtained. The carbohydrate antigen hapten:BSA ratios of these lactose-BSA glycoconjugate vaccines were determined by MALDI-TOF/RTOF-MS and the glycation sites in the neoglycoconjugates were determined using nano-LC/ESI-QqTOF-MS/MS analysis of the trypsin and GluC V8 digests of the conjugates.
RESULTS
We have identified a total of 15 glycation sites located on the BSA lysine residues for the neoglycoconjugate vaccine formed with a hapten:BSA ratio of 5.1:1, However, the tryptic and GluC V8 digests of the hapten-BSA glycoconjugate with a hapten:BSA ratio of 19.0:1 allowed identification of 30 glycation sites located on the BSA. These last results seem to indicate that this conjugation results in formation of various glycoforms.
CONCLUSIONS
It was observed that the number of identified glycation sites increased when the hapten:BSA ratio of glycoconjugate formation increased, and that the location of the glycation sites appears to be mainly on the outer surface of the BSA carrier molecule which is in line with the assumption that the sterically more accessible lysine residues, namely those located on the outer surface of the BSA, would be conjugated preferentially.
Synthesis of carbohydrate-protein neoglycoconjugates has become an important avenue towards treatment of infectious diseases, as exemplified by the synthesis of carbohydrate based vaccines.[1–4] Different methods have been applied for the synthesis of neoglycoconjugate vaccines. One of the most efficient synthesis of glycoconjugates is provided by the squaric acid chemistry which allows single point attachment of the antigen to the protein carrier; it avoids the formation of cross-linking reactions, and allows recovery of the non-reacted squaric acid derivative (usually used in excess at the onset of the conjugation).[5] Kamath et al. were the first to monitor the conjugation using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS).[6] The first step in the conjugation process is the synthesis of the hapten in the form of a glycoside whose aglycone contains a primary amino group. The latter can react at pH 7.0 with a squaric acid diester to produce the corresponding amide ester. Subsequent reaction of the monoester with amino groups in the carrier protein at pH 9.0 forms the carbohydrate-protein glycoconjugate. More recently, Kováč's group developed a strategy that enables a one-pot preparation of a series of neoglycoconjugates with predetermined carbohydrate-protein ratios.[7–9] They were then able to make conjugates from fragments of the O-specific polysaccharide (O-PS) of Vibrio cholerae O1 serotype Ogawa[9,10] and Inaba[11,12] and bovine serum albumin (BSA) and investigated the immunogenicity of these hapten-BSA glycoconjugates.
While the hapten-protein ratio has been generally measured using MALDI-TOF or surface-enhanced laser desorption/ionization (SELDI)-TOF-MS,[10,13] the localization of the carbohydrate occupancies and the sites of conjugation has been more difficult to achieve.
We reported our first attempt to localize the conjugation sites in the hapten-BSA glycoconjugate made from the monosaccharide antigen of the O-PS of V. cholerae serotype Ogawa.[14] The glycoconjugate was first digested with trypsin and the suspected glycated digests were then analyzed by MALDI tandem mass spectrometry (MS/MS). However, the presence of only three glycation sites were identified, despite the average molecular mass of the conjugate evidenced that it contained ~5 moles of the ligand per mole of the carrier, as determined by MALDI-MS. This discrepancy was attributed to the poor ionization of the glycated peptides. Analysis of the tryptic digests by MS/MS showed also the presence of numerous carbohydrate fragment ions which markedly complicated the MS/MS sequencing of the glycopeptides. Thus, we have previously proposed that the trypsin digestion of the glycoconjugate is not efficient to cleave the glycated lysines and this may affect the total digestion of the glycoconjugate.[15]
In a separate study, we have investigated the location of glycation sites in an experimental neoglycoconjugate vaccine for anthrax by MALDI-MS/MS and liquid chromatography (LC)/MS/MS.[16] The carbohydrate portion of the neoglycoconjugate was the synthetic tetrasaccharide side chain of the Bacillus anthracis exosporium, 2-O-methyl-β-D-glucopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→2)-α-L-rhamnopyranoside, and the protein carrier was BSA.[16,17] The hapten-BSA ratio in the neoglycoconjugate was 5.4:1, as showed independently by MS. The conjugate was digested with either trypsin or GluC V8 before MS analysis. The advantage of using two different proteases allowed us to obtain higher sequence coverage. A total of 30 glycation sites were discovered when performing LC/MS/MS analysis of the digests, which is many more than was indicated by MALDI-MS/MS analysis of the digests. Additionally, we also observed carbohydrate fragment ions characteristic of the tetrasaccharide portion.
Based on these observations, and to further explore the power of mass spectrometry in combination with enzymatic digestion in glycoconjugate analysis, here we use a glycoconjugate composed of a simple model carbohydrate, the milk sugar lactose, composed of two D-hexosyl residues, and BSA as carrier. We hypothesize that the gas-phase reactivity of the β-lactose portion of the neoglycoconjugate should be different than that of the 4-(3-deoxy-L-glycero)-2-O-methyl-α-D-perosamine and of the tetrasaccharide 2-O-methyl-β-D-glucopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→2)-α-L-rhamnopyranoside which we used previously.[14,16] In addition, here we have used a combination of two different proteases, namely trypsin and GluC V8, for the digestion of various glycoconjugates. It is well known that, at pH 7.8, GluC V8 endoprotease cleave peptide bonds at the C-terminus of glutamic acid and aspartic acid residues.[18,19] Finally, the protease digests were analyzed using nano-LC/electrospray ionization quadrupole orthogonal time-of-flight (ESI-QqTOF)-MS/MS.
EXPERIMENTAL
Hapten-BSA glycoconjugate preparation
Conjugation of β-D-galactopyranosyl-(1→4)-β-D-glucopyranose (β-lactose) to BSA was carried out by the standard protocol.[8] Briefly, commercial β-lactose octaacetate was converted into 5-methoxycarbonylpentyl β-lactoside which was transformed to a squaric acid monoester before conjugation. The conjugations were carried out in 0.5 M borate buffer (pH 9.0) at the initial hapten-BSA ratio of 20:1 and using BSA (Sigma Aldrich, St. Louis, MO, USA) as carrier protein. The conjugation times were 3 and 24 h.[7] Isolation of the neoglycoconjugate was carried out by centrifugal ultrafiltration using filtration devices with a cut-off 30 000 Da.[7]
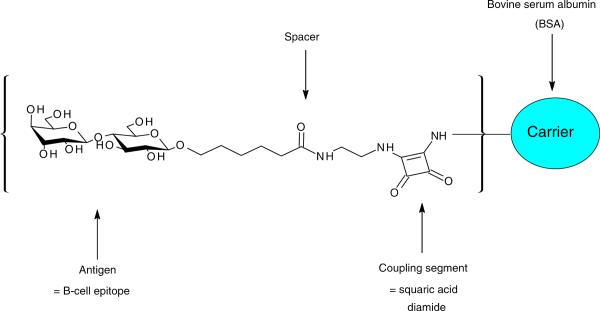
It has already been established that when the foregoing protocol is applied the carbohydrate-spacer moiety covalently attaches to the ε-amino group of the lysine residue in BSA.[14,16] Figure 1 shows a representation of the synthesized carbohydrate hapten-BSA glycoconjugate.
Figure 1.
Schematic representation of the lactose-BSA glycoconjugate.
Digestion
The digestions of the lactose-BSA glycoconjugates were carried out with trypsin and GluC V8 protease (Sigma Aldrich, St. Louis, MO, USA). Thus, 100 μg of the glycoconjugate was dissolved in a mixture of 0.1% RapiGest SF surfactant (1 μg, Waters, USA) in 50 mM of NH4HCO3 (100 μL) at a pH of 8.0 and reduced by treatment with 2 μL of 10 mM dithiothreitol (Sigma Aldrich, St. Louis, MO, USA) for 30 min at room temperature, followed by alkylation with 2 μL of a 50 mM iodoacetamide (Sigma Aldrich, St. Louis, MO, USA) for 1 h at room temperature. A portion (50 μg) of the glycoconjugate was digested with trypsin using 20 ng/mL of trypsin dissolved in NH4HCO3 (50 mM, 1 mL) at a trypsin:glycoprotein ratio of 1:25 (w/w) and incubated at 37 °C overnight with shaking. The other 50 μg of the glycoconjugate was digested using the GluC V8 endoprotease protease:glycoprotein ratio of 1:25 (w/w) and incubated at 37 °C overnight with shaking. The sample was then dried under vacuum and the residue was dissolved in 20 μL of 1% acetic acid (Sigma-Aldrich, Oakville, ON, Canada). An aliquot of each sample (10 μL) was then cleaned up using ZipTip C18 (Millipore, Bedford, MA, USA) before mass spectral analysis.
MALDI-TOF-MS analysis
Mass spectrometric analysis were carried out on a 4700 Proteomics analyzer with TOF-TOF optics (Applied Biosystems Foster City, CA, USA) and a 200-Hz frequency-tripled Nd: YAG laser. α-Cyano-4-hydroxycinnamic acid (α-CHCA) was used as matrix for the analysis of BSA and hapten-BSA conjugates with an average of 5000 to 8000 laser shots per spectra. Briefly, 1 μL of a 20 mg/mL solution of α-CHCA (dissolved in acetone, 0.1% trifluoroacetic acid (TFA); the use of acetone allowed good homogeneity of the matrix) was spotted on the MALDI plate and dried at room temperature. Then, an aliquot of 1 μL of sample was spotted on the top of the dried matrix and left to dry before the MALDI-MS experiments. The analysis was achieved in the linear mode and the MALDI-TOF mass spectrometer was calibrated using BSA.
LC-ESI-QqTOF-MS/MS analysis
The peptides were separated on a DIONEX UltiMate3000 Nano LC system (Germering, Germany). Digested glycoprotein (250 fmol) was dissolved in 0.1% TFA and loaded onto a precolumn (300 μm i.d. × 5 μm, C18 PepMap100, 5 μm; LC Packing, Sunnyvale, CA, USA) in order to desalt and concentrate the sample. After their elution from the precolumn, the peptide and glycopeptide mixtures were separated on a nanoflow analytical column (75 μm i.d. × 15 cm, C18 PepMap 100, 3 μm, 100 A; LC Packing, Sunnyvale, CA, USA) at a flow rate of 180 nL/min. The elution of the peptides and glycopeptides was achieved using the following mobile phases: 0.1% FA/0.01% TFA/2% ACN (A) and 0.08% FA/0.008% TFA/98% ACN (B). The elution started with 0% B for 10 min, followed by a gradient of 0–60% B in 55 min and 60–90% B in 3 min and was kept at 90% B for 3 min. The MS/MS analysis of the eluted peptides and glycopeptides was accomplished using an Applied Biosystems API-QSTAR XL quadrupole orthogonal time-of-flight (QqTOF)-MS/MS hybrid tandem mass spectrometer (Applied Biosystems International-MDS Sciex, Foster City, CA, USA) equipped with a nanoelectrospray source (Protana XYZ manipulator) which produces the electrospray through a PicoTip needle (10 μm i.d., New Objectives, Woburn, MA, USA) carrying a voltage of 2400 V. The TOF analyzer was calibrated using a renin solution (1 pmol/μL) and looking for the ions at m/z 586.9815 and.9723. The collision energies used during the collision-induced dissociation (CID)-MS/MS analyses were determined automatically using the information-dependent acquisition (IDA) method integrated in the Analyst software.
RESULTS
As previously indicated, the covalent attachment of lactose to BSA was carried out at an initial hapten-BSA ratio of 20:1 and the conjugation reaction was allowed to proceed for either 3 or 24 h. Thus, two β-lactoside-BSA glycoconjugate vaccine models were produced.
MALDI-MS analysis of the hapten-BSA glycoconjugates
In order to determine the average number of carbohydrate-spacer moieties linked to BSA (hapten-BSA ratio), the two glycoconjugates were analyzed using MALDI-MS. Molecular masses and hapten-BSA ratios were calculated by comparing the molecular weight of the conjugate to the molecular weight of the starting BSA. Thus, the protonated molecule [M + H]+ was observed at m/z 69307.96 for the glycoconjugate obtained after a reaction time of 3 h, whereas [M + H]+ was found at m/z 77258.62 after a conjugation time of 24 h.
The molecular weight calculations of these glycoconjugates were based on the presumption that squaric acid covalently links to the ε-amino groups of the lysine residues in BSA by loss of an ethanol molecule[13,15] and that the molecular weight of the fragment of disaccharide-spacer-squaric acid which will bond to the BSA is 577.22 Da (Table 1). As a consequence, we concluded that the MALDI-MS revealed two neoglycoconjugates formed with different hapten-BSA ratios of 5.1:1 (reaction time: 3 h) and 19.0:1 (reaction time: 24 h). These hapten-BSA ratios were found to be identical to those previously determined by SELDI-TOF-MS.[7]
Table 1.
Molecular ions identified during the MALDI-MS analysis of the carbohydrate lactose-BSA glycoconjugates obtained with different reaction times
| Observed ion m/z |
Calculated | ||
|---|---|---|---|
| Reaction time | [M + 2H]2+ | [M + H]+ | lactose-BSA ratio |
| BSA | 33161.81 | 66318.88 | – |
| 3 h | 34619.44 | 69307.96 | 5.1 |
| 24 h | 38683.93 | 77258.62 | 19.0 |
Supplementary Fig. S1 (Supporting Information) and Table 1 display the MS and the m/z values of (a) BSA, (b) the neoglycoconjugate obtained after a reaction time of 3 h, and (c) the neoglycoconjugate obtained after a reaction time of 24 h.
LC/MS/MS analysis of the tryptic and GluC V8 digests of the neoglycoconjugate with lactose:BSA ratios of 5.1:1and 19.0:1
LC/MS/MS analysis was carried out on both tryptic and GluC V8 digests of glycoconjugates with hapten:protein ratios of 5.1:1 and 19.0:1. The MS/MS spectra obtained were submitted to the MASCOT library in order to identify the peptides formed from BSA. The search parameters were as follows: enzyme = trypsin or GluC V8, fixed modification = carbamidomethyl (C), variable modification = oxidation (M), mass values = monoisotopic, peptide mass tolerance = ±0.2 Da, fragment mass tolerance = ±0.2 Da, and maximum missed cleavage = 1. The MASCOT reports of LC/MS/MS data of the tryptic digest of the glycoconjugates demonstrated the identification of two Serum Albumin isoforms from the Bos taurus species. The serum albumin precursor (gi|1351907) was identified with the following sequence coverage: 45% (25 peptides identified) and 47% (25 peptides identified), for the neoglycoconjugates with a lactose:BSA ratio of respectively 5.1:1 and 19.0:1. The serum albumin protein (gi|74267962) was also identified for the analysis of the tryptic digests of the glycoconjugates with a hapten-protein ratio of 5.1:1 (sequence coverage = 41%, 23 peptides identified) and 19.0:1 (sequence coverage = 43%, 23 peptides identified).
Similarly, the data of the LC/MS/MS analysis of the GluC V8 digests of the glycoconjugates with ratios of 5.1:1 and 19.0:1 were submitted to the MASCOT library which gave a match to the same two Serum Albumin proteins from the Bos taurus species mentioned previously: the serum albumin precursor (gi|1351907) and the serum albumin (gi|74267962). The sequence coverages for the albumin precursor (gi|1351907) for the neoglycoconjugates with hapten:BSA ratios of 5.1:1 and 19.0:1 were found to be respectively 39% (20 peptides identified) and 37% (20 peptides identified). Finally, serum albumin protein (gi|74267962) was identified with the following sequence coverage: 40% (19 peptides identified) for the hapten-BSA glycoconjugate 5.1:1 and 33% (19 peptides identified) for hapten-BSA glycoconjugate 19.0:1.
Most importantly, the reports obtained contained also a list of ions that did not match any protein; these were considered to be potentially glycated BSA peptides. These ions were selected and subjected to CID-MS/MS sequencing, in order to identify the correct sequences of the glycated peptides and their exact glycation sites.
LC/MS/MS analyses of the tryptic digest of the neoglycoconjugate with a hapten:BSA ratio of 5.1:1
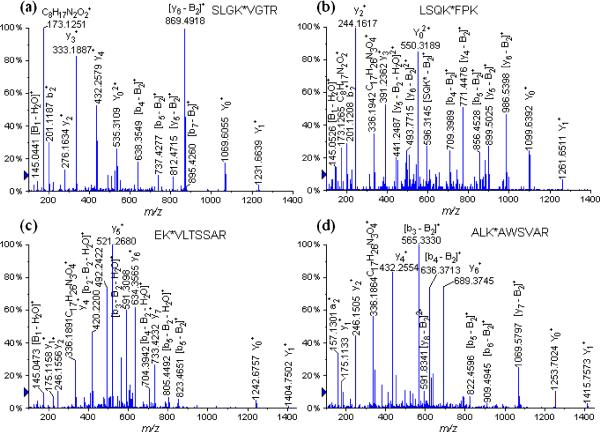
We have identified nine glycopeptides having lysine residues occupied on different glycation sites noted with an asterisk. These were identified as follows (Table 2): SLGK*VGTR (Lys 455) at m/z 697.3748 (+2), LSQK*FPK (Lys 245) at m/z 712.3812 (+2), EK*VLTSSAR (Lys 211) at m/z 783.8877 (+2), ALK*AWSVAR (Lys 235) at m/z 789.4228 (+2), LAK*EYEATLEECCAK (Lys 374) at m/z 797.7141 (+3), CASIQK*FGER (Lys 228) at m/z 886.4299 (+2), CCTK*PESER (Lys 463) at m/z 871.8738 (+2), VTK*CCTESLVNR (Lys 498) at m/z 1021.9941 (+2) and K*VPQVSTPTLVEVSR (Lys 437) at m/z 1108.5900 (+2) (Table 2). Figure 2 displays the following examples of different glycopeptides obtained by tryptic digests of the lactose-BSA glycoconjugate: (a) SLGK*VGTR (Lys 455) at m/z 697.3748 (+2) and (b) LSQK*FPK (Lys 245) at m/z 712.3812 (+2), (c) EK*VLTSSAR (Lys 211) at m/z 783.8877 (+2) and (d) ALK*AWSVAR (Lys 235) at m/z 789.4228 (+2). The CID-MS/MS fragmentation pathways obtained by the isolated precursor glycopeptide ions enabled us to unambiguously determine various glycation sites.
Table 2.
Tryptic glycopeptides identified in the bovine serum albumin protein by LC/MS/MS analysis of the lactose-BSA glycoconjugates
| Peptide |
Lactose-BSA 5.1:1 |
Lactose-BSA 19.0:1 |
||||
|---|---|---|---|---|---|---|
| Sequence (star = glycation site) | Calculated m/z (charge) | Missed cleavage | Observed m/z (charge) | Deviation (Da) | Observed m/z (charge) | Deviation (Da) |
| SLGK*VGTR (Lys 455) | 697.3568 (+2) | 1 | 697.3748 (+2) | 0.0181 | 697.3567 (+2) | −0.0001 |
| LSQK*FPK (Lys 245) | 712.3640 (+2) | 2 | 712.3812 (+2) | 0.0172 | 712.3673 (+2) | 0.0033 |
| EK*VLTSSAR (Lys 211) | 783.8912 (+2) | 1 | 783.8877 (+2) | −0.0035 | 783.8900 (+2) | −0.0012 |
| ALK*AWSVAR (Lys 235) | 789.4068 (+2) | 1 | 789.4228 (+2) | 0.0161 | 789.4125 (+2) | 0.0058 |
| LAK*EYEATLEECCAK (Lys 374) | 797.6874 (+3) | 2 | 797.7141 (+3) | 0.0267 | 797.6954 (+3) | 0.0080 |
| CASIQK*FGER (Lys 228) | 886.4066 (+2) | 1 | 886.4299 (+2) | 0.0233 | 886.4033 (+2) | −0.0033 |
| CCTK*PESER (Lys 463) | 871.8587 (+2) | 1 | 871.8738 (+2) | 0.0151 | 871.8000 (+2) | −0.0587 |
| VTK*CCTESLVNR (Lys 498) | 1021.9667 (+2) | 1 | 1021.9941 (+2) | 0.0274 | 1021.9741 (+2) | 0.0074 |
| K*FWGK (Lys 156) | 621.3007 (+2) | 1 | – | – | – | – |
| TPVSEK*VTK (Lys 495) | 782.8959 (+2) | 2 | – | – | 782.8995 (+2) | 0.0036 |
| K*VPQVSTPTLVEVSR (Lys 437) | 1108.5811 (+2) | 0 | – | – | 1108.5854 (+2) | 0.0043 |
| SLHTLFGDELCK*VASLR (Lys 100) | 841.4162 (+3) | 1 | – | – | 841.4000 (+3) | −0.0162 |
Figure 2.
LC/MS/MS spectra of the tryptic glycated peptides (a) SLGK*VGTR (Lys 455) at m/z 697.3567 (+2), (b) LSQK*FPK (Lys 245) at m/z 712.3673 (+2), (c) EK*VLTSSAR (Lys 211) at m/z 783.8900 (+2), and (d) ALK*AWSVAR (Lys 235) at m/z 789.4125 (+2).
During the CID-MS/MS analyses, we used the pre-existing peptide nomenclature involving the detection of the x-, y-, zions and the a-, b-, c-ions for the individual peptide sequence.[20,21] For the glycopeptide product ions, we used the nomenclature established by Domon and Costello for the carbohydrate portion as: A, B, C, X, Y and Z.[22]
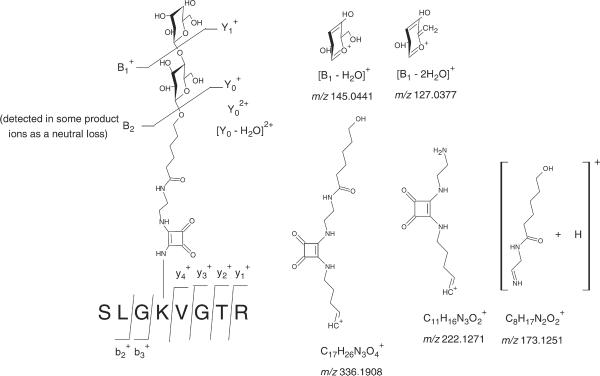
The CID-MS/MS analysis of the doubly charged glycated peptide SLGK*VGTR (Lys 455) at m/z 697.3748 (Fig. 2(a), Supplementary Table S1, see Supporting Information) afforded series of product ions corresponding to the entire peptide chain formed by loss of the β-D-galactopyranosyl moiety: at m/z 1231.6639, and the entire β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside: at m/z 1069.6055, at m/z 535.3109 and [Y0 − H2O]2+ at m/z 526.3004 (representation in Fig. 3). Additionally, we have noted the formation of product ions corresponding to the galactopyranosyl moieties: at m/z 163.0556, [B1 − H2O]+ at m/z 145.0441 and [B1 − 2H2O]+ at m/z 127.0377. Clearly, these carbohydrate fragments confirmed the identity of the carbohydrate conjugated to BSA and were an epitome of chemical stability of these cations in the gas phase. We also noted the formation of the product ions corresponding to the squaric acid spacer: at m/z 336.1908, at m/z 222.1271 and at m/z 173.1251 (product ions displayed in Fig. 3). As expected, we observed a propensity of the gas-phase loss of the carbohydrate disaccharide portion during the MS/MS analysis. For example, the following peptide product ions were observed and formed by the loss of the carbohydrate moiety: [b7 − B2]+ at m/z 895.4260, [y6 − B2]+ at m/z 869.4918, [y5 − B2]+ at m/z 812.4715, [b6 − B2]+ at m/z 794.4752, [b5 − B2]+ at m/z 737.4277, [b5 − B2 − H2O]+ at m/z 719.3841, [a5 − B2]+ at m/z 709.4391, [b4 − B2]+ at m/z 638.3549 and [y6 − B2 − H2O]2+ at m/z 426.2460. Finally, we also noted the presence of the conventional y- and b-product ions corresponding to the fragmentation of the peptide backbone, which allowed us to cover the sequence of the each entire glycated peptide.
Figure 3.
Product ions resulting from the tandem mass spectrometry analysis of the glyctated peptide SLGK*VGTR (Lys 455) at m/z 697.3567 (+2) extracted from the LC/MS/MS analysis of the tryptic digests of the hapten-BSA glycoconjugate with a lactose-BSA ratio of 5.1:1.
LC/MS/MS analyses of the GluC V8 digest of the neoglycoconjugate with a hapten:BSA ratio of 5.1:1
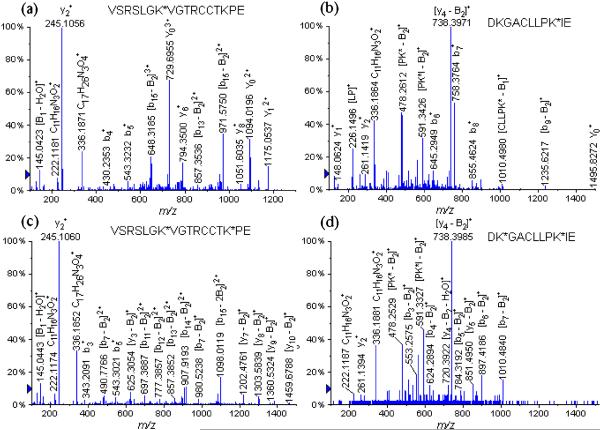
The manual sequencing of the spectra that were not assigned to a peptide of the Bos taurus protein on the MASCOT report enabled us to identify eight glycopeptides containing glycation sites at different lysine residue (Table 3): K*TPVSE (Lys 489) at m/z 618.7989, HVK*LVNE (Lys 65) at m/z 707.8591, K*LFTFHAD (Lys 528) at m/z 777.9566 (+2), LCK*VASLRE (Lys 100) at m/z 826.4216 (+2), VSRSLGK*VGTRCCTKPE (Lys 455) at m/z 837.7418 (+3), VSRSLGK*VGTRCCTKPESE (Lys 455) at m/z 909.7741 (+3), DKGACLLPK*IE (Lys 204) at m/z 910.4582 (+2), YAVSVLLRLAK*E (Lys 374) at m/z 969.5187 (+2), LLYYANK*YNGVFQE (Lys 183) at m/z 1149.5413 (+2) and K*LFTFHADICTLPDTE (Lys 528) at m/z 1242.5946 (+2) (Table 3). Figures 4(a) and 4(b) display several examples of the identified glycopeptides during the LC/ESI-QqTOF-MS/MS analysis of the GluC V8 digests: (a) VSRSLGK*VGTRCCTKPE (Lys 455) at m/z 837.7418 (+3) and (b) DKGACLLPK*IE (Lys 204) at m/z 910.4582 (+2).
Table 3.
Glycopeptides identified in the bovine serum albumin protein by LC/MS/MS analysis of the GluC V8 digests of the lactose-BSA glycoconjugates
| Peptide |
Lactose-BSA 5.1:1 |
Lactose-BSA 19.0:1 |
||||
|---|---|---|---|---|---|---|
| Sequence (star = glycation site) | Calculated m/z (charge) | Missed cleavage | Observed m/z (charge) | Deviation (Da) | Observed m/z (charge) | Deviation (Da) |
| K*TPVSE (Lys 489) | 618.7904 (+2) | 0 | 618.7989 (+2) | 0.0085 | 618.7940 (+2) | 0.0036 |
| HVK*LVNE (Lys 65) | 707.8513 (+2) | 0 | 707.8591 (+2) | 0.0078 | 707.8565 (+2) | 0.0052 |
| K*LFTFHAD (Lys 528) | 777.8644 (+2) | 0 | 777.9566 (+2) | 0.0922 | 777.9564 (+2) | 0.0920 |
| LCK*VASLRE (Lys 100) | 826.4087 (+2) | 0 | 826.4216 (+2) | 0.0129 | 826.4137 (+2) | 0.0050 |
| VSRSLGK*VGTRCCTKPE (Lys 455) | 837.7407 (+3) | 0 | 837.7418 (+3) | 0.0011 | 837.7468 (+3) | 0.0061 |
| VSRSLGK*VGTRCCTKPESE (Lys 455) | 909.7656 (+3) | 1 | 909.7741 (+3) | 0.0085 | 909.7714 (+3) | 0.0058 |
| DKGACLLPK*IE (Lys 204) | 910.4480 (+2) | 1 | 910.4582 (+2) | 0.0103 | 910.4539 (+2) | 0.0059 |
| YAVSVLLRLAK*E (Lys 374) | 969.5198 (+2) | 0 | 969.5187 (+2) | −0.0011 | 969.4790 (+2) | −0.0408 |
| LLYYANK*YNGVFQE (Lys 183) | 1149.5389 (+2) | 0 | 1149.5413 (+2) | 0.0024 | 1149.5565 (+2) | 0.0176 |
| K*LFTFHADICTLPDTE (Lys 528) | 1242.5726 (+2) | 2 | 1242.5946 (+2) | 0.0220 | 1242.5876 (+2) | 0.0150 |
| K*SHCIAE (Lys 309) | 710.8113 (+2) | 0 | – | – | 710.8039 (+2) | −0.0074 |
| K*SLHTLFGD (Lys 88) | 797.3804 (+2) | 0 | – | – | 797.3361 (+2) | −0.0443 |
| CCDK*PLLE (Lys 304) | 805.8445 (+2) | 1 | – | – | 805.8449 (+2) | 0.0004 |
| FAK*TCVADE (Lys 75) | 808.8481 (+2) | 1 | – | – | 808.8551 (+2) | 0.0070 |
| GPK*LVVSTQTALA (Lys 597) | 930.9883 (+2) | 0 | – | – | 931.0042 (+2) | 0.0159 |
| IAHRFK*DLGEE (Lys 36) | 945.9523 (+2) | 2 | – | – | 945.9632 (+2) | 0.0109 |
| AK*DAFLGSFLYE (Lys 346) | 968.9514 (+2) | 1 | – | – | 968.9538 (+2) | 0.0024 |
| VSRSLGK*VGTRCCTK*PE (Lys 455, Lys 463) | 1029.8153 (+3) | 0 | – | – | 1029.8239 (+3) | 0.0086 |
| K*DAIPE (Lys 318) | 624.7904 (+2) | 1 | – | – | 624.8000 (+2) | 0.0096 |
| VTK*LVTD (Lys 256) | 676.3402 (+2) | 0 | – | – | 676.3457 (+2) | 0.0055 |
| K*SHCIAEVE (Lys 309) | 824.8668 (+2) | 1 | – | – | 824.8706 (+2) | 0.0038 |
| LTK*VHK*E (Lys 263, Lys 266) | 1003.9750 (+2) | 0 | – | – | 1003.9831 (+2) | 0.0081 |
| K*VTK*CCTE (Lys 495, Lys 498) | 1089.4619 (+2) | 0 | – | – | 1089.4600 (+2) | −0.0019 |
| VSRSLGK*VGTRCCTK*PESE (Lys 455, Lys 463) | 1101.8402 (+3) | 1 | – | – | 1101.8476 (+3) | 0.0074 |
| DK*GACLLPK*IE (Lys 197, Lys 204) | 1198.5599 (+2) | 1 | – | – | 1198.5604 (+2) | 0.0005 |
| KK*FWGK*YLYE (Lys 156, 160) | 1257.5868 (+2) | 0 | – | – | 1257.5983 (+2) | 0.0115 |
Figure 4.
LC/MS/MS spectra of the GluC V8 digests glycated peptides (a) VSRSLGK*VGTRCCTKPE (Lys 455) at m/z 837.7418 (+3), (b) DKGACLLPK*IE (Lys 204) at m/z 910.4582 (+2), (c) VSRSLGK*VGTRCCTK*PE (Lys 455, Lys 463) at m/z 1029.8189 (+3), and (d) DK*GACLLPK*IE (Lys 197, Lys 204) at m/z 1198.5604 (+2).
The MS/MS of the precursor glycopeptide ion VSRSLGK*VGTRCCTKPE (Lys 455) at m/z 837.7418 (+3) shown in Fig. 4(a) (Supplementary Table S2, see Supporting Information) adopted the same CID fragmentation pathway, similar to the other tryptic digested glycopeptides ions. These CID-MS/MS fragmentations appear to be initiated by loss of the entire disaccharide moiety. Thus the following product ions were identified: the entire peptide ion produced by loss of the β-D-galactopyranosyl moiety; the product ion at m/z 1175.0537, and the intact disaccharide product (β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside) ion; at m/z 1094.0196 and at m/z 729.6955. It is important to point out the formation of the diagnostic product ions which were created specifically by the carbohydrate-spacer moiety: the mixed product ions containing a fragment of lysine and the squaric acid spacer assigned as [C17H26N3O4]+ at m/z 336.1871 and [C11H16N3O2]+ at m/z 222.1181. In addition, we were also able to detect the product ions corresponding to the monosaccharide galactopyranosyl moiety: at m/z 163.0527 and [B1 − H2O]+ at m/z 145.0423.
Additionally, some peptide fragment ions formed by loss of the entire β-lactose moiety were also detected: [b13 − B2]2+ at m/z 857.3536, [b16 − B2]3+ at m/z 680.9973 and [b15 − B2]3+ at m/z 648.3185. Furthermore, other product ions covering the sequence of the glycopeptide are displayed in Fig. 4(a) and Supplementary Table S2 (see Supporting Information). Another example of a glycopeptide identified during the LC/MS/MS analysis of GluC V8 digests of the precusor DKGACLLPK*IE ion (Lys 204) at m/z 910.4582 (+2) is shown in Fig. 4(b).
Thus, a total of 15 glycation sites were identified during the LC/MS/MS analysis of tryptic and GluC V8 digests of the lactose-BSA glycoconjugate, which are displayed in Fig. 5 (a). When we compare the average number of glycation sites determined by the foregoing analysis (15 glycation sites) to the ratio of 5.1:1 determined for the lactose-BSA conjugate, the difference can be explained by the formation of a mixture of glycoforms during conjugation. Finally, the total sequence coverage of the glycoconjugate obtained by the trypsin and GluC V8 digestion was found to be 86%.
Figure 5.
BSA sequence where the glycation sites are indicated by an asterix (red = identified on tryptic digests, blue = identified on GluC V8 digests and red and underlined = identified on both tryptic and GluC V8 digests) for the neoglycoconjugates with a lactose-BSA ratio of (a) 5.1:1 and (b)19.0:1.
LC/MS/MS analyses of the tryptic digest of the neoglycoconjugate with a hapten:BSA ratio of 19.0:1
The following glycated peptides were identified allowing the determination of 11 glycation sites (Table 2) and these were assigned as follows: SLGK*VGTR (Lys 455) at m/z 697.3567 (+2), LSQK*FPK (Lys 245) at m/z 712.3673 (+2), TPVSEK*VTK (Lys 495) at m/z 782.8995 (+2), EK*VLTSSAR (Lys 211) at m/z 783.8900 (+2), ALK*AWSVAR (Lys 235) at m/z 789.4125 (+2), LAK*EYEATLEECCAK (Lys 374) at m/z 797.6954 (+3), SLHTLFGDELCK*VASLR (Lys 100) at m/z 841.4000 (+3), CASIQK*FGER (Lys 228) at m/z 886.4033 (+2), CCTK*PESER (Lys 463) at m/z 871.8000 (+2), VTK*CCTESLVNR (Lys 498) at m/z 1021.9741 (+2) and K*VPQVSTPTLVEVSR (Lys 437) at m/z 1108.5854 (+2).
LC/MS/MS analysis of the GluC V8 digests of the neoglycoconjugate with a hapten:BSA ratio of 19.0:1
The LC/MS/MS analysis of the GluC V8 digests of the neoglycoconjugate with a hapten:BSA ratio of 19.0:1 permitted us to determine 24 glycation sites through the identification of the following glycated peptides (Table 3) shown as follows: K*TPVSE (Lys 489) at m/z 618.7940 (+2), K*DAIPE (Lys 318) at m/z 624.8000 (+2), VTK*LVTD (Lys 256) at m/z 676.3457 (+2), HVK*LVNE (Lys 65) at m/z 707.8565 (+2), K*SHCIAE (Lys 309) at m/z 710.8039 (+2), K*LFTFHAD (Lys 528) at m/z 777.9564 (+2), K*SLHTLFGD (Lys 88) at m/z 797.3361 (+2), CCDK*PLLE (Lys 304) at m/z 805.8449 (+2), FAK*TCVADE (Lys 75) at m/z 808.8551 (+2), K*SHCIAEVE (Lys 309) at m/z 824.8706 (+2), LCK*VASLRE (Lys 100) at m/z 826.4137 (+2), VSRSLGK*VGTRCCTKPE (Lys 455) at m/z 837.7468 (+3), VSRSLGK*VGTRCCTKPESE (Lys 455) at m/z 909.7714 (+3), DKGACLLPK*IE (Lys 204) at m/z 910.4539 (+2), GPK*LVVSTQTALA (Lys 597) at m/z 931.0042 (+2), IAHRFK*DLGEE (Lys 36) at m/z 945.9632 (+2), AK*DAFLGSFLYE (Lys 346) at m/z 968.9538 (+2), YAVSVLLRLAK*E (Lys 374) at m/z 969.4790 (+2), LLYYANK*YNGVFQE (Lys 183) at m/z 1149.5565 (+2) and K*LFTFHADICTLPDTE (Lys 528) at m/z 1242.5876 (+2). In this case, it is worth mentioning that we were able to scrutinize six bis-glycated peptides which carry two nearby glycation sites. These were assigned as follows: LTK*VHK*E (Lys 263, Lys 266) at m/z 1003.9831 (+2), VSRSLGK*VGTRCCTK*PE (Lys 455, Lys 463) at m/z 1029.8239 (+3), K*VTK*CCTE (Lys 495, Lys 498) at m/z 1089.4600 (+2), VSRSLGK*VGTRCCTK*PESE (Lys 455, Lys 463) at m/z 1101.8476 (+3), DK*GACLLPK*IE (Lys 197, Lys 204) at m/z 1198.5604 (+2) and KK*FWGK*YLYE (Lys 156, 160) at m/z 1257.5983 (+2). An interesting example to discuss is the diagnostic peptide VSRSLGKVGTRCCTKPE, which was detected with either one single glycation site on the Lys 455 residue affording the glycopeptide at m/z 837.7468 (+3) or with two glycation sites on the Lys 455 and Lys 463 residues, producing the glycated peptide at m/z 1029.8239 (+3). The CID-MS/MS analysis of the precursor ions containing two glycation sites VSRSLGK*VGTRCCTK*PE (Lys 455, Lys 463) at m/z 1029.8239 (+3) is shown in Fig. 4(c) and Supplementary Table S3 (see Supporting Information). The CID fragmentation pathway of this precursor glycated peptide ion afforded peptide product ions formed by loss of only one entire β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside. Thus, we detected the y-product ions obtained by the loss of the entire carbohydrate moiety on the Lys 463 residue. The majority of the remaining product ions were identified as follows: [y10 − B2]+ at m/z 1459.6788, [y9 − B2]+ at m/z 1360.5324, [y8 − B2]+ at m/z 1303.5839, [y7 − B2]+ at m/z 1202.4761, [y4 − B2]+ at m/z 726.3182 and [y3 − B2]+ at m/z 625.3054. However, it is interesting to note that the loss of the entire β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside on the Lys 455 residue was also observed on the following b-product ions: [b8 − B2]+ at m/z 1079.6271, [b7 − B2]+ at m/z 980.5238, [b14 − B2]2+ at m/z 907.9193, [b13 − B2]2+ at m/z 857.3852, [b12 − B2]2+ at m/z 777.3857, [b11 − B2]2+ at m/z 697.3887, [b9 − B2]2+ at m/z 568.7979 and [b7 − B2]2+ at m/z 490.7766. Finally, during the product ion scan of the precursor VSRSLGK*VGTRCCTK*PE ion (Lys 455, Lys 463) at m/z 1029.8239 (+3), we identified a product ion formed by the loss of the two entire β-D-galactopyranosyl-(1→4)-β-D-glucopyranosides carried on the Lys 455 and Lys 463 residues: [b15 − 2B2]2+ at m/z 1098.0119. In addition, another series of diagnostic product ions, which were described earlier, were detected and assigned as: [C17H26N3O4]+ at m/z 336.1852 and [C11H16N3O2]+ at m/z 222.1174. The galactopyranosyl [B1]+ product ion at m/z 163.0559 was also observed.
Thus, it is important to note that we observed peptides having the same sequence, which were glycosylated with either one and/or two carbohydrate haptens. To our knowledge, this important scientific fact is reported for the first time in the literature and need to be considered. In addition, the presence of peptides having the same sequence, which are glycosylated with one and/or two carbohydrate haptens, confirms the fact that the glycoconjugate was indeed a mixture of glycoforms.
Figure 4(d) displays the LC/MS/MS analysis of glycopeptide DK*GACLLPK*IE at m/z 1198.5604 (+2), derivatized at the Lys 197 and Lys 204 residues, and the product ions with their m/z values are reported in Supplementary Table S4 (see Supporting Information). Moreover, the peptides with the following sequences VSRSLGKVGTRCCTKPE and VSRSLGKVGTRCCTKPESE were also observed with one glycosylation on the Lys 455 residue and two glycosylations on the Lys 455 and Lys 463 residues (Table 3).
When combining the data of the LC/MS/MS analysis of the tryptic and GluC V8 digests, a total of 30 glycation sites were identified (displayed in Fig. 5(c)), and the total sequence coverage was evaluated to be 89%.
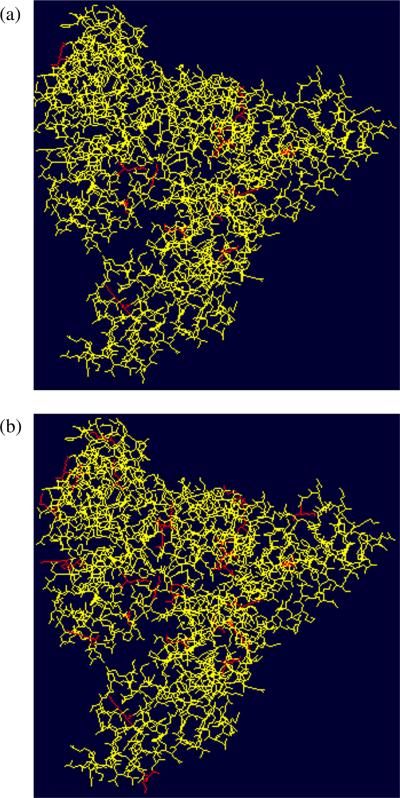
Figure 6 displays the three-dimensional structure (obtained using Swiss-Prot pdb viewer[23,24]) of the lactose-BSA glycoconjugates with hapten:BSA ratios of 5.1:1 (Fig. 6(a)) and 19.0:1 (Fig. 6(b)), where the glycated lysines are highlighted in red. We can thus observe that the majority of the glycated lysine residues are located on the outer surface of the protein.
Figure 6.
Three-dimensional structures of the lactose-BSA glycoconjugates with hapten-BSA ratios of (a) 5.1:1 and (b) 19.0:1.
CONCLUSIONS
The MALDI-MS analysis of the different lactose-BSA glycoconjugates allowed us to determine different hapten:BSA ratios formed during conjugation: 5.1:1 and 19.0:1. The LC/MS/MS analysis identified 15 glycation sites for the neoglycoconjugate with a hapten:BSA ratio of 5.1:1 and 30 glycation sites for the neoglycoconjugate with a hapten:BSA ratio of 19.0:1, showing that the conjugates were mixtures of glycoforms. This could be deduced from the higher number of glycation sites compared to the average molar number of haptens present in conjugates per mole of the carrier. The presence of glycoforms was also confirmed by the fact that for the same glycoconjugates with a hapten:BSA ratio of 19.0:1, glycopeptides of the same amino acid sequence were detected containing either one or two different glycation sites.
As expected, it was observed that the number of identified glycation sites increased when the hapten:BSA ratio of glycoconjugate formation increased, and that the location of the glycation sites appears to be mainly on the outer surface of the BSA carrier molecule (Fig. 6), which is in line with the assumption that the sterically more accessible lysine residues, namely those located on the outer surface of the BSA, would be conjugated preferentially.
As observed also during the LC/MS/MS analysis of the digested Bacillus anthracis exosporium-specific tetrasaccharide-BSA glycoconjugate,[16] we were able to detect various conventional diagnostic carbohydrate fragment ions ([B1]+, [B1 − H2O]+ and [B1 − 2H2O]+). We also noted that the formation of product ions and, in some glycopeptides, the loss of the neutral B2 product ions (formation of [y − B2]+ and [b – B2]+ ions). These findings seem to indicate the gas-phase fragility of these ions and inclination of the glycosylated oxygen-carbon spacer bond to rupture. Therefore, we conclude that, regardless of the nature of the carbohydrate portion in the synthetic neoglycoconjugates under investigation, the glycosylated O6-C1 bond, between the aglycone and the carbohydrate portion is easy to rupture in the gas phase.
Supplementary Material
Footnotes
SUPPORTING INFORMATION Additional supporting information may be found in the online version of this article.
REFERENCES
- [1].Kováč P, editor. Synthetic Oligosaccharides. Indispensable Probes in the Life Sciences. American Chemical Society; Washington, DC: 1994. (ACS Symposium Series 560). [Google Scholar]
- [2].Lee YC, Lee RT. In: Synthetic Glycoconjugates. Allen HJ, Kisailus EC, editors. Marcel Dekker; New York: 1992. pp. 121–165. [Google Scholar]
- [3].Galonic DP, Gin DY. Chemical glycosylation in the synthesis of glycoconjugate antitumor vaccines. Nature. 2007;446:1000. doi: 10.1038/nature05813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dick WE, Jr, Beurret M. Glycoconjugates of bacterial carbohydrate antigens. In: Cruise JM, Lewis RE Jr, editors. Conjugate Vaccines. vol. 10. Krager; Basel, Switzerland: 1989. pp. 48–114. [PubMed] [Google Scholar]
- [5].Tietze LF, Arlt M, Beller M, Glusenkamp KH, Jahde E, Rajewsky MF. Anticancer agents, 15. Squaric acid diethyl ester: a new coupling reagent for the formation of drug bio-polymer conjugates. Synthesis of squaric acid ester amides and diamides. Chem. Ber. 1991;124:1215. [Google Scholar]
- [6].Kamath VP, Diedrich P, Hindsgaul O. Use of diethyl squarate for the coupling of oligosaccharide amines to carrier proteins and characterization of the resulting neoglycoproteins by MALDI-TOF mass spectrometry. Glycoconjugate J. 1996;13:315. doi: 10.1007/BF00731506. [DOI] [PubMed] [Google Scholar]
- [7].Hou SJ, Saksena R, Kováč P. Preparation of glycoconjugates by dialkyl squarate chemistry revisited. Carbohydr. Res. 2008;343:196. doi: 10.1016/j.carres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang J, Yergey A, Kowalak J, Kováč P. Studies towards neoglycoconjugates from the monosaccharide determinat of Vibrio chlerae O:1 serotype Ogawa, using the diethyl squarate reagent. Carbohydr. Res. 1998;313:15. doi: 10.1016/s0008-6215(98)00261-4. [DOI] [PubMed] [Google Scholar]
- [9].Saksena R, Ma X, Kováč P. One-pot preparation of a series of glycoconjugates with predetermined antigen-carrier ratio from oligosaccharides that mimic the O-PS of Vibrio cholerae O:1, serotype Ogawa. Carbohydr. Res. 2003;338:2591. doi: 10.1016/s0008-6215(03)00273-8. [DOI] [PubMed] [Google Scholar]
- [10].Chernyak A, Karavanov A, Ogawa Y, Kováč P. Conjugating oligosaccharides to proteins by squaric acid diester chemistry: rapid monitoring of the progress of conjugation, and recovery of the unused ligand. Carbohydr. Res. 2001;330:479. doi: 10.1016/s0008-6215(01)00018-0. [DOI] [PubMed] [Google Scholar]
- [11].Ma X, Saksena R, Chernyak A, Kováč P. Neoglycoconjugates from synthetic tetra- and hexasaccharides that mimic the terminus of the O-PS of Vibrio cholerae O:1, serotype Inaba. Org. Biomol. Chem. 2003;1:775. doi: 10.1039/b211660j. [DOI] [PubMed] [Google Scholar]
- [12].Saksena R, Chernyak A, Karavanov A, Kováč Methods Enzymol. 2003;362:125. doi: 10.1016/S0076-6879(03)01010-3. [DOI] [PubMed] [Google Scholar]
- [13].Hegedűs G, Bélai I, Székács A. Development of an enzyme-linked immunosorbent assay (ELISA) for the herbicide trifluralin. Anal. Chim. Acta. 2000;421:121. [Google Scholar]
- [14].Jahouh F, Saksena R, Aiello D, Napoli A, Sindona G, Kováč P, Banoub JH. Glycation sites in neoglycoconjugates from the terminal monosaccharide antigen of the O-PS of Vibrio cholerae O1, serotype Ogawa, and BSA revealed by matrix-assisted laser desorption-ionization tandem mass spectrometry. J. Mass Spectrom. 2010;45:1148. doi: 10.1002/jms.1796. [DOI] [PubMed] [Google Scholar]
- [15].Rawlings ND, Barrett AJ. Families of serine peptidases. Methods Enzymol. 1994;244:19. doi: 10.1016/0076-6879(94)44004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jahouh F, Hou SJ, Kováč P, Banoub JH. Determination of the glycation sites of Bacillus anthracis neoglycoconjugate vaccine by MALDI-TOF/TOF-CID-MS/MS and LC-ESI-QqTOF-tandem mass spectrometry. J. Mass Spectrom. 2011;46:993. doi: 10.1002/jms.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Daubenspeck JM, Zeng H, Chen P, Dong S, Stei-chen CT, Krishna NR, Pritchard DG, Turnbough CL. J. Biol. Chem. 2004;279:30945. doi: 10.1074/jbc.M401613200. [DOI] [PubMed] [Google Scholar]
- [18].Drapeau GR, Boily Y, Houmard J. Purification and properties of an extracellular protease of Staphylococcus aureus. J. Biol. Chem. 1972;247:6720. [PubMed] [Google Scholar]
- [19].Birktoft JJ, Breddam K. Proteolytic enzymes: Glutamyl endopeptidases. Methods Enzymol. 1994;244:114. doi: 10.1016/0076-6879(94)44010-7. [DOI] [PubMed] [Google Scholar]
- [20].Roepstorff P, Fohlman J. Letter to the editors. Biol. Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- [21].Johnson RS, Martin SA, Biemann K, Stults JT, Watson JT. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass spectrometer: differentiation of leucine and isoleucine. Anal. Chem. 1987;59:2621. doi: 10.1021/ac00148a019. [DOI] [PubMed] [Google Scholar]
- [22].Domon B, Costello C. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate J. 1988;5:397. [Google Scholar]
- [23].Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- [24].Peitsch MC. Protein modeling by E-mail. Biol. Technol. 1995;13:658. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.