Herein we report an immobilized pH gradient (IPG) CIEF-MALDI MS platform designed for the separation of complex neuropeptides. This platform features a poly (glycidyl methacrylate-divinylbenzene) (GMA-DVB) based monolithic column for CIEF separation. Different from regular CIEF, carrier ampholytes are pre-immobilized on the monolithic surface instead of being added into the sample. An off-line coupling of IPG-CIEF to MALDI MS has been established. Optimizations and comparisons with regular CIEF are performed with BSA tryptic peptides and extracted neuropeptide mixtures from crustacean Callinectes sapidus. It has been demonstrated that separation of complex peptide mixtures in neutral and basic range can be achieved in less than 10 min with comparable separation efficiency with regular CIEF, while MS signal is significantly enhanced when employing IPG-CIEF. Enhanced neuropeptide detection is also observed after coupling IPG-CIEF with MALDI MS.
Neuropeptides are a class of short chain amino acids. They are expressed in neurons as signaling molecules which are involved in many physiological and pathological processes. Identification and characterization of neuropeptides are important for deciphering their functions, but their discovery and identifications are often challenging due to their extreme low quantities and complex chemical and physical properties. In recent decades, mass spectrometry (MS) has been extensively applied to the study of neuropeptides. Compared with traditional methods including Edman degradation and immunocytochemistry, MS features high sensitivity and mass accuracy, which enable de novo sequencing and identification of extracted neuropeptides in a complex mixture [1–3]. In order to reduce sample complexity and interference among molecules prior to mass spectrometric detection, a number of well-established separation techniques have been coupled with MS, most commonly including electrophoresis and liquid chromatography [4–5]. However, with the needs for more in-depth characterization of low abundance neuropeptides, these traditional separation methods are often limited by their separation efficiency, large sample consumption and long analysis time.
Electrophoretic-based separations in capillary format have been widely employed and coupled with MS for the analysis of proteins and peptides due to their high separation efficiency with low sample consumption [6–7]. Among these techniques, capillary isoelectric focusing (CIEF) has been considered as one of the most promising techniques for MS-based proteomics and peptidomics [5]. Different from capillary zone electrophoresis (CZE), CIEF employs a pH gradient which is provided by carrier ampholytes within the capillary, and separates analytes according to their isoelectric points (pI), enabling highly efficient separation and focusing in a short time. Applications have been reported including coupling CIEF to both matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI) MS [8–12]. We have previously reported a membrane-assisted CIEF (MA-CIEF) system which can be coupled with MALDI-Fourier transform mass spectrometry (FTMS) for the analysis of neuropeptides and other complex peptide mixtures with enhanced efficiencies [13]. Acidic peptides are found to be more favored in the CIEF system; however, the pI values for most crustacean neuropeptide families are in the basic range of 13 to 14, which cannot be separated or detected. Therefore, it is critical to establish a platform that can be applied to basic neuropeptides.
The monolith has been demonstrated as a promising alternative material for LC separation in 1990s [14]. Since then it has been widely applied to a variety of separation approaches, including both electrophoretic-driven [15–16] and pressure-driven formats [17–18]. In this study, we report an immobilized pH-gradient CIEF (IPG-CIEF) system coupling with MALDI MS. In comparison to previously reported IPG-CIEF systems [19–22], a poly (glycidyl methacrylate-divinylbenzene) (GMA-DVB) monolithic column with optimized rigidity and permeability is fabricated with carrier ampholytes immobilized inside. This platform has been tested and applied to the analysis of both bovine serum albumin (BSA) tryptic peptides and neuropeptides extracted from the crustacean nervous system, with high separation efficiency and high sensitivity observed.
Acetic acid, hydrochloric acid, sodium hydroxide, ammonium hydroxide, acetone, acetonitrile, ammonium bicarbonate and urea were purchased from Fisher Scientific (Pittsburgh, PA). Glycidyl methacrylate (GMA, 97%), ethylene dimethacrylate (EDMA, 98%), divinylbenzene (DVB, 80%), 1-propanol (99.5%), 1,4-butandiol (99%), 2,2′-Azobis(2-methylpropionitrile) (AIBN, 98%), 3-(trimethoxysilyl) propyl ethacrylate (98%), α-Cyano-4-hydroxycinnamic acid (CHCA, 99%), trifluoroacetic acid, Pharmalyte 3–10, iodoacetamide (IAA) and bovine serum albumin (BSA) were from Sigma-Aldrich (St. Louis, MO). Ethanol was purchased from Decon Laboratories (King of Prussia, PA). 2,5-dihydroxybenzoic acid (DHB) was obtained from Alfa Aesar (Ward Hill, MA). Sequencing grade modified trypsin and D/L-dithiothreitol (DTT) were from Promega (Madison, WI). Millipore C18 Ziptip column was used for sample cleaning, and all water used in this study was doubly distilled on a Millipore filtration system (Bedford, MA). The physiological saline consisted of 440 mM NaCl, 11mM KCl, 26 mM MgCl2, 13 mM CaCl2, 11 mM Trizma base, and 5 mM maleic acid in pH 7.45.
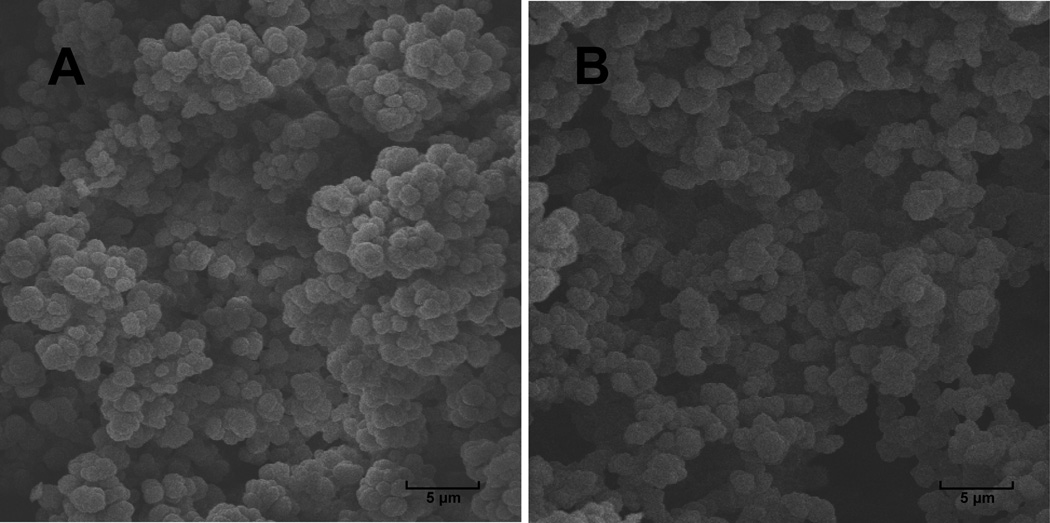
The procedure of monolithic column fabrication was modified from previous reports [20, 23]. A fused-silica capillary with 75 µm i.d. and 360 µm o.d. (Polymicro Technologies, Phoenix, AZ) was rinsed with acetone and water, and then flushed with 0.2 M NaOH and 0.2 M HCl for 30 min at a flow rate of 0.5 µL/min, respectively, followed by water and ethanol. A 20% solution of 3-(trimethoxysilyl) propyl methacrylate prepared in 95% ethanol was pumped through the capillary at a flow rate of 0.5 µL/min for 90 min. The capillary was then washed with acetone, dried in a stream of nitrogen, and left at room temperature overnight. A polymerization mixture consisting of GMA, DVB, 1-propanol, 1,4-butandiol (8%:12%:45%:35%, w/w), and 1% AIBN was purged with nitrogen for 10 min. The capillary was filled with this mixture and thermally initiated polymerization was carried out in a water bath at 70 °C for 24 h. After polymerization, the capillary was flushed with acetonitrile, water and 5% Pharmalytes 3–10 for 30 min, respectively. It was then connected to TriSep-2100 HV power supplier from Unimicro Technologies (Pleasanton, CA) through anolyte (1% HAc) and catholyte (1% NaOH) reservoirs, and focused under 5 kV for 10 min to form pH gradient within the capillary. The pH gradient was further immobilized by heating the capillary in a water bath for 24 hours at 70 °C, followed by flushing with water for 30 min. Figure 1A shows the scanning electron microscope (Hitachi S570, Pleasanton, CA) image of GMA-DVB monolith. A homogenous and continuous monolith structure could be observed after polymerization and washing. For a 35 cm long 75 µm i.d. fused-silica capillary, the pressure drop on HPLC was less than 300 psi when purged with ACN/H2O at 0.3 µL/min. We have compared GMA-DVB monolithic column with poly (glycidyl methacrylate- ethylene dimethacrylate) (GMA-EDMA) column which is employed in many other studies [24–25]. When EDMA is employed as cross-linker, in order to reduce non-specific binding, very low monomer to porogen ratios must be used which consequently results in poor rigidity of the monolithic structure [26–27], as shown in Figure 1B. We found that the GMA-EDMA structure collapsed and formed clogs within the capillary after one or two CIEF runs due to the high pressure added for fraction collection. However, by employing DVB which only has a phenol ring in the monolithic structure after polymerization as cross-linker, rigid monolith structure can be obtained while still maintaining a low backpressure.
Figure 1. Scanning electron microscope images of monolith structures in a 75 µm i.d. capillary.
A: GMA-DVB monolith. A rigid and uniform structure was found when employed for IPG-CIEF. B: GMA-EDMA monolith with poor uniformity and larger pore size which will result in collapsed structure after CIEF.
We tested the performance of GMA-DVB based IPG-CIEF system with BSA tryptic peptides. Briefly, BSA was dissolved in 25 mM ammonium bicarbonate solution, reduced by D/L-dithiothreitol and then reacted with iodoacetamide before overnight digestion by trypsin in room temperature. After digestion, BSA tryptic peptide mixture was dissolved in water and loaded onto the monolithic capillary after cleaning up with Millipore C18 Ziptip column. A high voltage of 10 kV was applied to the capillary via anolyte and catholyte reservoirs as described above, and focusing was kept for 8 to 10 min until current became stable. After focusing, CIEF fractions were migrated by air pressure and collected on a MALDI plate. Profiling of peptides in the range of 500–3000 Da was performed using an autoflex III MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) equipped with a Smartbeam 2 laser and a ground MALDI target. Detailed information on the instrument settings have been described in a previous report [13].
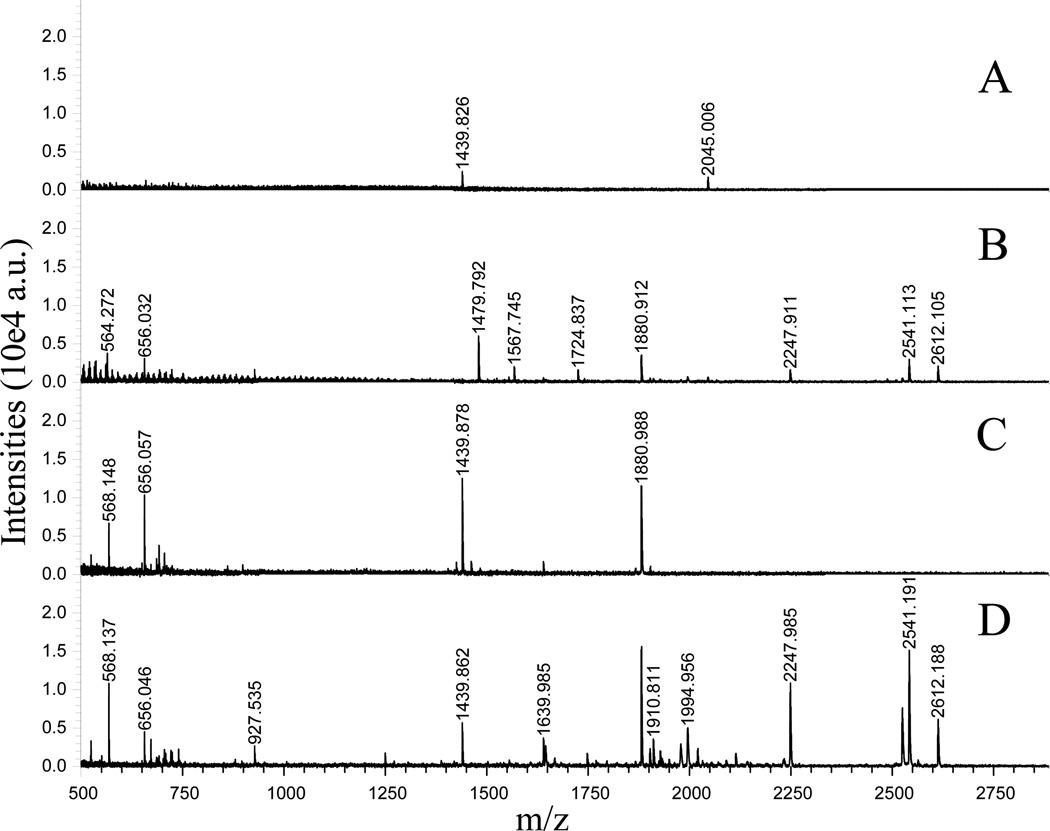
Figure 2 shows the comparison of regular CIEF and IPG-CIEF separations of BSA tryptic peptides with MALDI-TOF/TOF detection. A previously developed regular CIEF [13] was employed here to compare its separation efficiency and sensitivity to the IPG-CIEF. Similar to our previous report, a regular CIEF separated the BSA tryptic peptides into fractions in accordance with their pI values. Figure 2A and 2B show representative basic and neutral fractions after regular CIEF separation. However, low peak intensities were observed due to the existence of carrier ampholytes that has suppressed the MS ionization efficiency of the peptides. Report has shown that even only 0.5% of carrier ampholytes could significantly decrease a MS signal, and the suppression will become much more severe when carrier ampholyte concentration increases [8]. For IPG-CIEF, separation is also conducted based on pI differences of analytes. However, distinct from the conventional CIEF, carrier ampholytes are immobilized on the monolithic surface rather than being added to the sample solution. In this case, when high voltage is applied, separation can be achieved by the pre-immobilized pH gradient, while the interference of carrier ampholytes in the effluent fractions has been eliminated. Figs. 2C and 2D show the corresponding fractions to Figs. 2A and 2B after IPG-CIEF separation. A comparison of the same peptides fractionated under both CIEF modes reveals that the peak intensities are significantly increased by using IPG-CIEF, resulting in increased number of peptides being detected. The results also indicated that a number of peptides with high pI values, e.g., carbamidomethyl-modified tryptic peptides are highly enriched in the later fractions using the present IPG-CIEF setup. While some of the acidic peptides that can be detected with regular CIEF (Figure 1A), including m/z 1567.75 (DAFLGSFLYEYSR) and m/z 1724.79 (MPCTEDYLSLILNR) are not detectable in IPG-CIEF. A possible explanation is that due to the lack of capillary coating most acidic carrier ampholytes band migrates and brings the acidic peptides into the anolyte solution with a velocity determined by the local field strength and anion effective mobility [28], while the un-reacted silica surface will generate a small electroosmotic flow that retains basic peptides to the cathode direction. Table S1 (supplementary information) shows a detailed list of tryptic peptides separated by both regular CIEF and IPG-CIEF. These results suggest that IPG-CIEF offers superior performance as compared to regular CIEF with enhanced MS signals especially for peptides in the high pI ranges.
Figure 2. Comparison of regular CIEF and IPG-CIEF separations with BSA tryptic peptides.
Panels A and B: Representative mass spectra showing the peptide profiles collected from an earlier and a later fraction after regular CIEF separation. Panels C and D: Representative mass spectra showing the peptide profiles collected from corresponding fractions after IPG-CIEF separation.
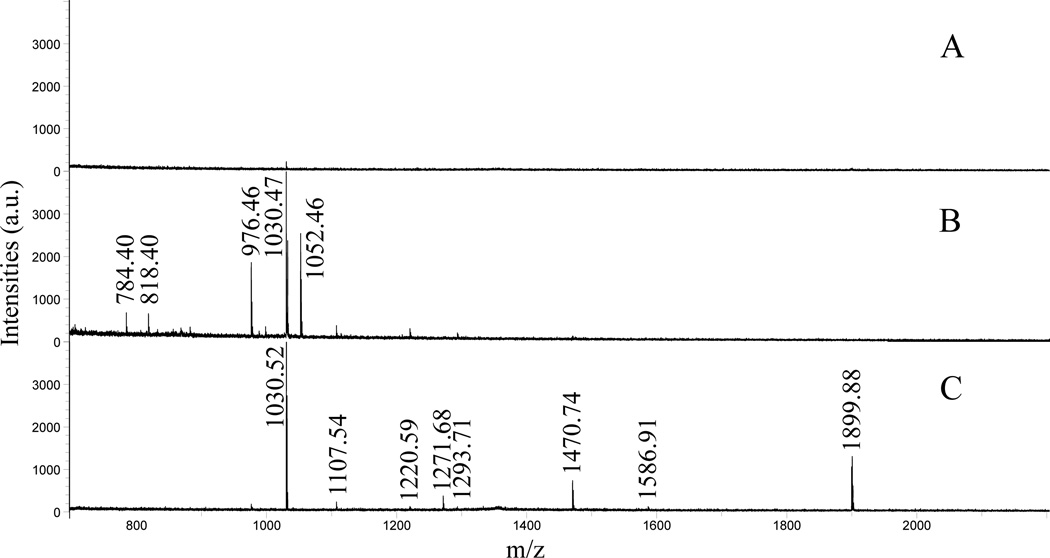
Upon validation of the IPG-CIEF/MALDI MS platform, the new platform was further employed to the analysis of neuropeptide mixtures extracted from blue crab (C. sapidus) pericardial organs. The sample preparation method is previously described [7]. In brief, animals were kept in artificial seawater in 10–12 °C and anesthetized with ice before dissection. The dissected neural tissues were kept in physiological saline, and then extracted with cold acidified methanol with 90:9:1 ratio of methanol, water and acetic acid. The extraction was performed three times, and the supernatants were combined, dried and dissolved in 0.1% of TFA. In order to remove proteins, lipids and salts, the samples were cleaned with Ziptip C18 column before CIEF-MALDI MS analysis. CIEF conditions and MS settings were kept consistent with the study of BSA tryptic peptides. Figure 3 compares the mass spectra of direct profiling of crude extract (Figure 3A) and two representative fractions after separation by IPG-CIEF (Figure 3B, 3C). As shown in Figure 3A, a direct profiling of extracted neuropeptides with MALDI-TOF/TOF observed almost no neuropeptide signals due to severe ion suppression from the high complexity of the tissue extract. The same sample was then loaded onto a GMA-DVB monolithic column for IPG-CIEF separation. Because the majority of crustacean neuropeptide families except for orcokinins are highly basic with pI values around 13 to 14, none of them can be detected with regular CIEF separation as we have previously investigated. Figure 3B shows mass spectrum acquired from a representative basic fraction equals to ~pI 10. Some basic neuropeptides, including m/z 784.40 (FVGGSRYa), m/z 976.46 (SGFYANRYa), m/z 1030.47 (pEGFYSQRYa), multiple peptide isoforms from the RFamide and A-type allatostatin families with pI values around 10 are found in this fraction (the m/z are labeled on the spectrum). Again, it is worth noting that none of these peptides could be observed in a regular CIEF fraction due to their extremely low abundances. No neutral peptides were found, mainly due to the lack of neutral neuropeptides in the extracted samples as previously investigated [1]. Since the IPG-CIEF fractions were collected from catholyte to anolyte direction in this study (high pI then low pI), it is expected to see acidic peptides in later fractions. However, interestingly, in later fractions, a series of putative B-type allatostatin peptides with pI values at 14 were detected, including m/z 1107.54 (AGWSSMRGAWa), m/z 1220.59 (SGDWSSLRGAWa), m/z 1293.71 (STNWSSLRSAWa), m/z 1470.74 (VPNDWAHFRGSWa), and m/z 1586.91 (MFAPLAWPKGGARWa). A putative C-type allatostatin at m/z 1899.88 (pQVRFRQCYFNPISCF), which was recently reported by our lab [29], was also co-eluted with the B-type allatostatins (ASTs). We also performed the experiment by using EDMA as cross-linker for monolith columns, but no ASTs were observed. Based on the fact that all the AST family neuropeptides we observed are tryptophan-rich neuropeptides, a possible explanation about observing these peptides in later fractions of the IPG-CIEF separation could be due to the potential π-π interactions between the indole groups on the tryptophan and the phenol ring on DVB. This unique interaction between the Trp-containing AST family peptides and the phenol ring on DVB could contribute to stronger retention of these peptides on the DVB column, leading to their later elution during the fractionation process. This observation suggests a possible different separation mechanism other than regular CIEF separation based on pI differences of analytes.
Figure 3. Separation of neuropeptides extracted from C. sapidus pericardial organ.
A: un-separated neuropeptide extract directly analyzed with MALDI-TOF/TOF. B: A representative mass spectrum collected from a basic fraction after CIEF separation. C: Mass spectrum showing a fraction containing the allatostatin family neuropeptides.
Collectively, we developed a GMA-DVB monolithic column for IPG-CIEF separation of peptides. With carrier ampholytes immobilized on the column, suppression of ionization is eliminated which results in significantly enhanced MS signal. Evaluation with BSA tryptic peptides demonstrates that the IPG-CIEF system can separate and focus tryptic peptides with similar efficiency compared with regular CIEF in neutral and basic ranges, while much higher peak intensities are observed in mass spectra. This system has been applied to the analysis of neuropeptides in a complex mixture extracted from the blue crab. In addition to the putative basic neuropeptides observed, the allatostatin family neuropeptides with higher pI values were separated and detected. These results reveal the potential of using GMA-DVB based IPG-CIEF as a promising tool for the study of proteomics and peptidomics.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation grant (CHE-0967784) and National Institutes of Health grant (1R01DK071801). The authors thank the Biological and Biomaterials Preparation, Imaging and Characterization Facility at UW-Madison for taking scanning electron microscope image. We also thank Bruker Daltonics for graciously loaning the autoflex III MALDI TOF/TOF mass spectrometer. L. Li acknowledges a Vilas Associate Award and an H.I. Romnes Faculty Research Fellowship.
Abbreviations
- GMA
glycidyl methacrylate
- DVB
divinylbenzene
- IPG
immobilized pH gradient
- CIEF
capillary isoelectric focusing
Footnotes
The authors have declared no conflict of interest.
Supplementary information is available.
References
- 1.Ma M, Wang J, Chen R, Li L. J. Proteome Res. 2009;8:2426–2437. doi: 10.1021/pr801047v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu Q, Li L. Anal. Chem. 2005;77:7783–7795. doi: 10.1021/ac051324e. [DOI] [PubMed] [Google Scholar]
- 3.Li L, Kelley WP, Billimoria CP, Christie AE, Pulver SR, Sweedler JV, Marder E. J. Neurochem. 2003;87:642–656. doi: 10.1046/j.1471-4159.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Sweedler JV. Ann. Rev. Anal. Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 5.Simpson D, Smith RD. Electrophoresis. 2005;26:1291–1305. doi: 10.1002/elps.200410132. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Ye H, Zhang Z, Girdaukas G, Li L. Anal. Chem. 2011;83:3462–3469. doi: 10.1021/ac200708f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Zhang Y, Xiang F, Zhang Z, Li L. J. Chromatogr. A. 2010;1217:4463–4470. doi: 10.1016/j.chroma.2010.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuroda Y, Yukinaga H, Kitano M, Noguchi T, Nemati M, Shibukawa A, Nakagawa T, Matsuzaki K. J. Pharm. Biomed. Anal. 2005;37:423–428. doi: 10.1016/j.jpba.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 9.Silvertand LLH, Torano J, de Jong G, Bennekom W. Electrophoresis. 2008;29:1985–1996. doi: 10.1002/elps.200700434. [DOI] [PubMed] [Google Scholar]
- 10.Yu W, Li Y, Deng C, Zhang X. Electrophoresis. 2006;27:2100–2110. doi: 10.1002/elps.200500820. [DOI] [PubMed] [Google Scholar]
- 11.Balgley BM, Wang WJ, Song T, Fang XP, Yang L, Lee CS. Electrophoresis. 2008;29:3047–3054. doi: 10.1002/elps.200800050. [DOI] [PubMed] [Google Scholar]
- 12.Wang YJ, Balgley BM, Rudnick PA, Evans EL, DeVoe DL, Lee CS. J. Proteome Res. 2005;4:36–42. doi: 10.1021/pr049876l. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Wang J, Hui L, Li L. J. Chromatogr. A. 2011;1218:5336–5343. doi: 10.1016/j.chroma.2011.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svec F, Fréchet JM. Anal. Chem. 1992;64:820–822. doi: 10.1021/ac00035a008. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Wu R, Wu M, Zou H. Electrophoresis. 2010;31:1457–1466. doi: 10.1002/elps.200900672. [DOI] [PubMed] [Google Scholar]
- 16.Ivanov AR, Horvath C, Karger BL. Electrophoresis. 2003;24:3663–3673. doi: 10.1002/elps.200305620. [DOI] [PubMed] [Google Scholar]
- 17.Wang FJ, Dong J, Ye M, Wu R, Zou H. J. Chromatogr. A. 2009;1216:3887–3894. doi: 10.1016/j.chroma.2009.02.082. [DOI] [PubMed] [Google Scholar]
- 18.Luo Q, Gu Y, Wu S, Rejtar T, Karger BL. Electrophoresis. 2008;29:1604–1611. doi: 10.1002/elps.200700741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang Y, Cong Y, Liang Z, Zhang L, Zhang Y. Electrophoresis. 2009;30:4034–4039. doi: 10.1002/elps.200900209. [DOI] [PubMed] [Google Scholar]
- 20.Zhu G, Yuan H, Zhao P, Zhang L, Liang Z, Zhang W, Zhang Y. Electrophoresis. 2006;27:3578–3583. doi: 10.1002/elps.200600189. [DOI] [PubMed] [Google Scholar]
- 21.Wang T, Ma J, Zhu G, Shan Y, Liang Z, Zhang L, Zhang Y. J. Sep. Sci. 2010;33:3194–3200. doi: 10.1002/jssc.201000324. [DOI] [PubMed] [Google Scholar]
- 22.Wang T, Fekete A, Gaspar A, Ma J, Liang Z, Yuan H, Zhang L, Schmitt-Kopplin P, Zhang Y. J. Sep. Sci. 2011;34:422–427. doi: 10.1002/jssc.201000720. [DOI] [PubMed] [Google Scholar]
- 23.Mallik R, Jiang T, Hage DS. Anal. Chem. 2004;76:7013–7022. doi: 10.1021/ac049001q. [DOI] [PubMed] [Google Scholar]
- 24.Svec F, Huber CG. Anal. Chem. 2006;78:2100–2107. doi: 10.1021/ac069383v. [DOI] [PubMed] [Google Scholar]
- 25.Mallik R, Hage D. J. Sep. Sci. 2006;29:1686–1704. doi: 10.1002/jssc.200600152. [DOI] [PubMed] [Google Scholar]
- 26.Yang C, Zhu G, Zhang L, Zhang W, Zhang Y. Electrophoresis. 2004;25:1729–1734. doi: 10.1002/elps.200405916. [DOI] [PubMed] [Google Scholar]
- 27.Zhu G, Yang C, Zhang L, Liang Z, Zhang W, Zhang Y. Talanta. 2006;70:2–6. doi: 10.1016/j.talanta.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 28.Mack S, Cruzado-Park I, Chapman J, Ratnayake C, Vigh G. Electrophoresis. 2009;30:4049–4052. doi: 10.1002/elps.200800690. [DOI] [PubMed] [Google Scholar]
- 29.Ma M, Szabo TM, Jia C, Marder E, Li L. Peptides. 2009;30:1660–1668. doi: 10.1016/j.peptides.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.