Living organisms are continually exposed to DNA damage arising from reactive species inside the cell and from environmental sources. Probably the most dangerous form of damage is the DNA doublestrand break (DSB), which interrupts both strands of the molecule. If it is not rapidly resealed, a DSB can cause aberrant chromosomal rearrangements, mutations, or cell death. To protect themselves, organisms from bacteria to humans have developed two major pathways to heal DSBs, homologous recombination (HR) and nonhomologous end-joining (NHEJ). HR utilizes an intact copy (homolog or sister chromatid) of the broken chromosome as a template for repair, whereas the NHEJ pathway joins the two ends of a DSB directly, with little or no requirement for sequence homology. Defects in either of these pathways can compromise genomic integrity and increase the potential for tumorigenesis. Recent studies indicate that, in addition to proteins that directly mediate enzymatic DNA repair, factors that organize specialized chromatin structures surrounding a DSB may facilitate DNA damage signaling and repair. It is well known that histone acetylases and histone deacetylases (HDACs) are important modifiers of chromatin and that they play a central role as transcriptional regulators. The work of Jazayeri et al. (1) in this issue of PNAS demonstrates that the Sin3p/Rpd3p deacetylase complex is required for efficient repair by NHEJ in Saccharomyces cerevisiae. This study sheds light on how chromatin, traditionally viewed as a barrier to DNA-templated processes, can be modified in a manner that increases the faithful transmission of genetic information.
Highly localized changes (expansion and contraction) in chromatin structure are necessary to drive many DNA-directed processes, including transcription, replication, recombination, and repair. There are at least two classes of enzymes that promote regional changes in higher-order chromatin structure. The so-called chromatin-remodeling complexes use ATPase activity to catalyze nucleosome mobility. Chromatin structure is also modulated by enzymes that add chemical groups, such as methyl, phosphate, acetyl, and ubiquitin, to residues in the N- and/or C-terminal histone tails. It has been proposed that such patterns of histone modifications form a “histone code” (2). According to this hypothesis, the combination of all histone modifications within a genomic locus (input) determines the flow of genetic information arising from this site (biological output).
There are clues from the recent literature that both chromatin-remodeling and histone-modifying enzymes alter chromatin topology in the response to DNA damage. For example, the HR protein Rad54 possesses an ATPase activity that is needed to remove nucleosomes and other DNA-binding proteins from the DNA-target site during recombination (3-5). By contrast, the S. cerevisiae linker histone Hho1p has been found to be inhibitory to HR-dependent events (6), possibly by limiting the activity of chromatin-remodeling proteins.
The first histone modification that was discovered to be specifically associated with DSBs is the phosphorylation H2AX (termed γ-H2AX) (7), a chromatin mark induced by the phosphatidyl-inositol-3-OH kinase-related kinases. γ-H2AX encompasses a chromatin region spanning thousands to millions of base pairs surrounding the lesion. The analysis of yeast H2A mutants unable to undergo phosphorylation demonstrated that γ-H2AX is required for efficient NHEJ (8). Similarly, the analysis of H2AX-deficient mice revealed a role for this histone variant in several aspects of DNA repair (9). Furthermore, H2AX phosphorylation (10) and, recently, H3 lysine methylation (11) have been shown to be essential for protecting genome integrity, illustrating that pathological outputs may arise from a compromised histone epigenetic profile.
Evidence for a DNA Repair-Specific Histone Code
The group led by Stephen Jackson (1) has now uncovered a second histone modification in S. cerevisiae that is induced in response to DNA damage. They discovered that the lysine 16 residue of histone H4 is differentially deacetylated in the proximity of a DSB. Importantly, this DNA damage-dependent hypoacetylation was found to genetically depend on the Sin3p protein, a component of a multisubunit complex (that includes Rpd3p) that catalyzes histone deacetylation in yeast. Disruption of either Sin3p or Rpd3p resulted in hypersensitivity to phleomycin, which causes DSBs, but not to other types of DNA-damaging agents, such as ultraviolet light. The DSB repair defect in the sin3Δ/rpd3Δ strains appears to be very specific, given that mutants were not sensitive to camptothecin or hydroxyurea, which produce DSBs in the S-phase of the cell cycle, where HR is highly active. Strikingly, Jazayeri et al. found that Sin3p/Rpd3p was required for NHEJ but did not influence HR. In line with these findings, a recent genome-wide screen in Caenorhabditis elegans identified several HDACs, including Sin3p, that contribute to the maintenance of genomic stability (12).
Interestingly, not only histone deacetylation but also histone acetylation has been linked to DSB repair (13-19). In a recent study, substitution of four conserved lysine residues (including lysine 16) in the N-terminal tail of H4 by Gln, which mimics the charge of the acetylated state, caused a defect in NHEJ (13). Moreover, disruption of the acetyl transferase activity of the nucleosome acetyltransferase of histone H4 complex conferred a very similar repair defect. Thus, we have learned that proteins that mediate histone phosphorylation (phosphatidylinositol-3-OH kinase-related kinases) and proteins that apparently have opposing effects on histone acetylation regulate the NHEJ pathway in yeast.
A strong correlation exists between increased histone acetylation and transcriptionally active chromatin and, conversely, between histone deacetylation and repressive chromatin. Because deletion of Sin3p/Rpd3 is known to have a widespread effect on numerous promoters (20), it was therefore conceivable that impaired NHEJ in sin3Δ strains was an indirect effect of this mutation on transcription. Indeed, the silencing proteins Sir2, Sir3, and Sir4 (components of a NAD-dependent HDAC) affect NHEJ indirectly by the repression of specific NHEJ transcripts (21). In contrast, however, Jazayeri et al. report that Sin3p disruption did not affect the expression of any of the known NHEJ factors. Moreover, in contrast to the global effects that Sin3p/Rpd3 has on acetylation levels, chromatin immunoprecipitation revealed that the deacetylation of H4 lysine 16 was differentially targeted to DSBs.
In addition, careful phenotypic analyses of the sin3Δ strains revealed several interesting differences with the canonical NHEJ pathway. For example, phleomycin hypersensitivity, characteristic of the sin3Δ strain, was not a property of the NHEJ-deficient cells. Secondly, telomere-length maintenance, which requires components of the NHEJ pathway, was not compromised in the absence of Sin3p. Finally, although the deletion of Sin3p affected plasmid end-joining as severely as in NHEJ strains, there was an important difference in the nature of residual junctions in the few transformants recovered. NHEJ-deficient strains employ an alternative-joining pathway to repair broken DNA that utilizes microhomologies to produce different-sized deletions. In marked contrast, the residual repair of linearized plasmids in sin3Δ strains was found to be accurate. This finding indicates that NHEJ factors are active in the absence of Sin3p and are even able to catalyze the ligation of DNA ends, albeit with reduced efficiency.
Changes in Chromatin Structure Near a DNA Break
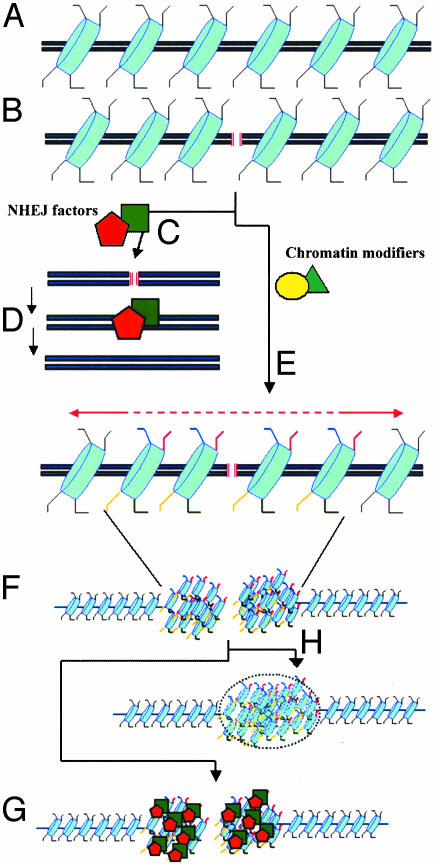
Taken together, these data suggest that the Sin3p/Rpd3 complex is not likely to be required for the targeting of NHEJ components to DSBs. However, Sin3p/Rpd3-dependent deacetylation of H4 lysine 16 may generate a highly localized region of chromatin that facilitates the synapsis of broken ends. It is possible that the removal of acetyl groups from histones would result, either directly or indirectly, in a chromatin configuration near the lesion that is more condensed (Fig. 1). The increase in the local concentration of proteins in the vicinity of the DSB would enhance the likelihood of assembling an efficient DNA repair complex (Fig. 1). At the same time, both ends of the broken DNA molecule would be tethered together, decreasing their propensity to undergo aberrant recombination reactions. This mode of reorganization of chromatin structure has also been proposed for histone H2AX phosphorylation (9), which modulates NHEJ and protects genomic integrity. In the absence of H2AX, mammalian cells show multiple chromosomal aberrations. However, like Sin3p-deficient strains, the junctions associated with recombination events in H2AX-deficient mouse cells revealed no anomalies in the end-joining process itself (10, 22-24). In light of these similarities, it is tempting to propose that a DSB triggers various histone modifications (including acetylation, deacetylation, and phosphorylation) that function combinatorially to control the dynamic remodeling of the chromatin microenvironment surrounding a DSB. In other words, DSB-induced posttranslational modifications of histone tails may represent a DNA repair-specific histone code. In linking histone deacetylation to NHEJ, the work of Jazayeri et al. provides us with one of the first epigenetic readouts of the cellular response to DSBs. The quest for mapping the full spectrum of histone modifying marks and defining the precise spatial configuration of chromatin triggered by DSBs will be a major challenge.
Fig. 1.
Histone modifications during NHEJ. (A) Intact DNA molecule showing the nucleosomes (in blue) and the protruding histone tails (black lines). (B) Generation of a DSB (red lines). During the repair process, at least two types of factors are recruited to the DSB: catalytic factors that heal the lesion by NHEJ (C and D) and enzymes that remodel chromatin by posttranslational changes of residues within the histone tail (e.g., red, phosphorylation; blue, acetylation; yellow, deacetylation) (E). The new pattern of epigenetic marks may trigger the compaction of chromatin in the microenvironment surrounding the break (F). This chromatin reconfiguration would serve to increase the local concentration of NHEJ-catalytic factors (G) and/or limit the diffusion of the broken DNA ends until the break is repaired (H).
See companion article on page 1644.
References
- 1.Jazayeri, A., McAinsh, A. D. & Jackson, S. P. (2004) Proc. Natl. Acad. Sci. USA 101, 1644-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074-1080. [DOI] [PubMed] [Google Scholar]
- 3.Alexiadis, V. & Kadonaga, J. T. (2002) Genes Dev. 16, 2767-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexeev, A., Mazin, A. & Kowalczykowski, S. C. (2003) Nat. Struct. Biol 10, 182-186. [DOI] [PubMed] [Google Scholar]
- 5.Jaskelioff, M., Van Komen, S., Krebs, J. E., Sung, P. & Peterson, C. L. (2003) J. Biol. Chem. 278, 9212-9218. [DOI] [PubMed] [Google Scholar]
- 6.Downs, J. A., Kosmidou, E., Morgan, A. & Jackson, S. P. (2003) Mol. Cell 11, 1685-1692. [DOI] [PubMed] [Google Scholar]
- 7.Pilch, D. R., Sedelnikova, O. A., Redon, C., Celeste, A., Nussenzweig, A. & Bonner, W. M. (2003) Biochem. Cell Biol. 81, 123-129. [DOI] [PubMed] [Google Scholar]
- 8.Downs, J. A., Lowndes, N. F. & Jackson, S. P. (2000) Nature 408, 1001-1004. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Capetillo, O., Lee, A., Nussenzweig, M. C. & Nussenzweig, A. (2004) DNA Repair, in press. [DOI] [PubMed]
- 10.Celeste, A., Difilippantonio, S., Difilippantonio, M. J., Fernandez-Capetillo, O., Pilch, D. R., Sedelnikova, O. A., Eckhaus, M., Ried, T., Bonner, W. M. & Nussenzweig, A. (2003) Cell 114, 371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters, A. H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A., et al. (2001) Cell 107, 323-337. [DOI] [PubMed] [Google Scholar]
- 12.Pothof, J., van Haaften, G., Thijssen, K., Kamath, R. S., Fraser, A. G., Ahringer, J., Plasterk, R. H. & Tijsterman, M. (2003) Genes Dev. 17, 443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bird, A. W., Yu, D. Y., Pray-Grant, M. G., Qiu, Q., Harmon, K. E., Megee, P. C., Grant, P. A., Smith, M. M. & Christman, M. F. (2002) Nature 419, 411-415. [DOI] [PubMed] [Google Scholar]
- 14.Ikura, T., Ogryzko, V. V., Grigoriev, M., Groisman, R., Wang, J., Horikoshi, M., Scully, R., Qin, J. & Nakatani, Y. (2000) Cell 102, 463-473. [DOI] [PubMed] [Google Scholar]
- 15.Barlev, N. A., Poltoratsky, V., Owen-Hughes, T., Ying, C., Liu, L., Workman, J. L. & Berger, S. L. (1998) Mol. Cell. Biol. 18, 1349-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukamoto, Y., Kato, J. & Ikeda, H. (1997) Nature 388, 900-903. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt, D. R. & Schreiber, S. L. (1999) Biochemistry 38, 14711-14717. [DOI] [PubMed] [Google Scholar]
- 18.Cai, R. L., Yan-Neale, Y., Cueto, M. A., Xu, H. & Cohen, D. (2000) J. Biol. Chem. 275, 27909-27916. [DOI] [PubMed] [Google Scholar]
- 19.Kao, G. D., McKenna, W. G., Guenther, M. G., Muschel, R. J., Lazar, M. A. & Yen, T. J. (2003) J. Cell Biol. 160, 1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robyr, D., Suka, Y., Xenarios, I., Kurdistani, S. K., Wang, A., Suka, N. & Grunstein, M. (2002) Cell 109, 437-446. [DOI] [PubMed] [Google Scholar]
- 21.Valencia, M., Bentele, M., Vaze, M. B., Herrmann, G., Kraus, E., Lee, S. E., Schar, P. & Haber, J. E. (2001) Nature 414, 666-669. [DOI] [PubMed] [Google Scholar]
- 22.Celeste, A., Petersen, S., Romanienko, P. J., Fernandez-Capetillo, O., Chen, H. T., Sedelnikova, O. A., Reina-San-Martin, B., Coppola, V., Meffre, E., Difilippantonio, M. J., et al. (2002) Science 296, 922-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassing, C. H., Chua, K. F., Sekiguchi, J., Suh, H., Whitlow, S. R., Fleming, J. C., Monroe, B. C., Ciccone, D. N., Yan, C., Vlasakova, K., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 8173-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassing, C. H., Suh, H., Ferguson, D. O., Chua, K. F., Manis, J., Eckersdorff, M., Gleason, M., Bronson, R., Lee, C. & Alt, F. W. (2003) Cell 114, 359-370. [DOI] [PubMed] [Google Scholar]