Abstract
Deletion of the transcriptional modulator Cited2 in the mouse results in embryonic lethality, cardiovascular malformations, adrenal agenesis, cranial ganglia fusion, exencephaly, and left–right patterning defects, all seen with a varying degree of penetrance. The phenotypic heterogeneity, observed on different genetic backgrounds, indicates the existence of both genetic and environmental modifiers. Mice lacking the LIM domain-containing protein Lmo4 share specific phenotypes with Cited2 null embryos, such as embryonic lethality, cranial ganglia fusion, and exencephaly. These shared phenotypes suggested that Lmo4 may be a potential genetic modifier of the Cited2 phenotype. Examination of Lmo4-deficient embryos revealed partially penetrant cardiovascular malformations and hypoplastic thymus. Examination of Lmo4;Cited2 compound mutants indicated that there is a genetic interaction between Cited2 and Lmo4 in control of thymus development. Our data suggest that this may occur, in part, through control of expression of a common target gene, Tbx1, which is necessary for normal thymus development. Developmental Dynamics 239:1988–1994, 2010. © 2010 Wiley-Liss, Inc.
Keywords: Cited2, Lmo4, thymus development, Tbx1, modifiers
INTRODUCTION
The transcription factor CITED2, which binds CREBBP with high affinity (Bhattacharya et al., 1999), acts as a co-factor for transcription factors such as TFAP2, LHX2, PPARA, and SMAD2/3 (Glenn and Maurer, 1999; Braganca et al., 2003; Tien et al., 2004; Chou and Yang, 2006), as well as acting as a repressor of hypoxia-activated transcription (Bhattacharya et al., 1999). Mice null for Cited2 die in utero from phenotypically heterogeneous cardiac malformations including atrial, ventricular, and atrioventricular septal defects (ASD, VSD, AVSD); outflow tract defects (double-outlet right ventricle [DORV], common arterial trunk [CAT], tetralogy of Fallot [TOF], transposition of great arteries [TGA]); and interrupted and right-sided aortic arch (Bamforth et al., 2001, 2004; Yin et al., 2002; Weninger et al., 2005). Loss of Cited2 also gives rise to exencephaly, cranial ganglia fusion, abnormal migration of cardiac neural crest cells, absent adrenal glands, and placental abnormalities (Bamforth et al., 2001, 2004; Barbera et al., 2002; Yin et al., 2002; Weninger et al., 2005; Withington et al., 2006; Val et al., 2007). On a congenic C57Bl/6J background, Cited2-null mice show left–right patterning defects characterized by right atrial and pulmonary isomerism, and abnormal ventricular topology (Bamforth et al., 2004; Weninger et al., 2005). We have recently shown that the phenotypically heterogenous and penetrant cardiac malformations in Cited2 deficiency arise from a primary requirement in epiblast derivatives for left–right patterning, with a secondary cell-autonomous role in the mesoderm (MacDonald et al., 2008).
Cited2 binds the LIM homeodomain transcription factor, Lhx2, by means of LIM domains (Glenn and Maurer, 1999). Proteins containing LIM domains function as transcriptional regulators, and this domain may act as a molecular adaptor for multiprotein complexes. The subfamily of LIM-only (LMO) proteins comprises four members (Lmo1–Lmo4). These non-DNA binding transcription factors may affect transcription by protein–protein interaction with DNA-binding transcription factors or chromatin modeling proteins (Hahm et al., 2004). Lmo4 comprises two zinc-binding LIM domains and is widely expressed in the developing embryo, including the pharyngeal arches, thymus, and neural crest (Grutz et al., 1998; Kenny et al., 1998; Sugihara et al., 1998). Mouse embryos lacking Lmo4 display a phenotype that overlaps with Cited2−/− embryos. This includes exencephaly, in utero or perinatal lethality, and cranial nerve fusions (Hahm et al., 2004; Tse et al., 2004; Lee et al., 2005).
The shared specific phenotypes observed in Cited2 and Lmo4 mutant mice suggest that there may be a genetic interaction. In this study, we show that embryos lacking Lmo4 have partially penetrant cardiac malformations including VSD, DORV, and right-sided aortic arch, which have not previously been reported. The shared specific phenotypes are further extended by demonstrating cervical fusions and abnormal thymus in Cited2 and in Lmo4 mutant embryos. The thymus is significantly smaller in compound mutant embryos, and completely absent in embryos lacking both Lmo4 and Cited2, indicating that there is a genetic interaction. Expression of Tbx1, which is important for thymus development, was significantly reduced in Lmo4 and Cited2 mutant embryos. This suggests a novel role for Cited2 and Lmo4 acting, likely in part through Tbx1, to control development of the thymus.
RESULTS
Cervical Vertebrae Abnormalities in Cited2−/− Embryos
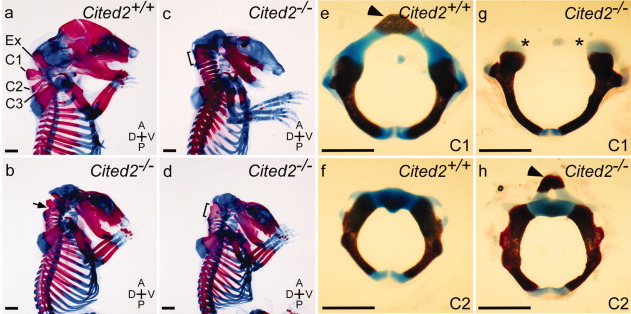
Lmo4−/− embryos show homeotic-like transformations in the rib cage and cervical vertebrae, although at low penetrance (Hahm et al., 2004). To investigate whether similar defects were present in the Cited2−/− embryos, skeletal preparations were made at embryonic day (E) 17.5. This revealed that Cited2−/− embryos (on a mixed genetic background) have fusions of the cervical vertebrae (Fig. 1). The cervical vertebrae may be completely fused (Fig. 1c,d) from the exoccipital bone through to the atlas (C1), axis (C2), and the third cervical vertebra (C3). Less severe fusions are also observed, which may even be asymmetric (Fig. 1c). The cervical skeleton from one of the less severely affected Cited2−/− embryos was disarticulated revealing an anterior transformation (Fig. 1g,h). The axis had a structure resembling the anterior arch of the atlas, and the atlas was missing this. The rib cages, however, were unaffected. Similar cervical vertebrae fusions were also observed in embryos on congenic 129Sv and C57Bl/6J genetic backgrounds (data not shown).
Figure 1.
Skeletal defects in embryonic day (E) 17.5 Cited2−/− embryos. a: Wild-type (Cited2+/+) embryo skeleton showing the normal appearance of the cervical vertebrae. b–d:Cited2−/− skeletons showing various degrees of fusion between the cervical vertebrae (indicated). All embryos depicted had exencephaly and are missing the occipital and frontal skull bones. e–h: Disarticulated cervical vertebrae from a Cited−/− skeleton with less severe fusions. e: Wild-type atlas, with anterior arch indicated (arrowhead). f: Wild-type axis. g:Cited−/− atlas which is missing the anterior arch (asterisks). h:Cited2−/− axis, which has a structure resembling the anterior arch of the atlas (arrowhead). Ex, exoccipital; C1, atlas; C2, axis. Scale bar = 1 mm.
Cardiovascular Defects and Thymus Hypoplasia in Lmo4−/− Embryos
Cardiovascular defects, a major aspect of the Cited2 phenotype, have not been reported in Lmo4 mutant mice. We therefore investigated whether the Lmo4−/− embryos suffered from any cardiac developmental abnormalities. Heterozygous Lmo4 mice (Tse et al., 2004) were intercrossed to analyze embryos at E15.5 by magnetic resonance imaging (MRI). Gross external analysis of Lmo4−/− embryos showed exencephaly (2/8; data not shown), as previously reported (Hahm et al., 2004; Tse et al., 2004; Lee et al., 2005), and edema (2/8; data not shown). Analysis by MRI revealed incomplete penetrance of cardiovascular defects in Lmo4−/− embryos, including VSD with DORV (2/8) and a right-sided aortic arch (1/8; Fig. 2).
Figure 2.
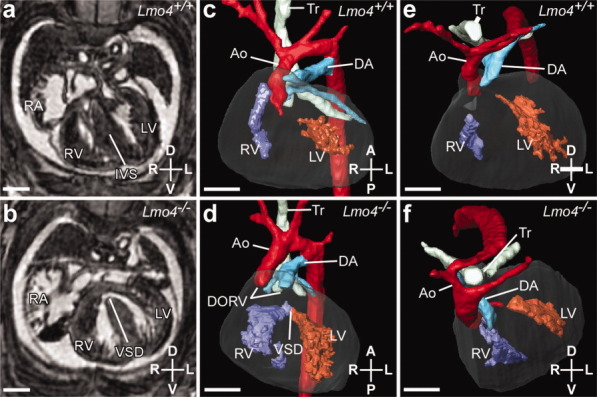
Cardiovascular defects observed in embryonic day (E) 15.5 Lmo4−/− embryos. a,b: Transverse magnetic resonance imaging sections of wild-type (a) and Lmo4−/− (b) embryo hearts. A ventricular septal defect (VSD) is visible in the Lmo4−/− heart. c–f: Three-dimensional reconstructions of embryo hearts and great vessels. c,d: Ventral views of wild-type embryo heart (c) showing normal topology, and Lmo4−/−embryo heart (d) showing a VSD and double-outlet right ventricle (DORV), where both the aorta (Ao) and ductus arteriosus (DA) arise from the right ventricle (RV). e,f: Anterior views of wild-type embryo heart (e) showing normal topology, and Lmo4−/−embryo heart (f) showing a right-sided aortic arch and ductus arteriosus. RA, right atrium; LA, left atrium; LV, left ventricle; IVS, intra-ventricular septum; PA, pulmonary arteries; Tr, trachea. Scale bar = 500 μm.
Lmo4−/− embryos also had consistently hypoplastic thymuses (8/8) compared with wild-type littermates which, in some cases, were malpositioned (3/8, Fig. 3b,d). Volume calculations showed a significant 2.3-fold decrease between wild-type (n = 6) and Lmo4−/−, thymuses (n = 8; P < 0.0005; Fig. 3e). Thymuses of Lmo4+/− littermates were not significantly different to those of wild-type littermates (n = 7; data not shown). Thymus size in Cited2−/− embryos at E15.5 was then assessed to see if they were affected. A significant 1.6-fold decrease in thymus volume was found (n = 6 wild-type, n = 6 Cited2−/−; P < 0.001; Fig. 3f). We also measured adrenal gland volume, because Cited2−/− embryos exhibit adrenal agenesis (Bamforth et al., 2001). However, we did not find any significant reduction in the size of adrenal glands in Lmo4−/− embryos compared with those of wild-type littermates (n = 6 for each genotype; data not shown).
Figure 3.
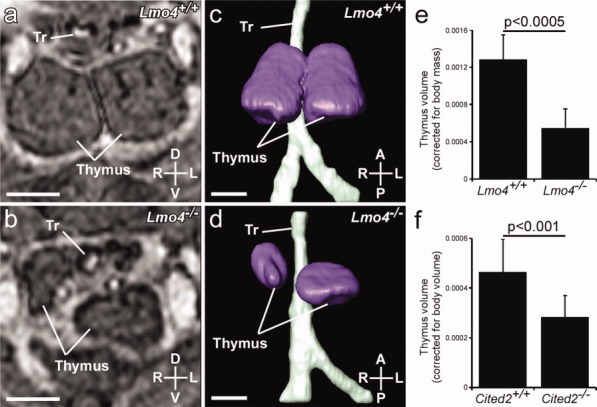
Thymus hypoplasia and malpositioning observed in embryonic day (E) 15.5 Lmo4−/− embryos. a,b: Transverse magnetic resonance imaging (MRI) sections of the thymus of wild-type (Lmo4+/+; a) and Lmo4−/− (b) littermate embryos. c,d: Three-dimensional (3D) reconstructions (ventral view) of thymus and trachea showing normal position and equal-sized lobes from wild-type (c) and hypoplasia, abnormal positioning and separation of the lobes in a Lmo4−/− embryo (d). e: Thymus volumes from Lmo4+/+ and Lmo4−/− embryos, corrected for embryo mass. f: Thymus volumes from Cited2+/+ and Cited2−/− embryos, corrected for whole embryo volume. (Graph values are means ± SD). Scale bar = 500 μm.
Cited2 and Lmo4 Genetically Interact In Vivo
As Cited2 and Lmo4 mutant mice share several specific phenotypes, we tested whether there was a genetic interaction in vivo by intercrossing Lmo4+/− mice with Cited2+/− mice (Supp. Table S1, which is available online). As Lmo4+/−;Cited2+/− mice were viable, we intercrossed them to produce compound and double knockout mice. Mice null for either Cited2 or Lmo4 were not present at weaning, as expected (Supp. Table S2). Embryos from Lmo4+/−;Cited2+/− intercrosses were collected at E15.5 and examined by MRI (n = 141). Supplementary Table S3 and Supp. Figures S1 and S2 summarize the defects observed in these embryos, the genotypes of which were all present in normal Mendelian ratios at this point in gestation. We observed no significant differences in the incidence of cardiovascular malformations in the compound heterozygous/knockout or double knockout embryos. Spina bifida was occasionally observed in embryos of the same genotype that exhibited exencephaly (Supp. Fig. 1), a phenotype not described before for either Lmo4 or Cited2 mutant embryos.
Although cardiovascular development did not seem to be affected by Lmo4 and Cited2 epistasis, the thymus was absent in Lmo4−/−;Cited2−/− embryos. Thymus defects were observed in most Lmo4;Cited2 compound mutant genotypes (Supp. Table S3), including thymus hypoplasia, separated thymus lobes, and one lobe absent (Fig. 4a,b). A single Lmo4−/−;Cited2+/− embryo was also lacking both thymus lobes. Thymuses from representative embryos of all genotypes were compared by volume (n = 5 each genotype; Fig. 4c). No significant difference was found between the thymuses of Lmo4+/−;Cited2+/+, Lmo4+/+;Cited2+/− or Lmo4+/−;Cited2+/− embryos when compared with thymuses from wild-type embryos. The thymuses of Lmo4−/−;Cited2+/+ (P = 0.0005) and Lmo4+/+;Cited2−/− (P = 0.0198) embryos were significantly reduced compared with those of wild-type embryos. Further loss of one copy of Lmo4 reduced the average thymus volume in Lmo4+/−;Cited2−/− embryos compared with Lmo4+/+;Cited2−/− embryos (P = 0.0187), but thymuses of Lmo4−/−;Cited2+/− embryos, when present, were not significantly smaller than in Lmo4−/−;Cited2+/+ embryos.
Figure 4.
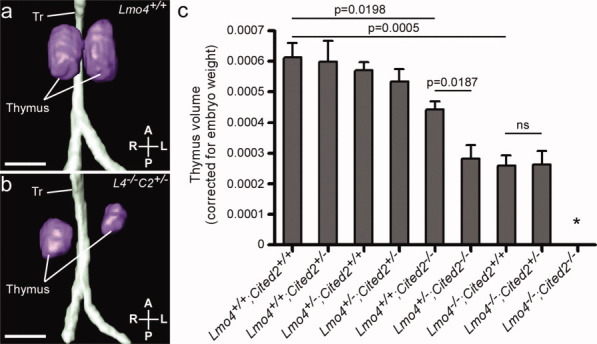
Impaired thymus development in Lmo4 and Cited2 compound mutant embryos. a,b: Three-dimensional (3D) reconstruction of thymuses from embryonic day (E) 15.5 embryos by MRI. a: Thymus and trachea from a wild-type (Lmo4+/+) embryo, showing normal position and equal-sized lobes. b:Lmo4−/−;Cited2+/− thymus illustrating hypoplasia, abnormal positioning, and separation of the lobes. c: Comparison of whole thymus volumes, adjusted for embryo weight, in embryos of all possible genotypes from Lmo4+/−;Cited2+/− intercrosses (Mean ± SEM). Lmo4−/−;Cited2−/− embryos do not have a thymus (*). Scale bar = 500 μm.
The complete lack of thymus in Lmo4−/−;Cited2−/− embryos is a novel phenotype, not previously seen in either individual knockout. This indicates that Cited2 and Lmo4 interact to control thymus development. To investigate whether there is a common target for Cited2 and Lmo4, embryos were harvested for quantitative reverse transcriptase-polymerase chain reaction (QRT-PCR) analysis. Pharyngeal arches and hearts were dissected from E10.5 Cited2−/− and Lmo4−/− embryos. Expression of Cited2 and Lmo4, as well as potential target candidate genes, Pitx2c and Tbx1, were investigated. Pitx2c was selected as it is a known target of Cited2 (Bamforth et al., 2004), and Tbx1 was selected as it has been implicated in thymus development (Jerome and Papaioannou, 2001).
Tbx1 Expression in Cited2−/− and Lmo4−/− Embryos
Gene expression levels were analyzed from the dissected hearts and pharyngeal arches of E10.5 embryos. Cited2−/− embryos showed a modest yet highly significant decrease in levels of Lmo4 mRNA when compared with wild-type littermates (1.35-fold decrease; P = 0.00006, n = 12 Cited2+/+ vs. n = 12 Cited2−/− embryos), but Lmo4−/− embryos did not show any difference in Cited2 levels (n = 12 Lmo4+/+ vs. n = 12 Lmo4−/− embryos; Fig. 5a,b), indicating that Lmo4 is downstream of Cited2. As we have previously shown (MacDonald et al., 2008), Cited2−/− embryos have lower levels of Pitx2c mRNA, compared with wild-type littermates (1.43-fold decrease, P = 0.01; Fig. 5c), but Lmo4−/− embryos did not show any difference in Pitx2c levels (n = 12 embryos for each genotype tested; Fig. 5d). Tbx1 levels were then investigated in the dissected pharyngeal arch regions from E10.5 Cited2−/− and Lmo4−/−embryos. A significant decrease in Tbx1 levels was found in both Cited2−/− (1.46-fold decrease, p = 0.009) and Lmo4−/− (2.3-fold decrease; P = 0.018, n = 12 embryos for each genotype tested; Fig. 5e,f). This suggests that Tbx1 could be a common target for Cited2 and Lmo4 in the developing pharyngeal arches.
Figure 5.
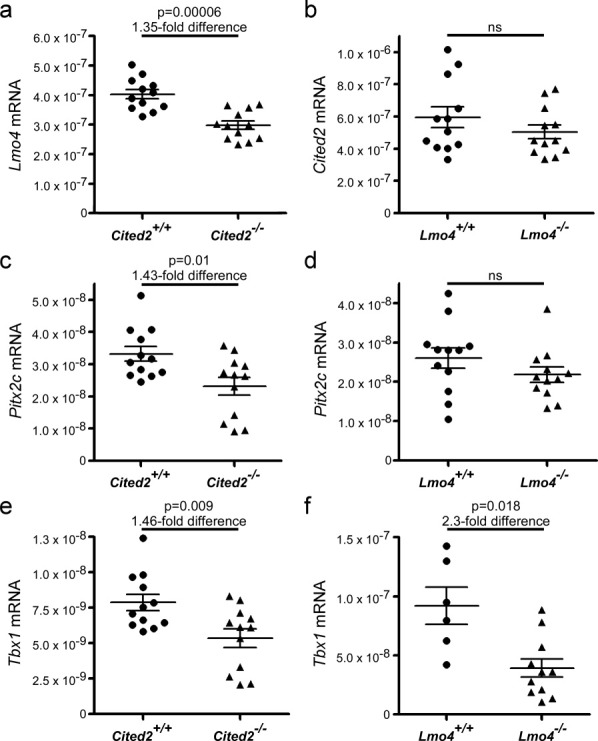
mRNA expression in embryos lacking Cited2 or Lmo4. Quantitative reverse transcriptase-polymerase chain reaction was conducted on pharyngeal arches and hearts harvested from embryonic day (E) 10.5 Cited2+/+, Cited2−/−, Lmo4+/+ and Lmo4−/− embryos. a:Lmo4 expression is significantly decreased in Cited2−/− embryos. b:Cited2 expression was not affected in Lmo4−/− embryos. c,d:Pitx2c expression was significantly decreased in Cited2−/− embryos (c) but was not affected in Lmo4−/− embryos (d). e,f:Tbx1 expression was reduced in Cited2−/− embryos (e) and in Lmo4−/− embryos (f). Mean ± SEM are indicated.
DISCUSSION
Genetic interactions can be identified through screens testing pairwise combinations of mutant genes that either suppress or enhance the original mutant phenotype (Hartman et al., 2001). In pairwise intervention experiments, the expected outcome of a double-intervention is the product of the two single-intervention effects; departure from this model indicates a functional relationship (i.e., either an aggravating or an alleviating effect, indicating compensatory function or gene-action in series, respectively) between the two target genes (St Onge et al., 2007). This approach has been used in Drosophila melanogaster and Caenorhabditis elegans to identify genes acting in the same pathway (Lehner et al., 2006; Sambandan et al., 2008). Also, genes that share similar or identical GO term annotations are more likely to interact genetically, as shown by the analysis of thousands of mutant Saccharomyces cerevisiae yeast strains (Wong et al., 2004). This is also true for genes that encode proteins found in the same subcellular location, or that act in the same protein complex. Another excellent predictor of a genetic interaction is a shared specific mutant phenotype (Tong et al., 2004). Mouse embryos lacking Cited2 or Lmo4 share specific phenotypes, both displaying exencephaly and cardiovascular, thymus, skeletal, and cranial ganglia defects. Experiments to identify a genetic interaction revealed that Lmo4−/−;Cited2−/− embryos completely lacked a thymus, a novel phenotype, not observed in either parental line. However, there was no significant increase in penetrance of cardiac malformation in Lmo4−/−;Cited2−/− embryos.
As discussed by Wong et al., a genetic interaction between two genes is indicated when the phenotype observed is more pronounced than the phenotype produced by the genes individually (Wong et al., 2004). Having re-analyzed previously published studies, Mani et al. (2008) concluded that the expected phenotype in offspring should be the product of the parental fitness, and not the average of parental fitness or that of the least fit parent as previously thought (Tong et al., 2001, 2004). This allows for greater expected deviation from parental fitness in offspring. For instance, if one parent had a fitness of 0.6 and one a fitness of 0.8, the expected fitness of the offspring is not 0.7 (average) or 0.6 (least fit parent), but 0.48 (product). Thus, applying this principle to these results, where Lmo4−/− embryos had thymuses 0.46 times the size of wild-type thymuses, and Cited2−/− thymuses were 0.78 times the size of wild-type thymuses, the expected thymus size of Lmo4−/−;Cited2−/− embryos should be 0.36 times the size of those in wild-type embryos. However, a total lack of thymus was discovered, dramatically exceeding the expected decrease. This indicates that a genetic interaction between Lmo4 and Cited2 is necessary for thymus development, suggesting that the genes act together in a common developmental process or molecular target.
Pitx2c is a known target of Cited2, but QRT-PCR data showed that it is not a target of Lmo4. Nor is Cited2 a target of Lmo4, although it appears that Lmo4 could be a target of Cited2. The data also revealed that Tbx1 expression levels are reduced in response to loss of both Cited2 and Lmo4 expression. This could indicate that Tbx1 is a common target of Cited2 and Lmo4, all of which are necessary for normal thymus formation. Indeed, Ivins et al. identified a reduction in Lmo4 expression in embryos lacking Tbx1 expression at E9.5 (Ivins et al., 2005). Tbx1 is expressed in the developing pharyngeal endoderm, which contributes to thymus formation. Absent thymus is a feature of DiGeorge / 22q11 deletion syndrome (Lischner, 1972; de la Chapelle et al., 1981), for which the majority of phenotypes observed are largely attributed to haploinsufficiency of TBX1 during development (Chieffo et al., 1997; Jerome and Papaioannou, 2001; Lindsay and Baldini, 2001). Moreover, in the mouse, morphology of the thymus is sensitive to Tbx1 dosage (Zhang and Baldini, 2007). Lmo4 is strongly expressed in the pharyngeal arches during mouse embryonic development (Hahm et al., 2004), specifically in the migratory cranial neural crest, and highly expressed in proliferating T lymphocytes in adult thymus (Kenny et al., 1998). Cited2 is also strongly expressed in the pharyngeal arches during development, in endoderm, mesenchyme and ectoderm (Weninger et al., 2005; MacDonald et al., 2008), and it is the third pharyngeal pouch from which the thymus is derived. The expression of Tbx1 in the fourth pharyngeal arch is also important for normal formation of the fourth pharyngeal arch arteries (Lischner, 1972; Jerome and Papaioannou, 2001), the development of which are affected in embryos lacking Cited2 and/or Lmo4.
Our data show that Lmo4−/− embryos have incomplete penetrance of cardiovascular defects in addition to those phenotypes already known, which has not been previously reported. Thymus defects have also not been previously reported and were detected by MRI, despite Tse et al. (2004) noting a normal cellular architecture of the thymus from histological sections, and no hypoplasia was recorded (Tse et al., 2004). Adding cardiovascular and thymus defects to the list of similarities between the recorded phenotypes of both Cited2 and Lmo4 knockout mice gave strong grounds for a physical and genetic interaction between Cited2 and Lmo4. Indeed, using GST pull-down assays, we were able to show that LMO4 physically interacts with residues 123–161 of CITED2, although we were unable to co-immunoprecipitate Lmo4 and Cited2 (data not shown).
To fully understand the genetic network in which Cited2, Lmo4, and Tbx1 are potentially interacting to control thymus development, further experiments will need to be performed. Although QRT-PCR data gives a quantitative measure of altered expression, qualitative assays will be needed to complement this, for example, by using in situ hybridization techniques to investigate whether the expression of Tbx1 is reduced specifically in the endoderm as may be expected. A more detailed investigation of candidate genes in wild-type and double heterozygote embryos and double homozygous null embryos will also need to be carried out, looking at the expression profile of genes specifically expressed in thymus such as Foxn1 (Nehls et al., 1996), and those expressed in the pharyngeal arches and known to affect thymus development, for example, Pax9, Eya1, and Six1 (Peters et al., 1998; Xu et al., 2002; Zou et al., 2006).
To summarize, in this study we have shown that mice lacking Lmo4 exhibit cardiovascular and thymus defects, which are novel phenotypes and add Lmo4 to the genetic network involved in cardiovascular development. Lmo4 interacts genetically with Cited2 in vivo to control thymus development, possibly through a common target gene in Tbx1.
EXPERIMENTAL PROCEDURES
Mice
Cited2+/− mice (Cited2tm1Bha) on a mixed or congenic C57BL/6J background (Bamforth et al., 2001, 2004) and Lmo4+/− mice (gift from T. Rabbitts, LMB Cambridge; Tse et al., 2004) were used in this study. Embryos were harvested at the indicated time points after detection of a vaginal plug (E0.5), and genotyped using allele-specific PCR (primer details are available on request). All animal experimentation was performed under UK Home Office authorization and regulations.
Imaging
Magnetic resonance imaging was performed on a horizontal 9.4T/21cm VNMRS Direct Drive MR system (Varian Inc., Palo Alto) essentially as described previously (Schneider et al., 2004) on embryos at E15.5. Skeletons were prepared and stained with alizarin red (bone) and Alcian blue (cartilage) by standard methods.
QRT-PCR
The hearts and pharyngeal arch regions were dissected free from E10.5 embryos as described (Prescott et al., 2005). RNA isolation, cDNA synthesis, and quantitative RT-PCR (QRT-PCR) reactions were carried out as described (MacDonald et al., 2008) using preoptimized TaqMan primer-probe sets from Applied Biosytems (Mus musculus assays Mm00516121_m1 (NM_010828 Cited2), Mm00495373_m1 (NM_010723 Lmo4), Mm00440826_m1 (NM_001042502 Pitx2c), Mm00448948_m1 (NM_011532 Tbx1), and eukaryotic 18S rRNA. Expression levels were normalized to 18S rRNA using the R0 method of analysis (Peirson et al., 2003).
Statistical Analyses
The chi-squared test was used to calculate the deviation from expected Mendelian ratios. QRT-PCR data was analyzed using a two-tailed, two-sample t-test assuming unequal variance.
Acknowledgments
We thank T. Rabbitts for Lmo4 mice. A.C.M is the recipient of a British Heart Foundation DPhil Studentship. S.B. is supported by a British Heart Foundation Chair Award [CH/09/003]. This work was funded by Wellcome Trust Program Grant Award [083228] to S.B., and by a Wellcome Trust Core Grant Award [075491/Z/04].
Supplementary material
Additional Supporting Information may be found in the online version of this article.
Supp. Fig. S1. Neural tube, cleft palate, and cardiac defects observed in mutant embryos. Embryos at embryonic day (E) 15.5 generated by intercrossing Lmo4+/−;Cited2+/− mice were examined. a: Normal wild-type embryo. b: Lmo4−/−;Cited2+/− embryo with exencephaly (Ex) and spina bifida (SB). Scale bar = 1 mm. c–l: Two-dimensional images generated by magnetic resonance imaging. c,d: Sagittal sections through wild-type and Lmo4−/−;Cited2+/− embryos. c: Wild-type embryo with complete palate (P) over the tongue (T). d: Lmo4−/−;Cited2+/− embryo with cleft palate (cP) and edema (E). e,f: Coronal sections of a normal wild-type (e) and abnormal Lmo4−/−;Cited2−/− (f) heart. g,h: Coronal sections of organs from normal wild-type (g) and abnormal Lmo4+/−;Cited2−/− (h) embryo. The mutant displays right pulmonary isomerism and right-sided stomach. i–l: Transverse sections of hearts. i: Normal wild-type heart. j: Lmo4+/−;Cited2−/− heart with a common atrium (A) and a primum atrial septal defect (ASDP). A common atrioventricular valve (AVV) opens into both ventricles. k: Lmo4+/−;Cited2−/− heart with a common atrium (A) and heart apex pointing to the right. l: Lmo4−/−;Cited2+/+ heart positioned severely to the left. IVS, interventricular septum; LV, left ventricle; MV, mitral valve; PAS, primary atrial septum; RV, right ventricle; TV, tricuspid valve; WT, wild-type. Scale bar = 500 μm.
Supp. Fig. S2. Cardiovascular defects observed in mutant embryos. Embryos at embryonic day (E) 15.5, generated by intercrossing Lmo4+/−;Cited2+/− mice, were examined by magnetic resonance imaging (MRI), and three-dimensional (3D) reconstructions were made. a–f: Ventral views. a: Wild-type (WT) heart showing normal topology. b: Lmo4−/−;Cited2+/− heart showing a ventricular septal defect (VSD) and double outlet right ventricle (DORV). The ductus arteriosus is right-sided. c: Lmo4−/−;Cited2+/− heart showing right-sided aortic arch (RAA) and ductus arteriosus (RDA), and a mirror image arrangement of the pharyngeal arch arteries. The heart is twisted severely to the left. d: Lmo4+/−;Cited2−/− heart showing an interrupted aortic arch (IAA) and DORV, with an aberrant right subclavian artery (ARSA). e: Lmo4−/−;Cited2−/− heart with a common arterial trunk (CAT). f: Lmo4+/−;Cited2−/− heart with CAT, RAA, and VSD. g,h: Dorsal views. g: Wild-type (WT) heart showing normal topology of venous return to the heart. h: Lmo4+/−;Cited2−/− heart showing a common atrium (A) with bilateral systemic venous sinuses (LSVS, RSVS) into which drain bilateral superior and inferior caval veins. Scale bar = 500 μm.
REFERENCES
- Bamforth SD, Braganca J, Eloranta JJ, Murdoch JN, Marques FI, Kranc KR, Farza H, Henderson DJ, Hurst HC, Bhattacharya S. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat Genet. 2001;29:469–474. doi: 10.1038/ng768. [DOI] [PubMed] [Google Scholar]
- Bamforth SD, Braganca J, Farthing CR, Schneider JE, Broadbent C, Michell AC, Clarke K, Neubauer S, Norris D, Brown NA, Anderson RH, Bhattacharya S. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat Genet. 2004;36:1189–1196. doi: 10.1038/ng1446. [DOI] [PubMed] [Google Scholar]
- Barbera JP, Rodriguez TA, Greene ND, Weninger WJ, Simeone A, Copp AJ, Beddington RS, Dunwoodie S. Folic acid prevents exencephaly in Cited2 deficient mice. Hum Mol Genet. 2002;11:283–293. doi: 10.1093/hmg/11.3.283. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Michels CL, Leung MK, Arany ZP, Kung AL, Livingston DM. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braganca J, Eloranta JJ, Bamforth SD, Ibbitt JC, Hurst HC, Bhattacharya S. Physical and functional interactions among AP-2 transcription factors, p300/CREB-binding protein, and CITED2. J Biol Chem. 2003;278:16021–16029. doi: 10.1074/jbc.M208144200. [DOI] [PubMed] [Google Scholar]
- Chieffo C, Garvey N, Gong W, Roe B, Zhang G, Silver L, Emanuel BS, Budarf ML. Isolation and characterization of a gene from the DiGeorge chromosomal region homologous to the mouse Tbx1 gene. Genomics. 1997;43:267–277. doi: 10.1006/geno.1997.4829. [DOI] [PubMed] [Google Scholar]
- Chou YT, Yang YC. Post-transcriptional control of Cited2 by transforming growth factor beta. Regulation via Smads and Cited2 coding region. J Biol Chem. 2006;281:18451–18462. doi: 10.1074/jbc.M601720200. [DOI] [PubMed] [Google Scholar]
- de la Chapelle A, Herva R, Koivisto M, Aula P. A deletion in chromosome 22 can cause DiGeorge syndrome. Hum Genet. 1981;57:253–256. doi: 10.1007/BF00278938. [DOI] [PubMed] [Google Scholar]
- Glenn DJ, Maurer RA. MRG1 binds to the LIM domain of Lhx2 and may function as a coactivator to stimulate glycoprotein hormone alpha-subunit gene expression. J Biol Chem. 1999;274:36159–36167. doi: 10.1074/jbc.274.51.36159. [DOI] [PubMed] [Google Scholar]
- Grutz G, Forster A, Rabbitts TH. Identification of the LMO4 gene encoding an interaction partner of the LIM-binding protein LDB1/NLI1: a candidate for displacement by LMO proteins in T cell acute leukaemia. Oncogene. 1998;17:2799–2803. doi: 10.1038/sj.onc.1202502. [DOI] [PubMed] [Google Scholar]
- Hahm K, Sum EY, Fujiwara Y, Lindeman GJ, Visvader JE, Orkin SH. Defective neural tube closure and anteroposterior patterning in mice lacking the LIM protein LMO4 or its interacting partner Deaf-1. Mol Cell Biol. 2004;24:2074–2082. doi: 10.1128/MCB.24.5.2074-2082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman JL, IV, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- Ivins S, Lammerts van Beuren K, Roberts C, James C, Lindsay E, Baldini A, Ataliotis P, Scambler PJ. Microarray analysis detects differentially expressed genes in the pharyngeal region of mice lacking Tbx1. Dev Biol. 2005;285:554–569. doi: 10.1016/j.ydbio.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Kenny DA, Jurata LW, Saga Y, Gill GN. Identification and characterization of LMO4, an LMO gene with a novel pattern of expression during embryogenesis. Proc Natl Acad Sci U S A. 1998;95:11257–11262. doi: 10.1073/pnas.95.19.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Jurata LW, Nowak R, Lettieri K, Kenny DA, Pfaff SL, Gill GN. The LIM domain-only protein LMO4 is required for neural tube closure. Mol Cell Neurosci. 2005;28:205–214. doi: 10.1016/j.mcn.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Baldini A. Recovery from arterial growth delay reduces penetrance of cardiovascular defects in mice deleted for the DiGeorge syndrome region. Hum Mol Genet. 2001;10:997–1002. doi: 10.1093/hmg/10.9.997. [DOI] [PubMed] [Google Scholar]
- Lischner HW. DiGeorge syndrome(s) J Pediatr. 1972;81:1042–1044. doi: 10.1016/s0022-3476(72)80575-4. [DOI] [PubMed] [Google Scholar]
- MacDonald ST, Bamforth SD, Chen CM, Farthing CR, Franklyn A, Broadbent C, Schneider JE, Saga Y, Lewandoski M, Bhattacharya S. Epiblastic Cited2 deficiency results in cardiac phenotypic heterogeneity and provides a mechanism for haploinsufficiency. Cardiovasc Res. 2008;79:448–457. doi: 10.1093/cvr/cvn101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani R, St. Onge RP, Hartman JL, Giaever G, Roth FP. Defining genetic interaction. Proc Natl Acad Sci U S A. 2008;105:3461–3466. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls M, Kyewski B, Messerle M, Waldschutz R, Schuddekopf K, Smith AJ, Boehm T. Two genetically separable steps in the differentiation of thymic epithelium. Science. 1996;272:886–889. doi: 10.1126/science.272.5263.886. [DOI] [PubMed] [Google Scholar]
- Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott K, Ivins S, Hubank M, Lindsay E, Baldini A, Scambler P. Microarray analysis of the Df1 mouse model of the 22q11 deletion syndrome. Hum Genet. 2005;116:486–496. doi: 10.1007/s00439-005-1274-3. [DOI] [PubMed] [Google Scholar]
- Sambandan D, Carbone MA, Anholt RRH, Mackay TFC. Phenotypic plasticity and genotype by environment interaction for olfactory behavior in Drosophila melanogaster. Genetics. 2008;179:1079–1088. doi: 10.1534/genetics.108.086769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JE, Bose J, Bamforth SD, Gruber AD, Broadbent C, Clarke K, Neubauer S, Lengeling A, Bhattacharya S. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev Biol. 2004;4:16. doi: 10.1186/1471-213X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara TM, Bach I, Kioussi C, Rosenfeld MG, Andersen B. Mouse deformed epidermal autoregulatory factor 1 recruits a LIM domain factor, LMO-4, and CLIM coregulators. Proc Natl Acad Sci U S A. 1998;95:15418–15423. doi: 10.1073/pnas.95.26.15418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien ES, Davis JW, Vanden Heuvel JP. Identification of the CREB-binding protein/p300-interacting protein CITED2 as a peroxisome proliferator-activated receptor alpha coregulator. J Biol Chem. 2004;279:24053–24063. doi: 10.1074/jbc.M401489200. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tse E, Smith AJ, Hunt S, Lavenir I, Forster A, Warren AJ, Grutz G, Foroni L, Carlton MB, Colledge WH, Boehm T, Rabbitts TH. Null mutation of the Lmo4 gene or a combined null mutation of the Lmo1/Lmo3 genes causes perinatal lethality, and Lmo4 controls neural tube development in mice. Mol Cell Biol. 2004;24:2063–2073. doi: 10.1128/MCB.24.5.2063-2073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Val P, Martinez-Barbera JP, Swain A. Adrenal development is initiated by Cited2 and Wt1 through modulation of Sf-1 dosage. Development. 2007;134:2349–2358. doi: 10.1242/dev.004390. [DOI] [PubMed] [Google Scholar]
- Weninger WJ, Floro KL, Bennett MB, Withington SL, Preis JI, Barbera JP, Mohun TJ, Dunwoodie SL. Cited2 is required both for heart morphogenesis and establishment of the left-right axis in mouse development. Development. 2005;132:1337–1348. doi: 10.1242/dev.01696. [DOI] [PubMed] [Google Scholar]
- Withington SL, Scott AN, Saunders DN, Lopes Floro K, Preis JI, Michalicek J, Maclean K, Sparrow DB, Barbera JP, Dunwoodie SL. Loss of Cited2 affects trophoblast formation and vascularization of the mouse placenta. Dev Biol. 2006;294:67–82. doi: 10.1016/j.ydbio.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Wong SL, Zhang LV, Tong AH, Li Z, Goldberg DS, King OD, Lesage G, Vidal M, Andrews B, Bussey H, Boone C, Roth FP. Combining biological networks to predict genetic interactions. Proc Natl Acad Sci U S A. 2004;101:15682–15687. doi: 10.1073/pnas.0406614101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Zheng W, Laclef C, Maire P, Maas RL, Peters H, Xu X. Eya1 is required for the morphogenesis of mammalian thymus, parathyroid and thyroid. Development. 2002;129:3033–3044. doi: 10.1242/dev.129.13.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Haynie J, Yang X, Han B, Kiatchoosakun S, Restivo J, Yuan S, Prabhakar NR, Herrup K, Conlon RA, Hoit BD, Watanabe M, Yang YC. The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc Natl Acad Sci U S A. 2002;99:10488–10493. doi: 10.1073/pnas.162371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Baldini A. In vivo response to high-resolution variation of Tbx1 mRNA dosage. Hum Mol Genet. 2007;17:150–157. doi: 10.1093/hmg/ddm291. [DOI] [PubMed] [Google Scholar]
- Zou D, Silvius D, Davenport J, Grifone R, Maire P, Xu PX. Patterning of the third pharyngeal pouch into thymus/parathyroid by Six and Eya1. Dev Biol. 2006;293:499–512. doi: 10.1016/j.ydbio.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.