The advent of gene targeting techniques has permitted the construction of specific genetic deficiencies to evaluate the biological contribution(s) of an individual protein. Mice lacking a precise DNA repair activity have been generated, and these mutants show various combinations of defective embryogenesis, tissue-specific dysfunction, hypersensitivity to DNA-damaging agents, premature senescence, genetic instability, and elevated cancer rates (1). That repair-deficient animals display such abnormalities underscores the fundamental importance of DNA repair in protecting against the mutagenic and cytotoxic effects of DNA damage.
Proteins participating in base excision repair (BER) cope with chromosomal damages that arise as spontaneous decomposition products or from reactions with metabolically or environmentally derived reactive chemicals (2)—namely oxygen free radicals and alkylating agents. Before now, attempts to generate mice that are defective in BER have led to embryonic lethality, as seen with the major apurinic/apyrimidinic (AP) endonuclease (APE) (3) and polymerase β (Polβ) (4), two factors recognized as central players in this pathway (see Table 1). Three additional genes having a less-well-defined relationship to BER, XRCC1, LIG1, and PARP, also have been knocked out (5–7). Whereas XRCC1 and LIG1 disruptions lead to lethality during embryogenesis, the poly(ADP-ribose) polymerase (PARP)-deficient embryos survive and produce relatively healthy appearing mice. In this issue of the Proceedings, Engelward, Weeda, and coworkers (8) report the construction of the first fully viable animal model that is deficient in a recognized BER component, AAG, a DNA glycosylase that initiates BER by excising damaged bases from DNA. We first will define the steps and players of the BER pathway and then will discuss issues brought out by these new findings, ranging from the determinants of cell survival to the biological contributions of BER.
Table 1.
Summary of human BER genes/proteins and related mouse knockout studies*
| Repair protein | Function | Human gene/cDNA isolation (references) | Mouse gene knockout features |
|---|---|---|---|
| AAG | Alkyladenine DNA glycosylase | 38–40 | Normal development |
| UDG | Uracil DNA glycosylase | 41, 42 | |
| TDG | Thymine (T:G)-mismatch DNA glycosylase | 43 | |
| hMUTY | Glycosylase for A opposite 8-oxo-guanine | 44 | |
| hNTH1 | Thymine glycol DNA glycosylase | 45, 46 | |
| hOGG1 | 8-oxo-guanine DNA glycoslyase | 47–51 | |
| APE/HAP1 | AP endonuclease | 11–13 | Embryonic lethal E5 to E9.5‡; no cell lines reported |
| PARP | Poly(ADP-ribose) polymerase | 61 | Reduced litter size and adult weight; increased mouse lethality by MNU and γ-rays; increased SCEs and chromatid breaks by MNU and γ-rays in bone marrow; increased G2/M arrest of fibroblasts by MNU; increased apoptosis by MNU |
| Pol↠| DNA polymerase | 52 | Embryonic lethal midgestation, E10.5; cell lines established |
| FEN1 | 5′-flap endonuclease | 53–55 | |
| XRCC1 | Ligase III partner | 56 | Embryonic lethal ∼E6.5; cell lines established in p53− background |
| LIG1 | DNA ligase | 57, 58 | Embryonic lethal ∼E11.5; disruption of fetal liver erythropoiesis |
| LIG3 | DNA ligase | 59, 60 |
For simplification, genes for the PCNA-dependent polymerases Polδ/ɛ and accessory factors are not included.
The rat cDNA was cloned before the human.
Results from two laboratories (Xanthoudakis; D. Ludwig, personal communication)
An Overview of Mammalian BER
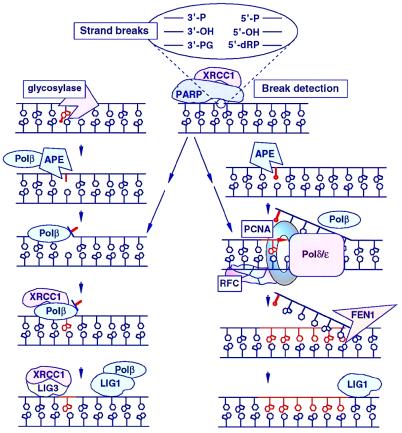
BER is a multistep process that involves the sequential activity of several proteins (see Table 1 and Fig. 1 for details). Typically, the repair sequence is initiated by a DNA glycosylase that recognizes and removes a damaged or improper base by hydrolyzing the N-glycosidic bond (reviewed in ref. 9). In humans, six DNA glycosylases have been identified (Table 1), and each excises an overlapping subset of deaminated (e.g., hypoxanthine), oxidized (e.g., 8-oxo-7,8-dihydroguanine), alkylated (e.g., 3-methyladenine, 3MeA), or mismatched (e.g., T:G) bases. The resulting abasic site is recognized by an AP endonuclease that incises the phosphodiester backbone immediately 5′ to the lesion, leaving behind a strand break with a normal 3′-hydroxyl group and an abnormal 5′-abasic terminus (reviewed in ref. 10). Humans possess a single major AP site-incision activity, the function of a protein with many aliases—APE, HAP1, APEX, and REF1 (11–14). The resulting 5′-abasic residue subsequently is removed, and the single nucleotide gap is filled (short-patch BER; Fig. 1, pathway on left), both of which are activities of DNA Polβ (15, 16). To complete this process, the nick is sealed by DNA ligase I (17) or a complex of XRCC1/DNA ligase III (18, 19). A similar short-patch BER process is present in prokaryotes as well (reviewed in ref. 20).
Figure 1.
Representation of BER. Subpathways involving single-nucleotide replacement (Left) or a “long-patch” replacement of less than 14 nucleotides (ref. 21; Right) are illustrated. Icons are defined in Table 1, and overlapping icons indicate reported interactions as referenced in text. The upper box depicts several possible termini that may arise from chemically induced or enzyme-catalyzed strand cleavage events. PG, phosphoglycolate; dRP, deoxyribose phosphate moiety.
An alternative BER pathway (Fig. 1, pathway on right) has been discovered in eukaryotes that involves the replacement of more than a single nucleotide (≈7 nucleotides) (21) and requires FEN1 to excise the flap-like structure produced by DNA polymerase strand displacement (22). This long-patch process may have evolved as a more efficient or redundant mechanism for the repair of 5′-termini (perhaps endonuclease-incised reduced or oxidized abasic sites) that are not substrates for the 5′-phosphodiesterase activity of Polβ (23, 24). The initial observation that this alternative pathway requires proliferating cell nuclear antigen (PCNA) suggested the involvement of Polɛ and Polδ in BER. However, it recently was shown in reconstitution assays that Polβ can carry out strand displacement and long-patch repair synthesis in vitro, and that PCNA functions to stimulate FEN1 endonuclease activity (22). Biochemical repair assays using crude cell extracts and PCNA-neutralizing antibodies also have found low levels of long-patch BER synthesis, supporting the idea of a Polβ-directed process (Y. Matsumoto, personal communication). Although the Polβ/FEN1 scenario may serve as a possible scheme for BER, it does not explain the PCNA-dependence originally described. The observations that Polβ (−/−) cells are fully proficient in repairing oxidative DNA damage (ref. 24; B. Rydberg and P. Cooper, personal communication) and that Polβ-neutralizing antibodies inhibit repair by only ≈70% (25) clearly demonstrate a role for other polymerases in this pathway as well. Taken together, these data suggest that long-patch BER be divided into two subpathways: (i) a PCNA-stimulated, Polβ-directed pathway, and (ii) a PCNA-dependent, Polδ/ɛ-directed pathway. As an added complexity, which will not be discussed here, it would appear that the substrate [i.e., the target damage or the form (circular or linear) of the DNA] also influences if short-patch or either of the long-patch repair pathways is used. What awaits in the wings is the determination of the overall contributions of the various BER pathways in vivo. The knockout animals and cell lines are a step in this direction.
The proteins of BER may act coordinately in complexes during the process as suggested by the interactions reported between APE and Polβ (26), Polβ and XRCC1 (27), XRCC1 and LIG3 (28), XRCC1 and PARP (29), and Polβ and LIG1 (17). PARP deserves attention as an innocent participant in BER (30) because of its reported interaction with XRCC1 (29), its role as a nick sensor (31), and because PARP knockouts show marked sensitivity to γ-rays and methylnitrosourea (MNU), agents that produce damages normally repaired by BER (ref. 7; Table 1).
Background on DNA Glycosylases
DNA glycosylases can be separated into two groups: those that possess only an N-glycosidic cleaving activity, and those that possess both an activity to remove substrate bases and an activity to incise the phosphodiester backbone immediately 3′ of the resulting AP site via a β-lyase mechanism (reviewed in ref. 9). The biological significance of the AP lyase activity, which produces a normal 5′-phosphate and an obstructive 3′-end (i.e., a 3′-deoxyribose moiety or a 3′-phosphate), is currently unclear. Furthermore, how, if at all, the type of initiating DNA glycosylase dictates downstream events during BER is unknown. It seems likely, however, that any glycosylase-initiated repair event would proceed through the short-patch pathway in which APE would act as the 3′-repair diesterase to remove the abnormal AP lyase-generated 3′-terminus before gap filling and ligation.
Engelward, Weeda, and colleagues (8) have genetically engineered animals deficient in AAG, a DNA glycosylase that removes a broad spectrum of base damages, including, but likely not limited to, 3MeA, 3-methylguanine, 7-methylguanine, 1,N6-ethenoadenine, hypoxanthine, and 8-oxo-7,8-dihydroguanine; AAG does not possess an AP lyase activity. It is worth mentioning that the mouse and human AAG proteins are only moderately conserved (≈80% identity at the amino acid level) and display some differences in their substrate preferences (32). Given this fact and considering the notable disparities that have been observed between certain repair-deficient mice and their counterpart human subjects, we must proceed with caution when interpreting data gathered from animal models. However, this caveat does not diminish the incredible wealth of information that is being obtained from these models (1).
The First Glycosylase-Deficient Animal Model
Protein extracts from tissues of AAG (−/−) animals display essentially no detectable repair activity for 3MeA, 1,N6-ethenoadenine, and hypoxanthine base modifications, although a hint of a minor lung-specific glycosylase activity for 1,N6-ethenoadenine lesions was reported (8). Furthermore, the knockout embryonic stem cells show hypersensitivity to a variety of alkylating agents and, surprisingly, to mitomycin C (33). Thus, AAG likely represents the major repair glycosylase for alkylation base damages, whereas its role in protection against mitomycin C is unclear. The finding that AAG-deficient animals survive embryogenesis raises several issues, particularly in light of the embryonic lethality of the other BER knockouts (Table 1).
First, the other BER components (i.e., APE, Polβ, XRCC1, and LIG1) may have roles outside of DNA repair that impact embryonic development. For instance, APE modulates the activity of several important transcription factors (its REF1 activity), such as Fos, Jun, and p53 (14, 34).
Second, the AAG (−/−) model may suggest that the base modifications that go unprocessed in these animals are less cytotoxic relative to AP sites or strand breaks (which can be formed by spontaneous, or AP lyase-catalyzed, β-elimination at AP sites, enzyme-mediated or free radical-induced cleavage, or by incomplete repair) that presumably accumulate in the other BER knockouts (Table 1). In other words, the base damages normally repaired by AAG accumulate but have no deleterious effect. This possibility seems unlikely because, for example, 3MeA has been shown to block DNA replication, and selective introduction of this lesion into chromosomal DNA with MeOSO2(CH2)2-lexitropsin induces killing of AAG-deficient cells (refs. 8 and 33 and references within).
Third, lethality may be dictated by the amount of DNA damage, which in normal cells would be influenced by the rates of formation and removal of the target lesion. Estimates for the spontaneous rates of formation of AP sites vs. 3MeA are ≈10,000 vs. ≈600, respectively, per cell per day (2). Therefore, from a quantitative standpoint, abasic sites should represent a greater burden than 3MeA lesions, and these numbers don’t take into account AP sites formed by DNA glycosylases or free radical-induced events. Thus, APE (−/−) cells would carry more damage than AAG (−/−) cells. For Polβ (−/−) and LIG1(−/−) embryos, we presume that all AP sites would be incised, resulting in a similar number of strand breaks in these embryos as compared with the number of AP sites in APE (−/−) embryos. If we imagine a threshold level of DNA damage that would induce cell death, this level would be attained in the APE and Polβ (−/−) embryos sooner than in AAG (−/−) embryos. However, embryonic death is likely to be influenced by more than just the amount of damage. This event is likely dictated by the response of the cell cycle checkpoint surveillance systems, which sense damage and mediate apoptosis. It will be interesting to learn more about cell cycle regulation during embryogenesis, which contains cell lineages undergoing extraordinarily rapid proliferation.
A fourth possibility for the survival of AAG (−/−) animals is that the rate of formation of the substrate bases in these mice is slower or equal to the rate of spontaneous loss of these bases in vivo, resulting in no significant accumulation of the lesions. The resulting AP sites would be repaired by the other BER components.
The fifth, and perhaps most likely, explanation for the survival of these animals is that one or more of the other DNA repair systems substitutes for AAG in its absence. There may, in fact, be a minor DNA glycosylase activity that can cope with the normal level of alkylation base damage, but that goes undetected in the repair assays used. The ability to cross different genetically engineered repair-defective backgrounds may uncover any potential overlap of the various corrective systems. For instance, if two repair systems possess redundancy for a common cytotoxic lesion, then breeding the appropriate repair-deficient animals would lead to embryonic lethality of the double knockout. Measuring the distribution of the repair patch lengths in AAG (−/−) also may provide clues as to which pathway is adopted.
We should note that the possibilities outlined above are not mutually exclusive.
Leaving Thoughts
In all, the generation of AAG-deficient animals provides a tremendous tool for investigating the biological contribution(s) of this repair enzyme and BER as a whole in relation to development, growth, cancer, aging, and disease. Specifically, having the knockout mice will permit measuring the in vivo levels of base damage over time, providing information about the steady-state levels. Particular attention should be paid to nondividing tissues where damage accumulation would be most acute. Moreover, continued monitoring of these animals will reveal if they develop a tissue-specific or age-dependent defect. It also will be interesting to see how these animals respond to DNA-damaging agents in terms of viability, development, and carcinogenesis. Although appearing relatively normal, PARP-deficient mice were abnormally sensitive to killing by γ-rays and by MNU, and in both the animal and culture, cells showed many traits consistent with a repair deficiency (7).
Forthcoming information from knockouts in other DNA glycosylases will determine if the characteristics observed with AAG (−/−) animals are the exception or a general phenomenon for DNA glycosylases. Deletion of OGG1 and NTH1 also may help elucidate the role of the AP lyase activity.
The potential to cross animals that are defective in different repair pathways, namely BER, nucleotide excision repair, and mismatch repair (35), will begin to unveil the relative contributions of the particular repair systems. The discovery that AAG (−/−) cells are hypersensitive to mitomycin C-induced DNA damage and the recent demonstrations that nucleotide excision repair corrects certain oxidative base damages (36, 37) indicates that we still have more to learn about the damage specificities of the various repair systems. Furthermore, as information on protein structure, catalytic mechanisms, and the interactive protein regions becomes available, we soon will be able to perform functional complementation studies with the repair-deficient cells to tease out the in vivo contribution of a particular protein component by using site-specific and deletion mutagenesis.
The distinction between embryonic vs. cellular lethality is noteworthy. Whereas several of the BER mouse knockouts lead to embryonic lethality (Table 1), the constraints on viability of cells in culture are considerably less demanding. Thus, investigators have been able to generate cell cultures from arresting (−/−) embryos of Polβ (24) and XRCC1 (R. Tebbs, J. Cleaver, R. Pedersen, and L. Thompson, unpublished results). The establishment of immortalized cultures from such mutant embryos likely involves the selective outgrowth of genetic variants that lose normal surveillance and regulatory systems that terminate embryonic growth in the face of excess DNA damage. Given the above cases of “life without repair,” what will be the outcome with APE (−/−) cells in culture?
Acknowledgments
We thank our colleagues for critical comments on the manuscript. This work was done under the auspices of the U.S. Department of Energy by the Lawrence Livermore National Laboratory under contract No. W-7405-ENG-48.
References
- 1.Friedberg E C, Meira L B, Cheo D L. Mutat Res. 1997;383:183–188. doi: 10.1016/s0921-8777(96)00057-2. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Xanthoudakis S, Smeyne R J, Wallace J D, Curran T. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu H, Marth J D, Orban P C, Mossmann H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 5.Tebbs, R. S., Meneses, J. J., Pedersen, R. A., Thompson, L. H. & Cleaver, J. E. (1996) Environ. Mol. Mutagen. 27, Suppl. 27, 68 (abstr.).
- 6.Bentley D, Selfridge J, Millar J K, Samuel K, Hole N, Ansell J D, Melton D W. Nat Genet. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- 7.Ménissier-de Murcia J, Niedergang C P, Trucco C, Ricoul M, Dutrillaux B, Oliver J, Masson M, Dierich A, LeMeur M, Walzinger C, Chambon P, de Murcia G. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelward B P, Weeda G, Wyatt M D, Broekhof J L M, de Wit J, Donker I, Allan J M, Gold B, Hoeijmakers J H J, Samson L D. Proc Natl Acad Sci USA. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krokan H E, Standal R, Slupphaug G. Biochem J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demple B, Harrison L. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 11.Demple B, Herman T, Chen D S. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robson C N, Hickson I D. Nucleic Acids Res. 1991;19:5519–5523. doi: 10.1093/nar/19.20.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seki S, Hatsushika M, Watanabe S, Akiyama K, Nagao K, Tsutsui K. Biochim Biophys Acta. 1992;1131:287–299. doi: 10.1016/0167-4781(92)90027-w. [DOI] [PubMed] [Google Scholar]
- 14.Xanthoudakis S, Miao G, Wang F, Pan Y C, Curran T. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto Y, Kim K. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 16.Singhal R K, Prasad R, Wilson S H. J Biol Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 17.Prasad R, Singhal R K, Srivastava D K, Molina J T, Tomkinson A E, Wilson S H. J Biol Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 18.Caldecott K W, McKeown C K, Tucker J D, Stanker L, Thompson L H. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. J Biol Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 20.Lindahl T, Karran P, Wood R D. Curr Opin Genet Dev. 1997;7:158–169. doi: 10.1016/s0959-437x(97)80124-4. [DOI] [PubMed] [Google Scholar]
- 21.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox L S, Lane D P, Abbondandolo A, Dogliotti E. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 22.Klungland A, Lindahl T. EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto Y, Kim K, Bogenhagen D F. Mol Cell Biol. 1994;14:6187–6197. doi: 10.1128/mcb.14.9.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobol R W, Horton J K, Kuhn R, Gu H, Singhal R K, Prasad R, Rajewsky K, Wilson S H. Nature (London) 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 25.Nealon K, Nicholl I D, Kenny M K. Nucleic Acids Res. 1996;24:3763–3770. doi: 10.1093/nar/24.19.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett R A O, Wilson D M, III, Wong D, Demple B. Proc Natl Acad Sci USA. 1997;94:7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubota Y, Nash R A, Klungland A, Schar P, Barnes D E, Lindahl T. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 28.Nash R A, Caldecott K W, Barnes D E, Lindahl T. Biochem. 1997;36:5207–5211. doi: 10.1021/bi962281m. [DOI] [PubMed] [Google Scholar]
- 29.Caldecott K W, Aoufouchi S, Johnson P, Shall S. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cleaver J E, Morgan W F. Mutat Res. 1991;257:1–18. doi: 10.1016/0165-1110(91)90016-o. [DOI] [PubMed] [Google Scholar]
- 31.de Murcia G, de Murcia J M. Trends Biochem Sci. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 32.Roy R, Kennel S J, Mitra S. Carcinogenesis. 1996;17:2177–2182. doi: 10.1093/carcin/17.10.2177. [DOI] [PubMed] [Google Scholar]
- 33.Engelward B P, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson L. EMBO J. 1996;15:945–952. [PMC free article] [PubMed] [Google Scholar]
- 34.Jayaraman L, Murthy K G, Zhu C, Curran T, Xanthoudakis S, Prives C. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 35.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 36.Satoh M S, Jones C J, Wood R D, Lindahl T. Proc Natl Acad Sci USA. 1993;90:6335–6339. doi: 10.1073/pnas.90.13.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reardon J T, Bessho T, Kung H C, Bolton P H, Sancar A. Proc Natl Acad Sci USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Connor T R, Laval F. EMBO J. 1990;9:3337–3342. doi: 10.1002/j.1460-2075.1990.tb07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samson L, Derfler B, Boosalis M, Call K. Proc Natl Acad Sci USA. 1991;88:9127–9131. doi: 10.1073/pnas.88.20.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chakravarti D, Ibeanu G C, Tano K, Mitra S. J Biol Chem. 1991;266:15710–15715. [PubMed] [Google Scholar]
- 41.Vollberg T M, Siegler K M, Cool B L, Sirover M A. Proc Natl Acad Sci USA. 1989;86:8693–8697. doi: 10.1073/pnas.86.22.8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen L C, Aasland R, Wittwer C U, Krokan H E, Helland D E. EMBO J. 1989;8:3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan J J, Wiebauer K, Jiricny J. J Biol Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 44.Slupska M M, Baikalov C, Luther W M, Chiang J H, Wei Y F, Miller J H. J Bacteriol. 1996;178:3885–3892. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aspinwall R, Rothwell D G, Roldan-Arjona T, Anselmino C, Ward C J, Cheadle J P, Sampson J R, Lindahl T, Harris P C, Hickson I D. Proc Natl Acad Sci USA. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hilbert T P, Chaung W, Boorstein R J, Cunningham R P, Teebor G W. J Biol Chem. 1997;272:6733–6740. doi: 10.1074/jbc.272.10.6733. [DOI] [PubMed] [Google Scholar]
- 47.Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kadama T, Takao M, Yasui A, Yamamoto K, Asano M, Fukasawa K, Yoshinari T, Inoue H, Ohtsuka E, Nishimura S. Can Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 48.Arai K, Mosrishita K, Shinmura K, Kohno T, Kim S-R, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 49.Radicella J P, Dherin C, Desmaze C, Fox M S, Boiteux S. Proc Natl Acad Sci USA. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roldán-Arjona T, Wei Y F, Carter K C, Klungland A, Anselmino C, Wang R P, Augustus M, Lindahl T. Proc Natl Acad Sci USA. 1997;94:8016–8060. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenquist T A, Zharkov D O, Grollman A P. Proc Natl Acad Sci USA. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zmudzka B Z, SenGupta D, Matsukage A, Cobianchi F, Kumar P, Wilson S H. Proc Natl Acad Sci USA. 1986;83:5106–5110. doi: 10.1073/pnas.83.14.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robins P, Pappin D J, Wood R D, Lindahl T. J Biol Chem. 1994;269:28535–28538. [PubMed] [Google Scholar]
- 54.Murray J M, Tavassoli M, al-Harithy R, Sheldrick K S, Lehmann A R, Carr A M, Watts F Z. Mol Cell Biol. 1994;14:4878–4888. doi: 10.1128/mcb.14.7.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harrington J J, Lieber M R. Genes Dev. 1994;8:1344–1355. doi: 10.1101/gad.8.11.1344. [DOI] [PubMed] [Google Scholar]
- 56.Thompson L H, Brookman K W, Jones N J, Allen S A, Carrano A V. Mol Cell Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barnes D E, Johnston L H, Kodama K, Tomkinson A E, Lasko D D, Lindahl T. Proc Natl Acad Sci USA. 1990;87:6679–6683. doi: 10.1073/pnas.87.17.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrini J H, Huwiler K G, Weaver D T. Proc Natl Acad Sci USA. 1991;88:7615–7619. doi: 10.1073/pnas.88.17.7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei Y F, Robins P, Carter K, Caldecott K, Pappin D J C, Yu G-L, Wang R P, Shell B K, Nash R A, Schär P, Barnes D E, Haseltine W A, Lindahl T. Mol Cell Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J, Tomkinson A E, Ramos W, Mackey Z B, Danehower S, Walter C A, Schultz R A, Besterman J M, Husain I. Mol Cell Biol. 1995;15:5412–5422. doi: 10.1128/mcb.15.10.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alkhatib H M, Chen D, Cherney B, Bhatia K, Notario V, Giri C, Stein G, Slattery E, Roeder R G, Smulson M E. Proc Natl Acad Sci USA. 1987;84:1224–1228. doi: 10.1073/pnas.84.5.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]