Abstract
Type I (e.g., IFN-α, IFN-β) and type II IFNs (IFN-γ) have antiviral, antiproliferative, and immunomodulatory properties. Both types of IFN signal through the Jak/STAT pathway to elicit antiviral activity, yet IFN-γ is thought to do so only through STAT1 homodimers, whereas type I IFNs activate both STAT1- and STAT2-containing complexes such as IFN-stimulated gene factor 3. In this study, we show that IFN-stimulated gene factor 3 containing unphosphorylated STAT2 (ISGF3II) also plays a role in IFN-γ–mediated antiviral activity in humans. Using phosphorylated STAT1 as a marker for IFN signaling, Western blot analysis of IFN-α2a–treated human A549 cells revealed that phospho-STAT1 (Y701) levels peaked at 1 h, decreased by 6 h, and remained at low levels for up to 48 h. Cells treated with IFN-γ showed a biphasic phospho-STAT1 response with an early peak at 1–2 h and a second peak at 15–24 h. Gene expression microarray following IFN-γ treatment for 24 h indicated an induction of antiviral genes that are induced by IFN-stimulated gene factor 3 and associated with a type I IFN response. Induction of these genes by autocrine type I and type III IFN signaling was ruled out using neutralizing Abs to these IFNs in biological assays and by quantitative RT-PCR. Despite the absence of autocrine IFNs, IFN-γ treatment induced formation of ISGF3II. This novel transcription factor complex binds to IFN-stimulated response element promoter sequences, as shown by chromatin immunoprecipitation analysis of the protein kinase R promoter. STAT2 and IFN regulatory factor 9 knockdown in A549 cells reversed IFN-γ–mediated IFN-stimulated response element induction and antiviral activity, implicating ISGF3II formation as a significant component of the cellular response and biological activity of IFN-γ.
Interferons are members of a family of cytokines that have antiviral, antiproliferative, and immunomodulatory properties (1). There are several types of IFNs, each of which interacts with a type-specific receptor complex. Type I IFNs, which include IFN-α, IFN-β, and IFN-ω, are ubiquitously expressed in mammals and interact with the IFN-α receptor (IFNAR) subunits 1 and 2 (2). Activated T lymphocytes, monocytes, and NK cells produce the single species of type II IFN (IFN-γ), which interacts with the IFN-γ receptor (IFNGR) subunits 1 and 2. Nearly every cell type expresses receptors for type I IFNs and IFN-γ (3, 4). The recently characterized type III IFNs include IFN-λ1 (IL-29), IFN-λ2 (IL-28A), and IFN-λ3 (IL-28B), which bind to the IFN-λ receptor (IFNLR1) and the IL-10Rβ subunit (IL-10Rβ). All IFNs exhibit species specificity (2, 5).
Each IFN initiates a biological response by binding to its cognate cellular receptor and activating the Jak/STAT pathway. Once bound, IFN-γ activates, by phosphorylation, Jak1 and Jak2, whereas IFN-α binding results in phosphorylation of Jak1 and Tyk2 (6, 7). Type III IFNs are also thought to activate Jak1 and Tyk2 (8). Subsequently, the activated protein kinases recruit and phosphorylate one or more of the cytoplasmic STAT proteins, which will then dimerize to form transcription factor complexes (4, 9–11).
The major transcription factor formed after IFN-γ stimulation, and to a lesser degree in response to type I IFNs, is a STAT1 homodimer (2). This complex, termed the γ activation factor/α activation factor, activates IFN-stimulated genes (ISGs) containing γ activation site promoter elements, including IFN regulatory factor 1 (IRF1) and guanylate-binding protein 1 (GBP1) (12–14). In contrast, the major complex formed after type I and type III IFN stimulation is ISG factor 3 (ISGF3), which is a heterotrimer composed of phosphorylated STAT1 and STAT2, and a third component, IRF9 (ISGF3γ/p48) (8, 15–17). ISGF3 binds to DNA containing IFN-stimulated response element (ISRE) promoter elements and stimulates transcription of ISGs such as 2′,5′-oligoadenylate synthetase 1 (OAS1), protein kinase R (PKR), myxovirus resistance protein A (MxA), and IRF7 (2, 18, 19).
Although the primary function of IFN-γ is modulating the immune response, it also has direct antiviral properties (3, 4, 20). However, most of the classical antiviral genes contain ISRE promoter motifs and are regulated through ISGF3 (21, 22). Several studies demonstrated ISGF3 complex activation following IFN-γ treatment in murine cells (23–25), but there is currently no evidence of this phenomenon in human cells. In this study, we provide evidence of the ISGF3 containing unphosphorylated STAT2 (ISGF3II) complex in human A549 cells after treatment with IFN-γ. Moreover, we provide evidence for the necessity of this transcription factor in IFN-γ–mediated antiviral activity.
Materials and Methods
Cell culture materials, viruses, neutralizing Abs, and IFNs
A549 human lung epithelial cells were obtained from American Type Culture Collection (Manassas, VA), maintained in RPMI 1640 (Invitrogen, Carlsbad, CA), and supplemented with 10% FBS (Invitrogen), 2 mM l-glutamine (Invitrogen), 50 U/ml penicillin G, and 50 µg/ml streptomycin (Invitrogen) at 37°C and 5% CO2 (complete RPMI 1640). IFN-α2a was obtained from Hoffman La Roche (Nutley, NJ), and human rIFN-γ was obtained from Genentech (South San Francisco, CA). Encephalomyocarditis virus (EMCV) was obtained from American Type Culture Collection, grown in murine-derived L929 cells (American Type Culture Collection), and its titer was determined by plaque assays on A549 cells. The neutralizing murine mAb (A10) for IFNAR2 was raised against rIFNAR2 extracellular domain by A&G Pharmaceutical (Columbia, MD), and the neutralizing mouse mAb for IFNGR1 was obtained from Santa Cruz Biotechnology (Z0-14; Santa Cruz, CA).
Western blot analysis
Three million A549 cells were seeded overnight in 10 ml complete RPMI 1640, treated as indicated, and harvested at the indicated times by trypsin/EDTA. Cells were lysed in mammalian protein extraction reagent lysis buffer with Halt protease inhibitor mixture and Halt phosphatase inhibitor mixtures (Thermo Fisher Scientific, Waltham, MA) for 30 min on ice. Equivalent amounts (10–20 µg) of total protein were subjected to electrophoresis in 10–20% Tris-glycine gels (Invitrogen) and transferred to polyvinylidene fluoride membranes (Bio-Rad, Hercules, CA) on ice for 2 h at 30 V. Membranes were probed using mouse monoclonal anti-STAT1, anti–phospho-STAT1 (pSTAT1) (Y701), anti-IRF9, and anti–IFN-induced protein with tetratricopeptide repeats 3 (IFIT3) from BD Biosciences (San Jose, CA); rabbit polyclonal anti-PKR from Santa Cruz Biotechnology (Santa Cruz, CA); and mouse monoclonal anti–β-actin from Abcam (Cambridge, MA). The mouse anti-MxA mAb was generously provided by O. Haller (University of Freiburg, Germany). Secondary goat anti-mouse and goat anti-rabbit Abs were purchased from Southern Biotechnology Associates (Birmingham, AL). Membranes were developed using the SuperSignal West Femto ECL kit (Thermo Fisher Scientific).
Coimmunoprecipitation
Three million A549 cells were seeded overnight in 10 ml complete RPMI 1640 and treated with 10 IU/ml IFN for the indicated times. Samples were harvested by trypsin/EDTA (Invitrogen) and lysed for 30 min on ice in 50 mM Tris (pH 8.0), 280 mM NaCl, 0.5% Nonidet P-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 1 mM DTT, 1mMPMSF, Halt protease inhibitor mixture, and Halt phosphatase inhibitor mixtures (Thermo Fisher Scientific) (10, 26). Five hundred micrograms of total protein was incubated with 0.5 µg STAT2 Ab (C-20; Santa Cruz Biotechnology) for 2.5 h and precipitated by protein A-agarose beads (Santa Cruz Biotechnology) for 1 h. Samples were resolved on 10–20% Tris-glycine gels (Invitrogen) and transferred onto polyvinylidene fluoride membranes (Bio-Rad). Membranes were probed using anti-STAT1, anti-STAT2, and anti-IRF9 Abs (BD Biosciences).
Chromatin immunoprecipitation
Three million A549 cells were seeded overnight in 10 ml complete RPMI 1640 and treated with 10 IU/ml indicated IFN for 24 h. Samples were lysed using the manufacturer’s directions of the MAGnify chromatin immunoprecipitation kit (Invitrogen). Chromatin was sheared at 21% power for 20 cycles of 5 s on and 5 s off using a Sonics VCX-500 sonicator. Chromatin immunoprecipitation (ChIP) was carried out according to manufacturer’s directions using STAT1 (E-23), STAT2 (C-20), and IRF9 (C-20) Abs from Santa Cruz Biotechnology or a nonimmune serum (IgG) Ab (Invitrogen). The ChIP-specific PKR (EIF2AK2) primer [(−)01Kb Assay Tile, catalog GPH020784(−)01A] was purchased from SABiosciences (Frederick, MD).
Microarray
Three million A549 cells were seeded overnight in 10 ml complete RPMI 1640 and treated with IFN, as indicated, for 24 h. Cells were harvested by trypsin/EDTA. Total RNA was purified using RNeasy mini kits (Qiagen, Valencia, CA). Total RNA (5 µg) was reverse transcribed using 5′-amino–modified oligo(dT) primer along with reagents from FairPlay III Microarray Indirect Labeling kit (Stratagene, La Jolla, CA). Dye coupling was performed using Cy3 or Cy5 obtained from Amersham Biosciences (Piscataway, NJ). Hybridization of sample onto arrays was carried out using a MAUI Hybridization System (BioMicro Systems, Salt Lake City, UT). Arrays consisted of ~21 K 70-mer oligonucleotides (Operon Biotechnologies, Huntsville, AL) printed onto epoxy-coated slides by the National Institute of Allergy and Infectious Diseases Microarray Research Facility (National Institutes of Health, Bethesda, MD). After overnight hybridization, arrays were washed, dried, and then scanned using an Axon GenePix 4200A microarray scanner along with GenePix Pro image analysis software (Molecular Devices, Sunnyvale, CA). Gene expression raw data were then uploaded to mAdb microarray database provided by Research Technologies Branch/National Institute of Allergy and Infectious Diseases (National Institutes of Health). Two group t tests using separate (unequal) variance analysis were performed on the data set by the mAdb microarray database software. Genes were considered significant by a symmetrical 1.8-fold upregulation or downregulation cutoff. The complete microarray data set is available at National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE25113.
RNA interference antiviral assays
A549 cells were seeded at a concentration of 2.5 × 105/well in 24-well plates in antibiotic-free RPMI 1640 containing 10% FBS and 2 mM l-glutamine. Cells were immediately transfected with 20 nM small interfering RNA (siRNA) oligos complexed in Lipofectamine 2000 (Invitrogen) in Opti-MEM (Invitrogen) for 24 h. IFN was then added for an additional 24 h. The media was then removed and replaced with RPMI 1640 containing 2% FBS, 2 mM l-glutamine, 50 U/ml penicillin G, and 50 µg/ml streptomycin with EMCV at a multiplicity of infection of 0.01. Cells were stained with crystal violet at 48 h and assessed for cytopathic effect. To determine efficiency of knockdown, cells were harvested prior to addition of EMCV, and lysates were prepared and analyzed, as described for Western blot analysis. Short interfering RNA oligo sequences were described previously (27), and were obtained from Invitrogen (25 bp) and Thermo Fisher Scientific Dharmacon (19 bp).
Quantitative RT-PCR
For siRNA-treated samples, cells were treated and harvested, as described in the previous section. For all other samples, 3 × 106 cells were seeded overnight in 10 ml RPMI 1640 and treated as indicated. Cells were harvested by trypsin/EDTA. Extraction of RNA from cells was performed using RNeasy mini kits (Qiagen). Twenty nanograms of total RNA was used for each reaction, and the assay was performed using the Brilliant SYBR Green QRT-PCR Master Mix Kit, 1-Step (Stratagene, La Jolla, CA). A three-step cycling protocol was performed consisting of one 30-min cycle at 50°C, followed by a 10-min cycle at 95°C, followed by 45 cycles of the following: 30 s at 95°C, 1 min at 55°C, and 30 s at 72°C on the Stratagene Mx3000p QPCR system. Relative mRNA expression was determined by normalization to GAPDH expression. Primers were obtained from Integrated DNA Technologies (San Diego, CA) and are listed in Table I (28).
Table I.
Primers used for qRT-PCR
| Gene Name |
Primer Sequence | |
|---|---|---|
| GAPDH | F | 5′-GAG TCA ACG GAT TTG GTC GT-3′ |
| R | 5′-TTG ATT TTG GAG GGA TCT CG-3′ | |
| HLA-A | F | 5′-GAC CAG GAG ACA CGG AAT GT-3′ |
| R | 5′-GAT GTA ATC CTT GCC GTC GT-3′ | |
| IFNA1 | F | 5′-GCA AGC CCA GAA GTA TCT GC-3′ |
| R | 5′-ACT GGT TGC CAT CAA ACT CC-3′ | |
| IFNA2 | F | 5′-AAA TAC AGC CCT TGT GCC TGG-3′a |
| R | 5′-GGT GAG CTG GCA TAC GAA TCA-3′a | |
| IFNB | F | 5′-AAG GCC AAG GAG TAC AGT C-3′a |
| R | 5′-ATC TTC AGT TTC GGA GGT AA-3′a | |
| IFNL1 | F | 5′-CGC CTT GGA AGA GTC ACT CA-3′a |
| R | 5′-GAA GCC TCA GGT CCC AAT TC-3′a | |
| IFNL2/3 | F | 5′-AGT TCC GGG CCT GTA TCC AG-3′a |
| R | 5′-GAG CCG GTA CAG CCA ATG GT-3′a | |
| IFNW | F | 5′-TGG TAA AAG GGA GCC AGT TG-3′ |
| R | 5′-TGC AGT TGC TGA TGA AGT CC-3′ | |
| IFIT3 | F | 5′-GAA CAT GCT GAC CAA GCA GA-3′ |
| R | 5′-CAG TTG TGT CCA CCC TTC CT-3′ | |
| PKR | F | 5′-ACG CTT TGG GGC TAA TTC TT-3′ |
| R | 5′-TTC TCT GGG CTT TTC TTC CA-3′ | |
| MxA | F | 5′-ACC ACA GAG GCT CTC AGC AT-3′ |
| R | 5′-CTC AGC TGG TCC TGG ATC TC-3′ | |
| OAS1 | F | 5′-ACA GGC AGA AGA GGA CTG GA-3′ |
| R | 5′-TAG AAG GCC AGG AGT CAG GA-3′ |
From Ref. 28.
F, forward; R, reverse.
Results
Distinct activation profiles of cells treated with IFN-α2a or IFN-γ
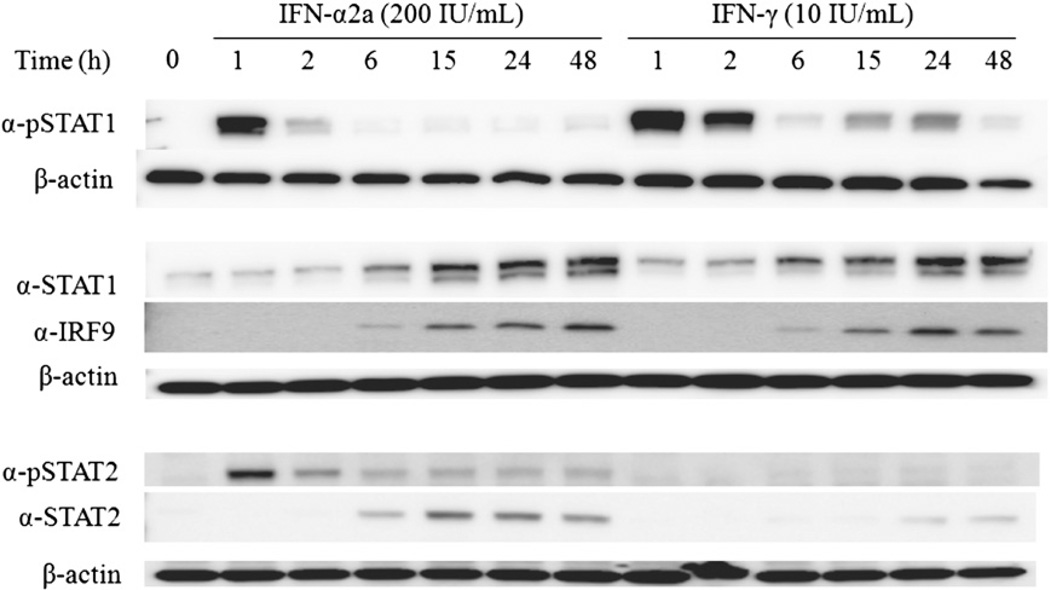
To determine any differences between the cellular response of IFN-α2a and IFN-γ, we first examined the activation of STAT1 after each IFN treatment. Both IFN-α2a and IFN-γ treatment increased production of total STAT1 protein with similar kinetics, starting at 6 h and continuing through 48 h (Fig. 1). However, they activate STAT1 differently; treatment with IFN-α2a leads to immediate phosphorylation of STAT1 that begins to decrease at 2 h and remains at low levels for up to 48 h, whereas IFN-γ treatment results in the same initial phosphorylation profile as IFN-α2a, with an additional increase in pSTAT1 at 15–24 h (Fig. 1).
FIGURE 1.
STAT1 signaling response in A549 cells treated with IFN-α2a and IFN-γ. Western blot of A549 cells treated with IFN-α2a and IFN-γ showing kinetics of STAT1 (via phosphorylation at Y701) and STAT2 (via phosphorylation at Y689) activation as well as total STAT1, STAT2, and IRF9 expression. Cells were treated with 200 IU/ml IFN-α2a or 10 IU/ml IFN-γ, and cell lysates were prepared at the indicated times. Equal amounts of total protein were subjected to SDS-PAGE and analyzed by Western blot using the indicated Abs. β-actin is shown as a control.
Upregulation of ISRE-containing genes by IFN-γ in A549 cells
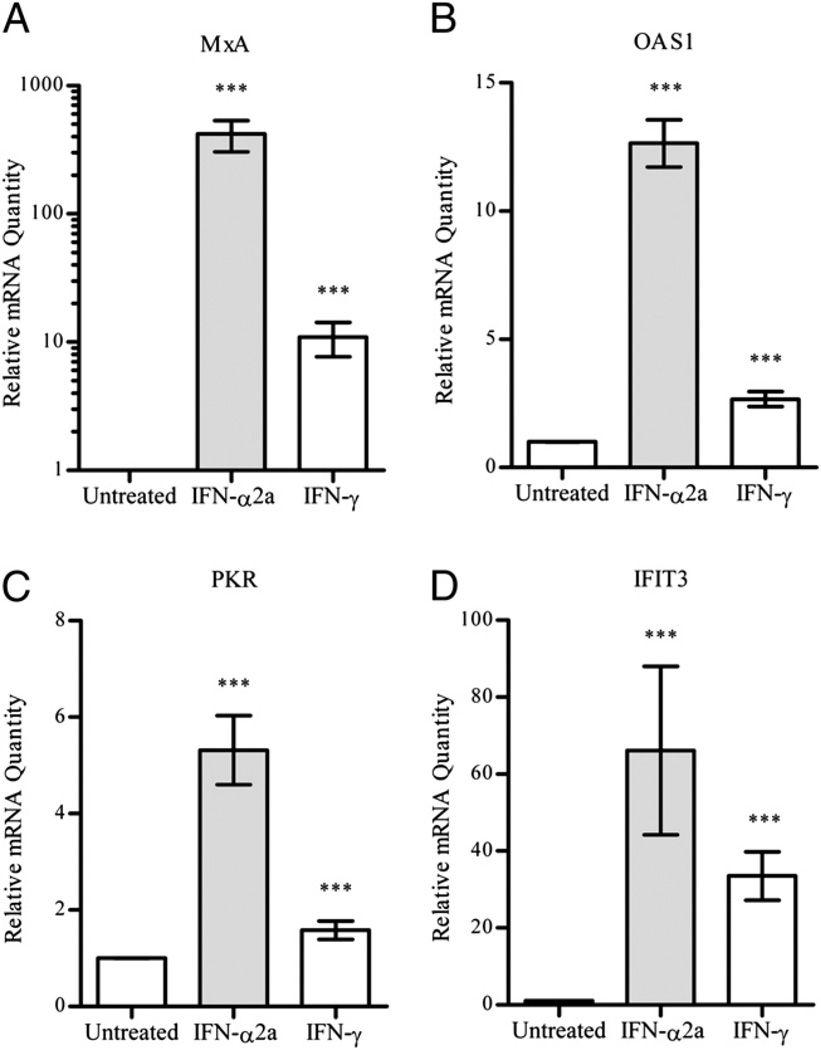
Because the profile of STAT activation between IFN-α2a and IFN-γ showed a major difference in STAT1 phosphorylation at 24 h, we used gene expression microarrays to examine differences in gene expression at this time point. Table II includes the data comparing statistically significant differences in gene expression (of at least 1.8-fold), as determined by Student t test, of A549 cells treated with 1 ng/ml IFN-α2a (200 IU/ml) or 1 ng/ml IFN-γ (10 IU/ml) for 24 h. Type I IFN (IFN-α2a) treatment resulted in distinct upregulation of several genes, including PLSCR1, TRIM6–TRIM34 (IFP1), LY6E (RIG-E), PNPT1, and HERC5 (Table II). Similarly, IFN-γ treatment resulted in upregulation of a specific subset of genes, including IRF1, GBP2, GBP3, WARS, and genes involved in Ag presentation such as CD74 (Table II). Interestingly, IFN-α2a and IFN-γ both upregulated a subset of genes that are traditionally associated with a type I IFN antiviral response and contain ISRE promoter motifs, including MxA (Mx1), PKR (EIF2AK2), and OAS1 (Table II). We used quantitative RT-PCR (qRT-PCR) to validate the microarray data for a small subset of genes involved in the antiviral response. As expected, IFN-α2a treatment resulted in upregulation of MxA, OAS1, PKR, and IFIT3. However, these genes were also upregulated after treatment with IFN-γ (Fig. 2). Addition of a neutralizing Ab to IFNAR1 (which blocks type I IFN signaling) had no effect on induction of these genes by IFN-γ (data not shown).
Table II.
Gene expression of select ISGs differentially regulated by either type I, type II, or both type I and type II IFNs
| IFN-α2a | IFN-γ | p Value | Entrez GeneID | Gene Name | Description |
|---|---|---|---|---|---|
| 3.21 | 1.60 | 0.0002 | 5359 | PLSCR1 | Phospholipid scramblase 1 |
| 3.11 | 1.65 | 0.0058 | 445372 | TRIM6–TRIM34 | IFN-responsive finger protein 1 (IFP1) |
| 3.62 | 1.82 | 0.0102 | 4061 | LY6E | Lymphocyte Ag 6 complex, locus E (Rig-E) |
| 2.91 | 0.98 | 0.0164 | 87178 | PNPT1 | Polyribonucleotide nucleotidyltransferase 1 |
| 4.40 | 1.31 | 0.0199 | 87178 | PNPT1 | Polyribonucleotide nucleotidyltransferase 1 |
| 2.78 | 1.74 | 0.0284 | 9830 | TRIM14 | Tripartite motif-containing 14 |
| 8.44 | 1.23 | 0.0288 | 51191 | HERC5 | Hect domain and RLD 5 |
| 1.16 | 20.36 | 0.0002 | 7453 | WARS | Tryptophanyl-tRNA synthetase |
| 1.27 | 13.62 | 0.0023 | 3659 | IRF1 | IFN regulatory factor 1 |
| 1.52 | 2.72 | 0.0110 | 840 | CASP7 | Caspase 7, apoptosis-related cysteine peptidase |
| 0.77 | 23.97 | 0.0122 | 10537 | UBD | Ubiquitin D |
| 1.03 | 6.87 | 0.0162 | 2635 | GBP3 | Guanylate-binding protein 3 |
| 1.25 | 2.72 | 0.0192 | 7098 | TLR3 | Toll-like receptor 3 |
| 1.24 | 1.90 | 0.0322 | 6774 | STAT3 | Signal transducer and activator of transcription 3 |
| 1.25 | 6.11 | 0.0353 | 2634 | GBP2 | Guanylate-binding protein 2 |
| 0.89 | 6.64 | 0.0437 | 972 | CD74 | CD74 molecule, MHC, class II |
| 0.84 | 5.98 | 0.0442 | 8651 | SOCS1 | Suppressor of cytokine signaling 1 |
| 27.37 | 4.93 | 0.0018 | 4939 | OAS2 | 2′-5′-Oligoadenylate synthetase 2, 69/71 kDa |
| 9.80 | 3.44 | 0.0045 | 4938 | OAS1 | 2′-5′-Oligoadenylate synthetase 1, 40/46 kDa |
| 4.66 | 7.50 | 0.0072 | 3105 | HLA-A | MHC, class I, A |
| 38.52 | 3.52 | 0.0082 | 3434 | IFIT1 | IFN-induced protein with tetratricopeptide repeats 1 |
| 45.95 | 10.56 | 0.0097 | 4599 | MX1 | Myxovirus resistance 1, IFN-inducible protein p78 |
| 19.67 | 3.60 | 0.0123 | 3429 | IFI27 | IFN, α-inducible protein 27 |
| 4.62 | 7.25 | 0.0134 | 3135 | HLA-G | MHC, class I, G |
| 3.69 | 2.15 | 0.0142 | 5610 | EIF2AK2 | Eukaryotic translation initiation factor 2-α kinase 2 |
| 8.57 | 2.23 | 0.0369 | 3665 | IRF7 | IFN regulatory factor 7 |
Relative transcript levels, as determined by gene expression microarray, of select ISG upregulated by type I IFN only, type II IFN only, and both type I and type II IFNs in IFN-α2a– and IFN-γ –treated cells. A549 cells were treated with 200 IU/ml IFN-α2a or 10 IU/ml IFN-γ for 24 h. Genes shown were selected by a 1.8-fold symmetric cutoff value. Fold upregulation or downregulation shown is the average of three independent experiments, arranged by decreasing statistical significance of at least 1.8-fold, as determined from an unpaired, two-tailed Student t test with separate (unequal) variance, with a cutoff of 0.05.
FIGURE 2.
Gene expression of select antiviral ISGs after IFN-α2a and IFN-γ treatment. Cells were treated with IFN for 24 h, after which total RNA was collected and assayed for relative mRNA transcript amounts of the ISGs MxA (A), OAS1 (B), PKR (C), and IFIT3 (D). The results are shown as the mean and SD of triplicate PCR reactions from three separate experiments. ***p < 0.001 by an unpaired, two-tailed t test for the indicated group compared with the untreated group.
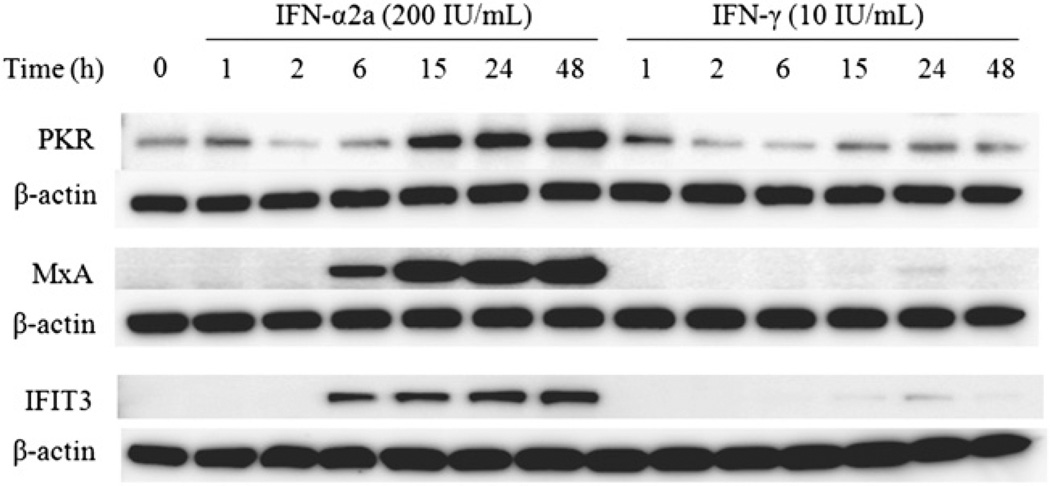
We also examined expression of MxA, PKR, and IFIT3 (which is also regulated by an ISRE-containing promoter element) at the protein level. PKR is both constitutively expressed and IFN inducible. There was a significant increase in PKR after treatment with IFN-α2a beginning at 6 h. We observed the same pattern of expression in IFN-γ–treated cells, albeit to a much lesser extent (Fig. 3). Additionally, IFN-γ treatment resulted in a delay of PKR expression compared with IFN-α2a: induction started at 6 h for IFN-α2a and 15 h for IFN-γ. MxA and IFIT3 had the same pattern of protein expression as PKR after treatment with both IFN-α2a and IFN-γ; however, in both cases, IFN-α2a induction of these proteins was greater than for IFN-γ.
FIGURE 3.
Expression of antiviral ISGs in A549 cells treated with IFN-α and IFN-γ. Cell lysates (from Fig. 1) were evaluated for expression of the ISGs MxA, PKR, and IFIT3. Cells were treated with 200 IU/ml IFN-α2a or 10 IU/ml IFN-γ, and cell lysates were prepared at the indicated times. Equal amounts of total protein were subjected to SDS-PAGE and analyzed by Western blot using the indicated Abs. β-actin is shown as a control.
IFN-γ induces formation of ISGF3II
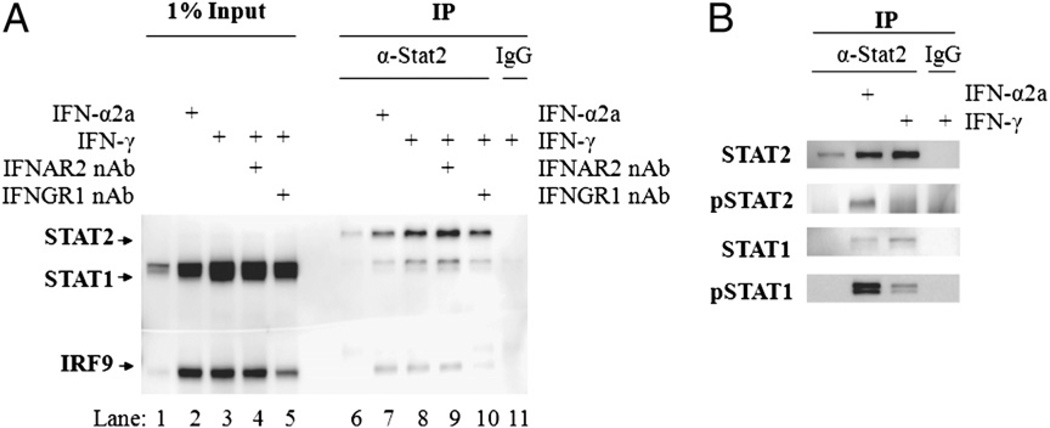
To determine how these ISRE-containing genes were being upregulated, we examined the possibility that IFN-γ induces ISGF3. We were able to isolate the three components of the ISGF3 complex by coimmunoprecipitation following treatment with IFN-α2a or IFN-γ for 24 h (Fig. 4A, lanes 7 and 8, respectively). The amount of the ISGF3 proteins precipitated did not decrease in the presence of a neutralizing Ab to IFNAR2 during the course of IFN-γ treatment (Fig. 4A, lane 9). However, the amount of STAT1 and IRF9 that precipitated with STAT2 at 24 h decreased when an Ab that neutralized IFN-γ signaling was added to the media after 15 h of IFN-γ treatment (Fig. 4A, lane 10). The complex was not present in untreated cells (Fig. 4A, lane 6) or in samples precipitated with nonspecific immune serum (Fig. 4A, lane 11). One percentage of the total cell lysate used for each sample was included to compare relative amounts of ISGF3/ISGF3II formation to the total amount of STAT1, STAT2, and IRF9 in each treatment group (Fig. 4A, lanes 1–5). STAT1 and IRF9 were upregulated with each treatment, but STAT2 was not. We also used coimmunoprecipitation to determine whether STAT1 and STAT2 were phosphorylated after IFN-α and IFN-γ treatment. As expected, STAT1 was phosphorylated after each treatment (Fig. 4B). Although IFN-γ treatment results in more STAT1 phosphorylation at 24 h, there is less pSTAT1 in the precipitated complex in response to IFN-γ compared with IFN-α2a. It is also interesting to note that whereas STAT2 was phosphorylated after IFN-α treatment, we were not able to detect phosphorylation of STAT2 in response to IFN-γ treatment (Fig. 4B). Therefore, to avoid confusion with the classical ISGF3 complex, the complex containing unphosphorylated STAT2 will be referred to as ISGF3II.
FIGURE 4.
Neutralization of STAT1 activation in IFN-γ–treated cells and accumulation of ISGF3 lacking STAT2 phosphorylation. A, STAT2 Abs were used to coimmunoprecipitate the ISGF3 complex in A549 cells. Cells were left untreated (lane 6) or treated with 10 IU/ml IFN-α (lane 7) or IFN-γ (lanes 8–11) and harvested at 24 h. The IFNAR1 Ab was added immediately prior to IFN addition (lanes 4, 9), and the IFNGR1 Ab was added after IFN-γ treatment for 15 h (lanes 5, 10). A Western blot for the ISGF3 components (STAT1, STAT2, and IRF9) was performed on the immunoprecipitated samples. One percent of the total amount of input protein for each immunoprecipitation treatment (untreated, lane 1; IFN-α2a, lane 2; IFN-γ, lanes 3–5) was added for comparison purposes. B, Western blot for pSTAT2 (Y689), pSTAT1 (Y701), and total STAT2 and STAT1 on immunoprecipitated samples, as described in A.
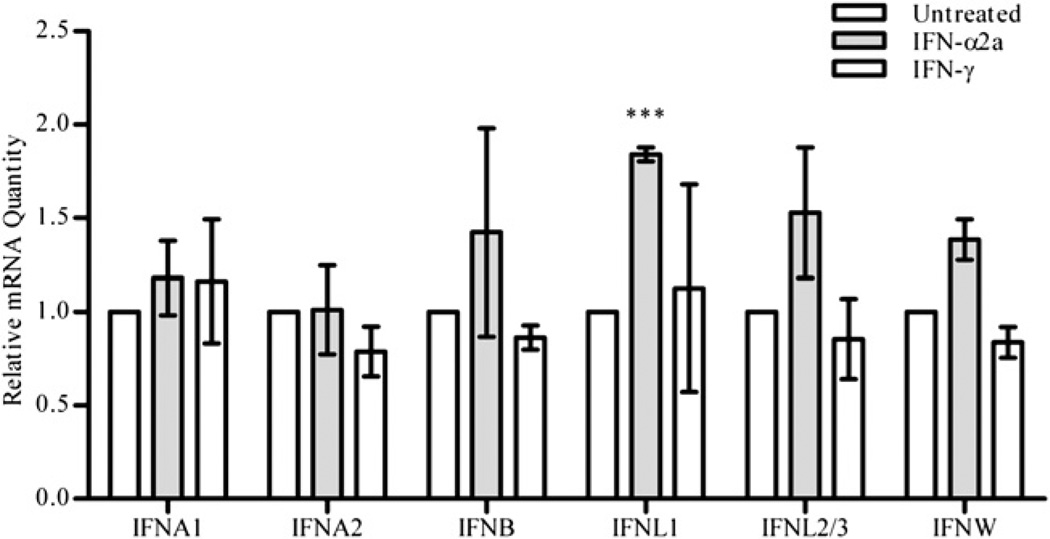
To exclude the possibility that autocrine type I IFN led to the formation of ISGF3II, we then used qRT-PCR to determine whether a 24-h IFN-γ treatment induced other IFNs. IFN-γ treatment did not upregulate IFN-α1, IFN-α2, IFN-β, IFN-λ1, IFN-λ2 or IFN-λ3, and IFN-ω at 24 h (Fig. 5). Although IFN-α2a treatment led to statistically significant upregulation of IFN-λ1 at 24 h, none of the other IFNs were upregulated.
FIGURE 5.
Gene expression of IFN genes after IFN treatment. Cells were left untreated or treated with 10 IU/ml IFN-α2a or IFN-γ for 24 h, after which total RNA was collected and assayed for relative mRNA transcript amounts of IFN-α1, IFN-α2, IFN-β, IFN-λ1, IFN-λ2 or IFN-λ3, or IFN-ω. The results are shown as the mean and SD of triplicate PCR reactions from three separate experiments. ***p < 0.001 by two-way ANOVA with a Bonferroni post-test comparing the indicated gene in the treatment group compared with the untreated group.
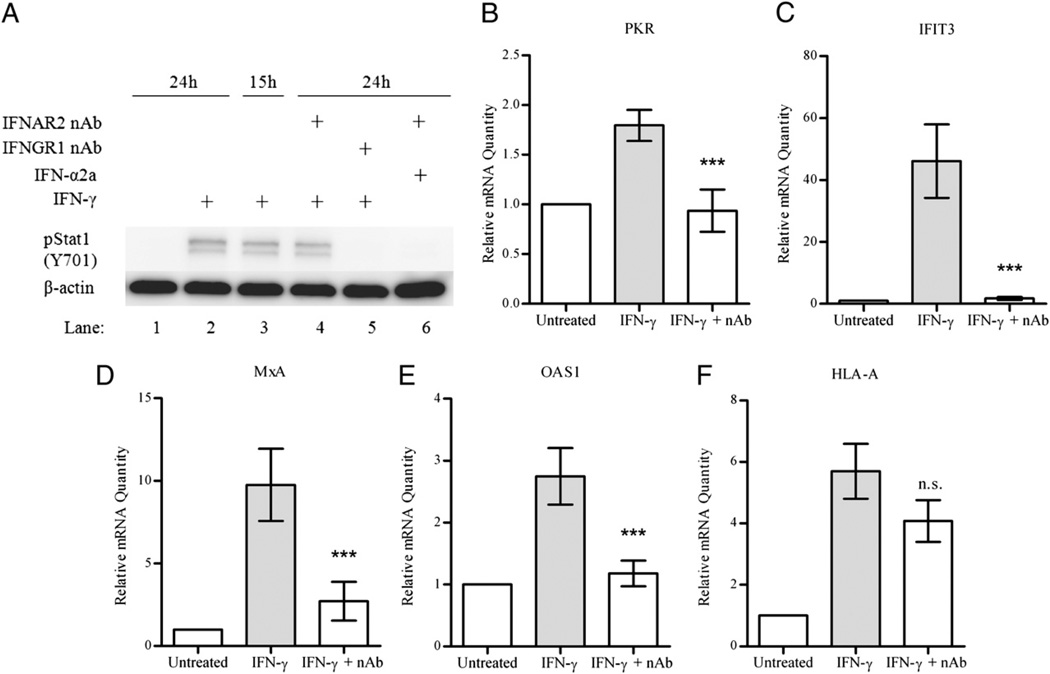
Next, we added a neutralizing Ab for IFNAR2 prior to treating cells with IFN-γ, then used pSTAT1 as a marker for IFN activation. There was no difference in the activation of pSTAT1 in IFN-γ–treated cells with neutralized IFNAR2 compared with those treated with IFN-γ alone (Fig. 6A, lanes 4 and 2, respectively). The Ab was capable of neutralizing the activity of any type I IFN present over the course of the treatment (Fig. 6A, lane 6). An Ab that neutralizes IFN-γ signaling was added to IFN-γ–treated cells at 15 h to allow for any possible cytokine production that might have corresponded to the induction of the second signaling peak from Fig. 1. The presence of this Ab completely abrogated pSTAT1 activity at later time points (Fig. 6A, lane 5). We also used a neutralizing Ab for IL-10Rβ to inhibit any possible type III IFN signaling. This also had no effect on pSTAT1 activity after treatment with IFN-γ (data not shown).
FIGURE 6.
Gene expression of select ISGs in IFN-γ–treated cells after neutralization with IFNGR1 Ab. A, IFN-treated cells were examined by Western blot for phosphorylation of STAT1 (at Y701) as an indicator of autocrine or paracrine IFN action. Cells were left untreated (lane 1) or treated with 10 IU/ml IFN-γ (lanes 2–5) or 10 IU/ml IFN-α2a (lane 6), and cell lysates were prepared at the indicated times. The IFNAR2 Ab was added prior to IFN treatment (lanes 4, 6), and the IFNGR1 Ab was added after 15-h IFN-γ treatment. nAb, neutralizing Ab. B–F, Cells were left untreated or treated with IFN-γ and harvested at 24 h, after which total RNA was collected and assayed for relative mRNA transcript amounts. The IFNGR1 Ab was added after 15 h of IFN-γ treatment and was present until harvest at 24 h. The relative mRNA levels of PKR (B), IFIT3 (C), MxA (D), OAS1 (E), and HLA-A (F) are shown as the mean and SD of triplicate PCR reactions from two separate experiments normalized to GAPDH. ***p < 0.001 and n.s., not significant by an unpaired, two-tailed t test comparing the IFN-γ and IFN-γ plus neutralizing Ab treatment groups.
To further demonstrate that no other cytokines were responsible for the second pSTAT1 signaling peak, we added an Ab to neutralize IFN-γ signaling at 15 h, then harvested the cells at 24 h. The upregulation of PKR, IFIT3, MxA, and OAS1 gene expression was also reversed upon neutralization of IFN-γ signaling after treatment for 15 h; each of these ISRE-inducible genes showed significant downregulation to steady state or near steady state levels at 24 h (Fig. 6B–E, respectively). In contrast, HLA-A, which is regulated by IRF1-containing transcription factors rather than ISGF3, showed no significant downregulation upon addition of the neutralizing Ab (Fig. 6F), demonstrating that IRF1-mediated transcription continued after neutralization of IFN-γ signaling, and PKR, IFIT3, MxA, and OAS1 transcription was independent of IRF1.
STAT2 binds to ISRE-containing antiviral gene promoters after IFN-γ treatment
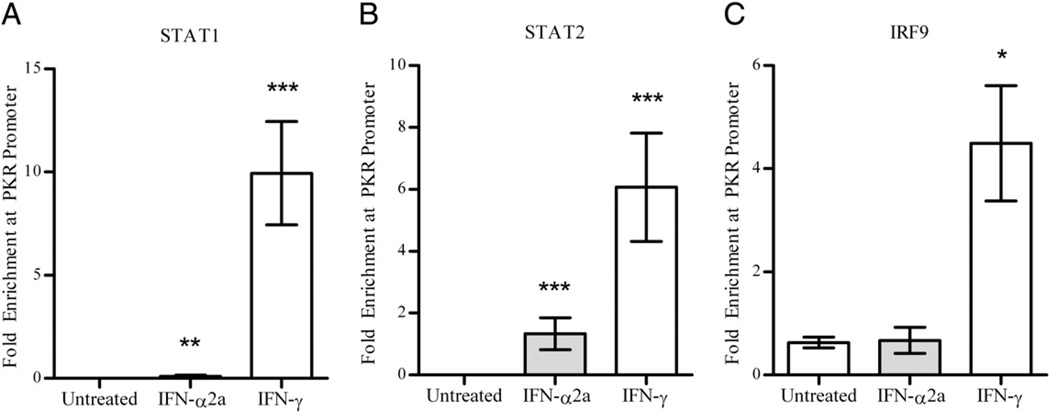
To show that ISGF3II is involved in transcribing IFN-γ–inducible genes, we used ChIP to demonstrate that STAT1, STAT2, and IRF9 physically bind to the promoters of the ISRE-containing genes following IFN-γ treatment. As expected, IFN-α2a induces recruitment of STAT1- and STAT2-containing complexes to the PKR promoter at 24 h (Fig. 7). However, STAT1-, STAT2-, and IRF9-containing transcription factor complexes are also recruited to the PKR promoter after treatment with IFN-γ (Fig. 7). Interestingly, IFN-α2a treatment did not result in statistically significant recruitment of IRF9 to the PKR promoter compared with untreated cells in this assay (Fig. 7C).
FIGURE 7.
Occupancy of the PKR promoter by (A) STAT1, (B) STAT2, and (C) IRF9 after IFN-γ and IFN-α2a treatment. Cross-linked, sheared chromatin ~1 kb in length from cells left untreated or treated with 10 IU/ml IFN-γ or 10 IU/ml IFN-α2a was immunoprecipitated with 1 µg nonimmune serum, 1 µg Ab against STAT1 and IRF9, or 2 µg Ab against STAT2. Quantitative PCR analysis was performed using a ChIP-specific primer for the PKR promoter. The results are shown as fold upregulation of occupancy at each site compared with background signal, and are the mean and SEM from triplicate PCR assays of a single biological experiment. The results are representative of three biological assays. ***p < 0.001; **p < 0.01; *p < 0.05 by two-tailed t test comparing the indicated group with the untreated control group.
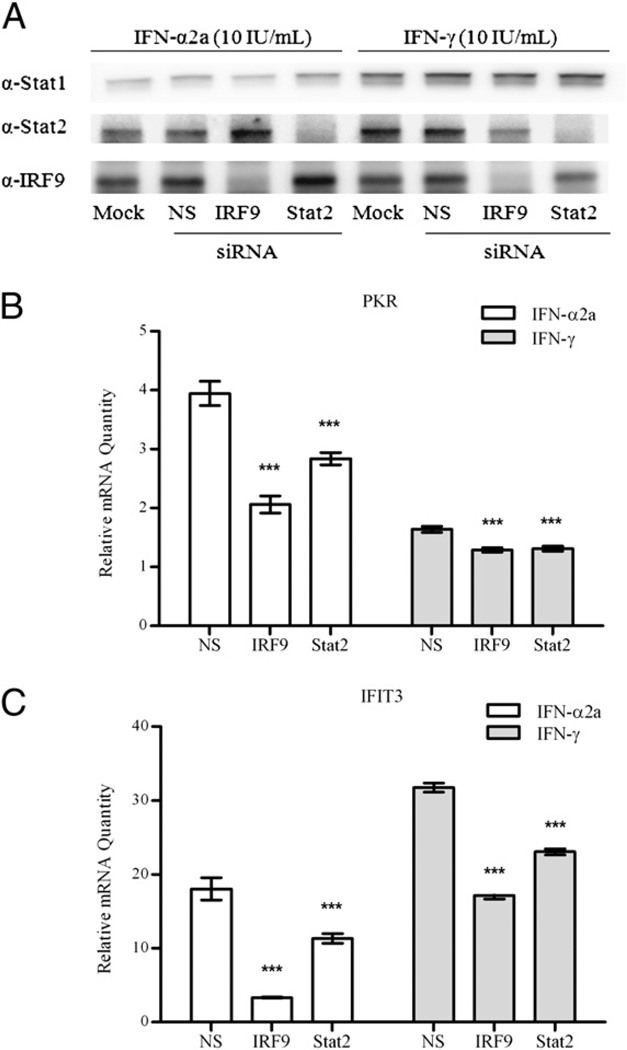
In addition, we analyzed the expression of ISRE-driven antiviral genes in IFN-γ–treated cells lacking STAT2 and IRF9. Both STAT2 and IRF9 siRNA treatment in A549 cells resulted in efficient inhibition of protein expression to nearly undetectable amounts (Fig. 8A). Knocking down one protein had no effect on expression of the other, and neither siRNA treatment resulted in decreased expression of STAT1. We used qRT-PCR to examine how absence of these proteins affected the gene expression of the ISGF3-inducible proteins PKR and IFIT3. Loss of both STAT2 and IRF9 resulted in significant downregulation of PKR mRNA expression when treated with IFN-α and IFN-γ (Fig. 8B). Similar results were obtained for IFN-mediated mRNA expression of IFIT3 following STAT2 and IRF9 siRNA treatment (Fig. 8C).
FIGURE 8.
Gene and protein expression of ISGs in STAT2- and IRF9-deficient cells after treatment with IFN-α2a or IFN-γ. A, STAT2 and IRF9 protein expression was knocked down by siRNA. Cells were either mock treated (lipofectamine only), treated with a nonspecific (NS) siRNA or siRNA for IRF9 or STAT2, and incubated with 10 IU/ml IFN-α or IFN-γ for 24 h. Cell lysates containing equivalent amounts of protein were added to each well and subjected to SDS-PAGE, after which Western blots were performed with STAT1, STAT2, or IRF9 Abs. B–C, Gene expression for (B) PKR and (C) IFIT3 was determined by qRT-PCR in cells transfected with siRNA targeting IRF9 and STAT2 and treated with 10 IU/ml IFN-α2a or IFN-γ. Data are presented as the mean and SD of three replicate assay wells from a single experiment. ***p < 0.001 by a separate one-way ANOVA for each IFN treatment, followed by a Bonferroni posttest for comparison between IRF9 siRNA or STAT2 siRNA treatment and the nonspecific (NS) siRNA treatment.
Biological role for ISGF3II in the antiviral activity of IFN-γ
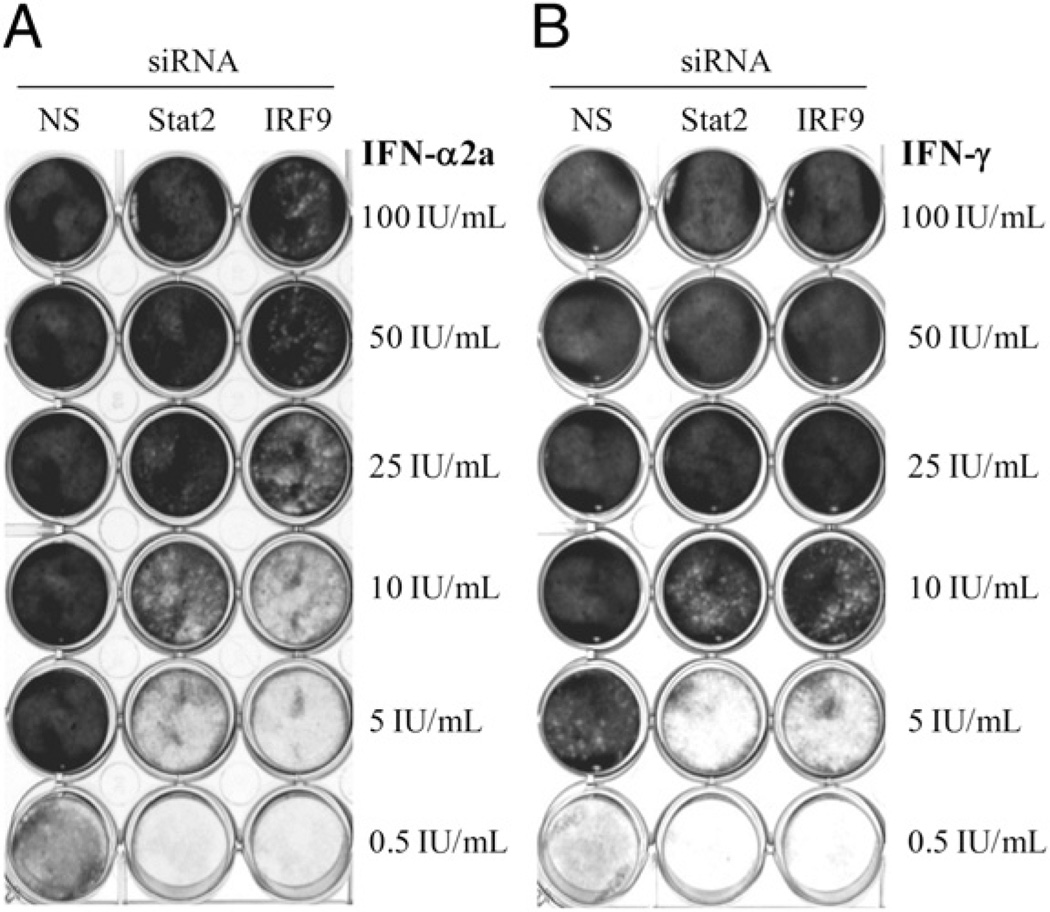
Although ISGF3II is present and classical antiviral genes driven through the ISRE promoter are expressed in A549 cells treated with IFN-γ, it is unclear whether this phenomenon has any biological function in these cells. Therefore, to disrupt ISGF3/ISGF3II formation after IFN treatment, we used siRNA to knock down STAT2 and IRF9. STAT2 and IRF9 siRNA reduced the ability of A549 cells to mount an antiviral response against ECMV after treatment with either IFN-α2a (Fig. 9A) or IFN-γ (Fig. 9B). In IFN-α2a–treated samples, antiviral activity was inhibited up to a concentration of 25 IU/ml in the STAT2 knockdowns, and activity was partially abrogated even at 100 IU/ml in the IRF9 knockdown cells. Either STAT2 or IRF9 knockdown inhibited the antiviral activity of up to 10 IU/ml IFN-γ.
FIGURE 9.
The absence of IRF9 or STAT2 abrogates the antiviral activity of IFN-γ in A549 cells. Specific siRNAs targeting IRF9, STAT2, or nonspecific (NS) control siRNAs were transfected at a concentration of 20 nM for 24 h before addition of the indicated amounts of IFN-α2a (A) or IFN-γ (B). Following 24 h of the indicated treatment, media was removed and EMCV was added at a multiplicity of infection of 0.01. Cells were stained with crystal violet at 48 h postinfection. The results are representative of greater than three separate experiments.
We also added neutralizing Abs for IFNAR2 and IL-10Rβ both during transfection and IFN treatment as well as upon addition of EMCV to block any possible production and signaling by type I or type III IFNs. These treatments did not result in increased abrogation of antiviral activity, demonstrating that these IFNs do not play a role in the antiviral activity of IFN-γ (data not shown).
Discussion
ISGF3 is firmly established as a critical component of type I IFN signaling, and it drives transcription of many classical antiviral ISGs (15, 29, 30). However, IFN-γ is thought to induce transcription and mount an antiviral response entirely without this transcription factor (15–17). Several studies indicate presence of ISGF3 after IFN-γ treatment in a murine system and impairment of antiviral activity in STAT2-null mice (23–25, 31). To our knowledge, the evidence in this study demonstrates both formation of ISGF3II and its role in IFN-γ–mediated antiviral activity for the first time in a human system.
Whereas most signaling occurs within minutes to hours of IFN-α2a and IFN-γ treatment, both the Western blot and coimmunoprecipitation data show that significant STAT activation also occurs much later in IFN-γ–treated cells. This may be due to the fact that type I IFN responses are primarily directed at early stages of an infection, whereas type II IFN plays a role in eliminating viral persistence and therefore requires prolonged Jak/STAT signaling for full biological activity (32, 33). Several studies have suggested that this might be explained by an autocrine type I IFN signaling loop, and that a very small amount of IFN continually activates the Jak/STAT pathway to provide a robust response upon encountering a pathogen (24, 31). However, the use of neutralizing Abs to both IFNAR1 and IL-10Rβ (which block type I and type III IFNs, respectively) and the qRT-PCR data for the type I and type III IFN genes ruled out the possibility of autocrine or paracrine IFN signaling as a cause of ISGF3II formation following IFN-γ treatment. Others have also demonstrated that IFN-γ activity is not dependent on type I IFNs (34).
At high IFN concentrations, continual signaling results in activation of many overlapping and redundant signaling pathways (2, 35, 36). For example, overexpression of STAT1 and IRF9 can lead to transient ISRE activation by homodimers of STAT1 or STAT2 complexed with IRF9, and STAT2 and IRF9 can associate with each other prior to IFN treatment (26, 37–39). In addition, several studies indicate that IRF1 complexes can bind to ISRE motifs and serve as a transcription factor for ISGs (40, 41). However, siRNA for STAT2 and IRF9 was able to abrogate the antiviral activity of both IFN-α2a and IFN-γ at physiological concentrations. If the alternative STAT/IRF transcription factors had a functional role in antiviral gene transcription, both IFNs should have induced an antiviral state in the cells lacking STAT2 or IRF9. In addition, relative mRNA quantities of classical antiviral genes in IFN-γ–treated cells neutralized at 15 h were significantly lower at 24 h in the absence of late IFN-γsignaling, whereas those that are transcribed using IRF-1–mediated gene transcription (e.g., MHC-I [HLA-A]) (42, 43) showed no significant change in expression.
The late time point at which the ISGF3II formation occurred in response to IFN-γ highlights the possible mechanism as to its formation. IFN signaling, over time, results in the accumulation of STAT1, STAT2, and IRF9 proteins in the cytoplasm. In these cells, prolonged signaling and phosphorylation of STAT1 allows these accumulated proteins to favorably interact to form ISGF3II even though STAT2 is not phosphorylated. There is growing evidence that the nonphosphorylated STATs, particularly STAT1 and STAT2, also play a role in IFN signaling (44–47). Therefore, the presence of unphosphorylated STAT2 in this complex combined with the low levels of this complex forming in the cytoplasm may account for the very low amount of ISGF3II imported into the nucleus and bound to the PKR promoter in response to IFN-γ. IFN-α2a treatment results in phosphorylation of both STAT1 and STAT2, resulting in formation of the classical ISGF3 complex that has high affinity for ISRE sequences. Because ISGF3II contains phosphorylated STAT1 only, it might be possible that it binds to ISRE sequences in a transient manner, whereas ISGF3 has a more stable interaction with antiviral ISG promoters. As such, the classical ISGF3 complex would be more efficient in inducing gene transcription than ISGF3II. The ability of classical ISGF3 to bind more efficiently to ISRE sequences could be one possible explanation for the increased ISG mRNA and protein levels in cells treated with IFN-α2a compared with IFN-γ.
Although ISGF3II does play a role in inducing ISGs and mediating antiviral activity in response to IFN-γ, it is important to note that the data suggest that it is not absolutely critical for the action of IFN at all concentrations; it appears to only play a major role at low concentrations of type I or type II IFNs. When either IRF9 or STAT2 was knocked down in A549 cells, the antiviral ISGs were still upregulated to a small degree. This is most likely due to residual levels of IRF9 and STAT2 upregulating the ISGs involved in antiviral activity, but other signaling pathways might still be involved in gene induction at low concentrations.
ISGF3 is one component of the myriad signaling pathways that regulate the hundreds of ISGs. Together, these results suggest that STAT2 and IRF9, via ISGF3II, play a significant role in IFN-γ signaling. Signaling through ISGF3II and ISGF3-like transcription factor complexes, in addition to those induced by the canonical STAT1 homodimer signaling pathway, may result in induction of a broader range of ISGs than typically thought of as IFN-γ inducible and explain the overlapping gene expression profiles and biological activities of type I and type II IFNs. ISGF3II formation played a crucial role in concentrations similar to those produced in a natural infection (48), suggesting that this transcription factor could be a critical component for establishing an antiviral state in vivo. The data presented in this study provide evidence of a new pathway by which IFN-γ can induce antiviral effects in humans.
Acknowledgments
We thank the members of the Zoon laboratory, particularly Dr. Corey Balinsky and Josef Mejido, for many helpful discussions and suggestions as well as Dr. John O’Shea, Dr. Alexander Schmidt, and the National Cancer Institute Fellows Editorial Board for reading and providing editorial notes on the manuscript. We also thank Dr. Otto Haller for generously providing the MxA Ab.
This work was supported by the Intramural Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Abbreviations used in this article
- ChIP
chromatin immunoprecipitation
- EMCV
encephalomyocarditis virus
- GBP
guanylate-binding protein
- IFIT3
IFN-induced protein with tetratricopeptide repeats 3
- IFNAR
IFN-α receptor
- IFNGR
IFN-γ receptor
- IRF
IFN regulatory factor
- ISG
IFN-stimulated gene
- ISGF3
ISG factor 3
- ISGF3II
ISGF3 containing unphosphorylated STAT2
- ISRE
IFN-stimulated response element
- MxA
myxovirus resistance protein A
- OAS1
2′,5′-oligoadenylate synthetase 1
- PKR
protein kinase R
- pSTAT
phospho-STAT
- qRT-PCR
quantitative RT-PCR
- siRNA
small interfering RNA
Footnotes
The sequence presented in this article has been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE25113.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Pestka S, Langer JA, Zoon KC, Samuel CE. Interferons and their actions. Annu. Rev. Biochem. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 2.Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell. Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 3.Maher SG, Romero-Weaver AL, Scarzello AJ, Gamero AM. Interferon: cellular executioner or white knight? Curr. Med. Chem. 2007;14:1279–1289. doi: 10.2174/092986707780597907. [DOI] [PubMed] [Google Scholar]
- 4.Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 5.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 6.Kotenko SV, Izotova LS, Pollack BP, Mariano TM, Donnelly RJ, Muthukumaran G, Cook JR, Garotta G, Silvennoinen O, Ihle JN, et al. Interaction between the components of the interferon gamma receptor complex. J. Biol. Chem. 1995;270:20915–20921. doi: 10.1074/jbc.270.36.20915. [DOI] [PubMed] [Google Scholar]
- 7.Gauzzi MC, Velazquez L, McKendry R, Mogensen KE, Fellous M, Pellegrini S. Interferon-alpha-dependent activation of Tyk2 requires phosphorylation of positive regulatory tyrosines by another kinase. J. Biol. Chem. 1996;271:20494–20500. doi: 10.1074/jbc.271.34.20494. [DOI] [PubMed] [Google Scholar]
- 8.Uzé G, Monneron D. IL-28 and IL-29: newcomers to the interferon family. Biochimie. 2007;89:729–734. doi: 10.1016/j.biochi.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 10.Heim MH, Kerr IM, Stark GR, Darnell JE., Jr Contribution of STAT SH2 groups to specific interferon signaling by the Jak-STAT pathway. Science. 1995;267:1347–1349. doi: 10.1126/science.7871432. [DOI] [PubMed] [Google Scholar]
- 11.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 12.Decker T, Lew DJ, Darnell JE., Jr Two distinct alpha-interferon-dependent signal transduction pathways may contribute to activation of transcription of the guanylate-binding protein gene. Mol. Cell. Biol. 1991;11:5147–5153. doi: 10.1128/mcb.11.10.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenlund AC, Farrar MA, Viviano BL, Schreiber RD. Ligand-induced IFN gamma receptor tyrosine phosphorylation couples the receptor to its signal transduction system (p91) EMBO J. 1994;13:1591–1600. doi: 10.1002/j.1460-2075.1994.tb06422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lew DJ, Decker T, Strehlow I, Darnell JE. Overlapping elements in the guanylate-binding protein gene promoter mediate transcriptional induction by alpha and gamma interferons. Mol. Cell. Biol. 1991;11:182–191. doi: 10.1128/mcb.11.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE., Jr ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. USA. 1990;87:8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 17.Levy DE, Kessler DS, Pine R, Darnell JE., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 18.Levy DE, Kessler DS, Pine R, Reich N, Darnell JE., Jr Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 19.Chang KC, Hansen E, Foroni L, Lida J, Goldspink G. Molecular and functional analysis of the virus- and interferon-inducible human MxA promoter. Arch. Virol. 1991;117:1–15. doi: 10.1007/BF01310488. [DOI] [PubMed] [Google Scholar]
- 20.Kontsek P, Karayianni-Vasconcelos G, Kontseková E. The human interferon system: characterization and classification after discovery of novel members. Acta Virol. 2003;47:201–215. [PubMed] [Google Scholar]
- 21.Pellegrini S, Schindler C. Early events in signalling by interferons. Trends Biochem. Sci. 1993;18:338–342. doi: 10.1016/0968-0004(93)90070-4. [DOI] [PubMed] [Google Scholar]
- 22.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto M, Tanaka N, Harada H, Kimura T, Yokochi T, Kitagawa M, Schindler C, Taniguchi T. Activation of the transcription factor ISGF3 by interferon-gamma. Biol. Chem. 1999;380:699–703. doi: 10.1515/BC.1999.087. [DOI] [PubMed] [Google Scholar]
- 24.Takaoka A, Mitani Y, Suemori H, Sato M, Yokochi T, Noguchi S, Tanaka N, Taniguchi T. Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science. 2000;288:2357–2360. doi: 10.1126/science.288.5475.2357. [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann A, Trilling M, Wagner M, Wilborn M, Bubic I, Jonjic S, Koszinowski U, Hengel H. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-gamma signaling and antiviral responses. J. Exp. Med. 2005;201:1543–1553. doi: 10.1084/jem.20041401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Moczygemba M, Gutch MJ, French DL, Reich NC. Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-alpha-stimulated transcription factor ISGF3. J. Biol. Chem. 1997;272:20070–20076. doi: 10.1074/jbc.272.32.20070. [DOI] [PubMed] [Google Scholar]
- 27.Tsuno T, Mejido J, Zhao T, Schmeisser H, Morrow A, Zoon KC. IRF9 is a key factor for eliciting the antiproliferative activity of IFN-alpha. J. Immunother. 2009;32:803–816. doi: 10.1097/CJI.0b013e3181ad4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kessler DS, Levy DE, Darnell JE., Jr Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc. Natl. Acad. Sci. USA. 1988;85:8521–8525. doi: 10.1073/pnas.85.22.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Kimura T, Kitagawa M, Yokochi T, Tan RS, Takasugi T, Kadokawa Y, et al. Regulation of IFN-alpha/beta genes: evidence for a dual function of the transcription factor complex ISGF3 in the production and action of IFN-alpha/beta. Genes Cells. 1996;1:995–1005. doi: 10.1046/j.1365-2443.1996.870287.x. [DOI] [PubMed] [Google Scholar]
- 31.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 32.van den Broek MF, Müller U, Huang S, Aguet M, Zinkernagel RM. Antiviral defense in mice lacking both alpha/beta and gamma interferon receptors. J. Virol. 1995;69:4792–4796. doi: 10.1128/jvi.69.8.4792-4796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van den Broek MF, Müller U, Huang S, Zinkernagel RM, Aguet M. Immune defence in mice lacking type I and/or type II interferon receptors. Immunol. Rev. 1995;148:5–18. doi: 10.1111/j.1600-065x.1995.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 34.Costa-Pereira AP, Williams TM, Strobl B, Watling D, Briscoe J, Kerr IM. The antiviral response to gamma interferon. J. Virol. 2002;76:9060–9068. doi: 10.1128/JVI.76.18.9060-9068.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonjardim CA, Ferreira PC, Kroon EG. Interferons: signaling, antiviral and viral evasion. Immunol. Lett. 2009;122:1–11. doi: 10.1016/j.imlet.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 37.Bluyssen HA, Muzaffar R, Vlieststra RJ, van der Made AC, Leung S, Stark GR, Kerr IM, Trapman J, Levy DE. Combinatorial association and abundance of components of interferon-stimulated gene factor 3 dictate the selectivity of interferon responses. Proc. Natl. Acad. Sci. USA. 1995;92:5645–5649. doi: 10.1073/pnas.92.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bluyssen HA, Levy DE. Stat2 is a transcriptional activator that requires sequence-specific contacts provided by stat1 and p48 for stable interaction with DNA. J. Biol. Chem. 1997;272:4600–4605. doi: 10.1074/jbc.272.7.4600. [DOI] [PubMed] [Google Scholar]
- 39.Majumder S, Zhou LZ, Chaturvedi P, Babcock G, Aras S, Ransohoff RM. p48/STAT-1alpha-containing complexes play a predominant role in induction of IFN-gamma-inducible protein-10 kDa (IP-10) by IFN-gamma alone or in synergy with TNF-alpha. J. Immunol. 1998;161:4736–4744. [PubMed] [Google Scholar]
- 40.Henderson YC, Chou M, Deisseroth AB. Interferon regulatory factor 1 induces the expression of the interferon-stimulated genes. Br. J. Haematol. 1997;96:566–575. doi: 10.1046/j.1365-2141.1997.d01-2057.x. [DOI] [PubMed] [Google Scholar]
- 41.Foss GS, Prydz H. Interferon regulatory factor 1 mediates the interferon-gamma induction of the human immunoproteasome subunit multicatalytic endopeptidase complex-like 1. J. Biol. Chem. 1999;274:35196–35202. doi: 10.1074/jbc.274.49.35196. [DOI] [PubMed] [Google Scholar]
- 42.Hobart M, Ramassar V, Goes N, Urmson J, Halloran PF. IFN regulatory factor-1 plays a central role in the regulation of the expression of class I and II MHC genes in vivo. J. Immunol. 1997;158:4260–4269. [PubMed] [Google Scholar]
- 43.Ramsauer K, Farlik M, Zupkovitz G, Seiser C, Kröger A, Hauser H, Decker T. Distinct modes of action applied by transcription factors STAT1 and IRF1 to initiate transcription of the IFN-gamma-inducible gbp2 gene. Proc. Natl. Acad. Sci. USA. 2007;104:2849–2854. doi: 10.1073/pnas.0610944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lou YJ, Pan XR, Jia PM, Li D, Xiao S, Zhang ZL, Chen SJ, Chen Z, Tong JH. IRF-9/STAT2 functional interaction drives retinoic acid-induced gene G expression independently of STAT1. Cancer Res. 2009;69:3673–3680. doi: 10.1158/0008-5472.CAN-08-4922. [DOI] [PubMed] [Google Scholar]
- 45.Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl. Acad. Sci. USA. 2009;106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson LR, McCormack SA, Yang CH, Pfeffer SR, Pfeffer LM. EGF induces nuclear translocation of STAT2 without tyrosine phosphorylation in intestinal epithelial cells. Am. J. Physiol. 1999;276:C419–C425. doi: 10.1152/ajpcell.1999.276.2.C419. [DOI] [PubMed] [Google Scholar]
- 48.Levin S, Hahn T. Interferon system in acute viral hepatitis. Lancet. 1982;1:592–594. doi: 10.1016/s0140-6736(82)91751-2. [DOI] [PubMed] [Google Scholar]