Abstract
Objectives
Insulin control of fatty acid metabolism has long been deemed dominated by suppression of adipose lipolysis. This study’s goal was to test the hypothesis that this single role of insulin is insufficient to explain observed fatty acid dynamics.
Methods and Results
Fatty acid kinetics were measured during a meal-tolerance test and insulin sensitivity assessed by IVGTT in overweight human subjects (n=15, BMI 35.8 ± 7.1 kg/m2). Non-steady state tracer kinetic models were formulated and tested using ProcessDB© software. Suppression of adipose release alone could not account for NEFA concentration changes postprandially, but when combined with insulin activation of fatty acid uptake was consistent with the NEFA data. The observed insulin Km for NEFA uptake was inversely correlated with both insulin sensitivity of glucose uptake (IVGTT Si) (r=−0.626, P=0.01), and whole body fat oxidation after the meal (r=−0.538, P=0.05).
Conclusions
These results support insulin regulation of fatty acid turnover by both release and uptake mechanisms. Activation of fatty acid uptake is consistent with the human data, has mechanistic precedent in cell culture, and highlights a new potential target for therapies aimed at improving the control of fatty acid metabolism in insulin-resistant disease states.
Keywords: non-esterified fatty acids, mathematical modeling, insulin, regulation
The intravenous glucose tolerance test (IVGTT), in all its incarnations, remains a staple analytical procedure in clinical endocrinology. Systems biology1 and modeling tools have a long history in this field and have yielded the clinically-important measure of insulin sensitivity, SI, widely used to measure whole-body insulin sensitivity in glucose metabolism1, 2. In recent years, pioneering efforts to extend this systems biology approach to insulin regulation of fatty acid metabolism have appeared3–6. These studies have addressed key issues of insulin-inhibited adipose lipolysis and parameter identifiability, but have not addressed the potential complication, also faced by models of glucose metabolism, that insulin regulates both entry and removal of plasma fatty acids. The relationship between insulin and NEFA clearance has been explored in pioneering studies by Carpentier and colleagues who provided evidence that “insulin per se does not stimulate plasma NEFA clearance at the whole body level.” Instead, these investigators demonstrated an inverse relationship between NEFA appearance rate and NEFA clearance, and concluded that, “…insulin stimulates plasma NEFA clearance by reducing the endogenous appearance rate of NEFA.” Studies of the specific uptake of fatty acids by different organs have been recently investigated by this group7.
The paucity of other data on physiologic control of fatty acid uptake is understandable in light of the widely held view that fatty acid removal is passive8. But passivity does not necessarily imply absence of regulation since it has been reported that, at least in rat adipocytes, long chain fatty acid uptake is predominantly transporter mediated9 and where a protein is involved, control is always possible. There are, for example, persistent hints that insulin accelerates fatty acid uptake. Early kinetic studies of fatty acid clearance in human subjects during glucose consumption revealed fatty acid fractional catabolic rates (FCRs) nearly 60% greater than FCRs measured 6–8h after glucose consumption ends10. More recently, elegant studies taking advantage of stable isotope tracer technology have reported fatty acid flux and mass data, or fatty acid metabolic clearance rates, in normal human subjects during insulin infusions of 2 or 3 h in duration11, 12. When FCRs are calculated from these results it can be determined that fatty acids are removed as much as 55% faster when insulin is 46 μU/ml compared to when insulin is 6 μU/ml11 and 42–56% faster when insulin is 44 μU/ml compared to 4.2 μU/ml12.
Another way to approach the question of insulin-mediated non-esterified fatty acid (NEFA) uptake is to contrast absolute values of fatty acid FCR as measured in different subject groups with different prevailing plasma insulin concentrations. Healthy subjects exhibited fatty acid FCRs of ~0.22 min−1 measured with radiotracer13 or stable isotope administration12. These low FCRs were reported while mean insulin concentrations were also low (4.2 ± 0.3 μU/ml). By contrast, in an obese, non-diabetic, Indian population, a much higher fatty acid FCR was reported (0.71 min−1) concurrent with a higher average fasting insulin concentration of 56 μU/ml14. Importantly, a potential cellular mechanistic basis for greater in vivo fatty acid FCR with higher insulin values is that fatty acid transporter content in the cell membrane is increased by insulin and that these transporters regulate a substantial fraction of cellular fatty acid uptake15–17. Physiological roles for membrane fatty acid transporters have recently been the subject of a comprehensive review18.
Insulin is already well-established as a potent regulator of glucose entry into plasma from liver and of fatty acid entry into plasma from adipose tissue. If insulin regulation of NEFA removal pathways could be convincingly demonstrated as a feature of human physiology, the prevailing paradigm for regulation of human fat metabolism would undergo a substantial revision. In return for the added complexity, investigators would gain new explanatory power and possibly new therapeutic targets. Moreover, the existence of insulin-sensitive NEFA uptake would increase the structural similarity between the insulin-glucose control system and the insulin-NEFA control system.
The primary goals of the present study were to test the hypothesis that insulin-activation of fatty acid uptake is a significant feature of human physiology, and to develop, in humans, a method for estimating the separate insulin sensitivities of two key determinants of plasma NEFA concentration: 1) adipose tissue release, and 2) peripheral fatty acid uptake. As a practical application of these methods, we quantified insulin sensitivities for the principal determinants of plasma fatty acid balance in 15 overweight subjects with metabolic syndrome during both a meal-tolerance test (MTT) and an IVGTT.
METHODS
Human subjects
The present project was part of a larger study of the effect of body weight on liver health in Hispanic and African-American subjects and a portion of the data (acylcarnitine concentrations) has been published recently19. Subjects (n=15) were recruited from health fairs and physician referral and the inclusion criteria were overweight or obese with elevated liver enzymes (ALT > 30, AST > 30) and/or characteristics of the metabolic syndrome20. Subjects were non-diabetic (fasting glucose <125 mg/dL), age 20–67 years, with stable body weight, and maintenance of pre-enrollment physical activity. The intensive nature of the tracer and modeling studies precluded a larger population study. Each subject participated in two metabolic tests, with the first designed to determine metabolic flux during an IVGTT and the second designed to measure the changes in metabolites following a standardized meal. This study was approved by the IRB at UT Southwestern Medical Center (approval # 062007-025).
Study design, IVGTT, and meal-tolerance test
Subjects consumed weight-maintaining diets formulated with comparison to 3-day dietary recall of usual intake for 3d before undergoing an IVGTT (admission #1) and for 7d before the MTT (admission #2). For admission #1, each subject reported to the Clinical and Translational Research Center (CTRC) at 0700 on the day of the study. One antecubital IV line was placed in each arm. The subject underwent an IVGTT and blood was collected at standard intervals up to 180 min (see fig. 4 for exact times) for measurement of non-steady state plasma glucose, NEFA, and insulin concentrations. The other IV line was used to administer a bolus of 50% dextrose (Hospira Inc., Lake Forest. IL) at 0.3 g/kg body weight at 0 min and a bolus of regular insulin (Humulin R U-100, Eli Lilly, Indianapolis, IN) at 0.03 units/kg body weight at 20 min. The IVGTT started between 0800 and 0945 after a 12h fast and after at least 30 min from IV line placement. Within 2 wks of the IVGTT, the subject was readmitted to the CTRC and underwent a constant infusion of 13C4-palmitate (7 μg/kg/min bound to albumin) to measure the total rate of appearance of plasma non-esterified fatty acids (RaNEFA) into the plasma compartment during both the fasted and fed states. Breakfast was withheld in the morning to bring the subjects to a significant fast in order to test adipose fatty acid release21. At noon, the subject consumed a meal consisting of a cocoa-flavored drink (cocoa, corn oil, heavy cream, sucrose, and skim milk), cereal, banana, and skim milk formulated in order to provide 38% of total daily energy needs. The test meal composition, analyzed using the Nutrition Data System for Research (NDSR, 2009), was (mean ± SD) 790 ± 119 kcal, 44.4 ± 9.5g fat, 15.8 ± 8.2 g protein, 90.1 ± 22.7 g carbohydrate, 4.22 ± 2.11 g fiber, and 83.3 ± 11.9 mg cholesterol. The subject was given 15 min to consume the meal. Blood samples were collected and fasting data are presented from 0600 (denoted −6h on fig. 1) and fed state data are 1200–1800 (time denoted 0h through 6h postprandially on fig. 1 and 2). Indirect calorimetry was performed using a metabolic cart (VMax Encore, Viasys Healthcare) in the hooded mode; data were obtained for 30 min while subjects were fasting (0800–0830) and again 2.5h (1430) after the initiation of the meal (1200).
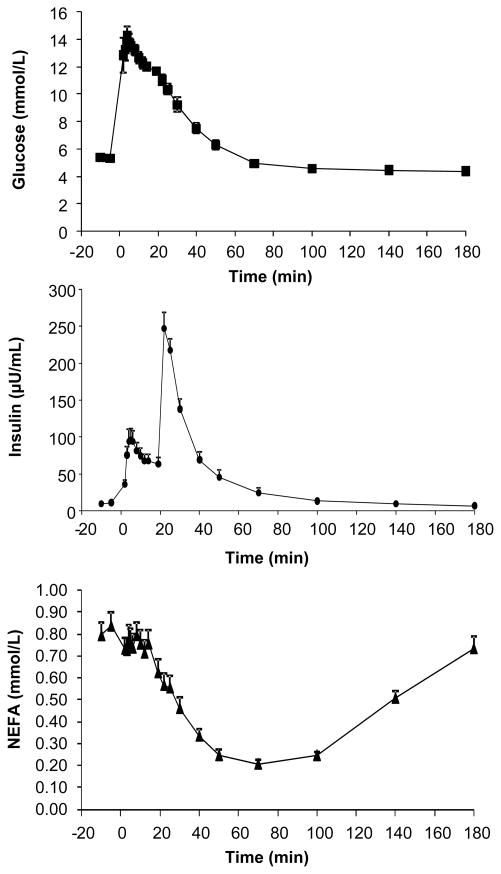
FIGURE 4. Concentrations of glucose, insulin and NEFA during the IVGTT.
Data are mean ± SE for 15 subjects.
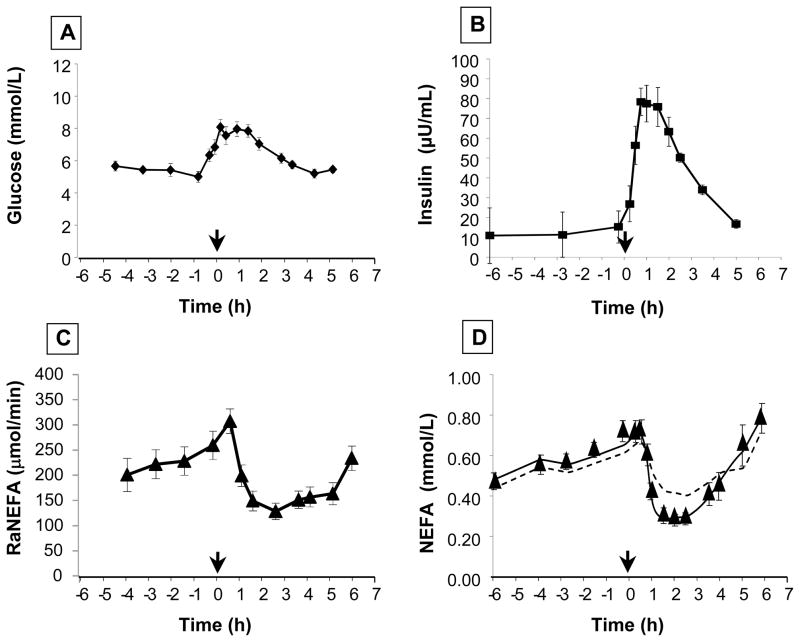
FIGURE 1. Concentrations of glucose and insulin, and RaNEFA before and after a standardized meal. Concentrations of NEFA with model fits.
Mean plasma concentrations of (A) glucose and (B) insulin, and (C) mean total fatty acid delivery to the plasma (RaNEFA) in subjects with metabolic syndrome (n = 15) fed a standardized meal at t=0 (12 noon). For details of meal, see Methods. The lines on graphs A–C represent simple linear interpolations between data points. By contrast, the lines on graph D are model solutions. The dotted line represents the model with constant plasma NEFA FCR and is incapable of accounting for the experimental data (filled triangles) representing the mean plasma NEFA concentration time course after the test meal. This model was constrained using the directly measured total RaNEFA as the only input to plasma NEFA and the value of the constant FCR was optimized during the fitting process. The solid line, by contrast, is the model solution that includes insulin-sensitive fatty acid uptake. Parameter values for this fit for the Puptake rate law (see text) are DelayFacUptake = 11 min; kmaxFacUptake = 0.063 min−1; KmFacUptake = 14 pmol/kgBW and hFacUptake = 4.2; kdiff = 0.112 min−1.
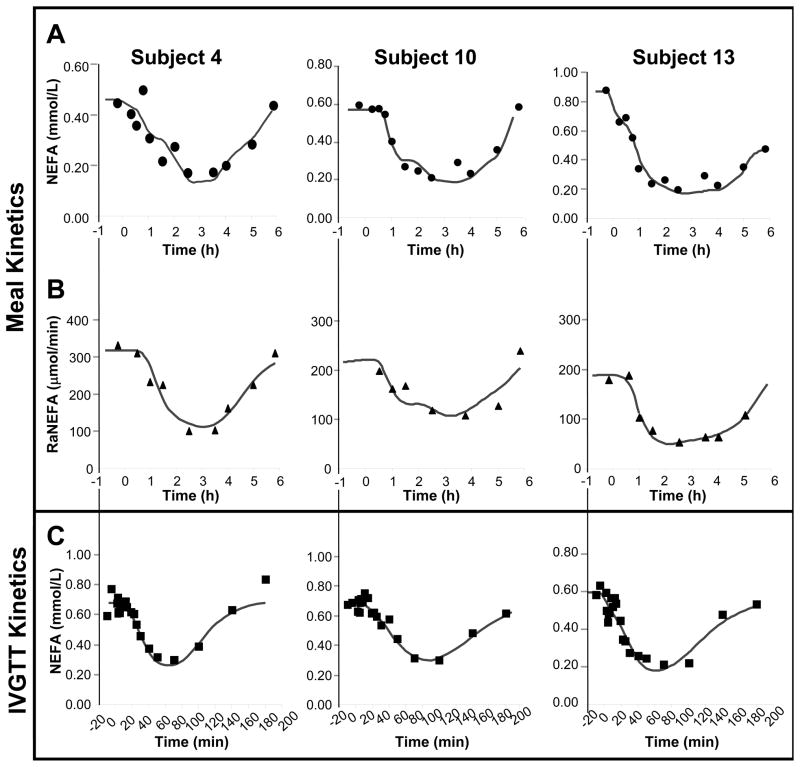
FIGURE 2. NEFA model fits to the measured meal-induced plasma NEFA concentrations and RaNEFA fluxes and IVGTT-induced plasma NEFA transients in 3 representative subjects.
Symbols represent measured data; the lines represent the model fits. Panels A and B are derived from the MTT while panel C represents data for the IVGTT. Note differing y-axes between the three subjects within each row. NEFA data were collected for 6h after the MTT and for 3h after the IVGTT. For all 15 subjects, mean parameter values are listed in table 1. Individual fits for each of the 15 subjects’ data can be found in supplementary figures I, II, and III.
Sample analysis
Blood was collected in tubes containing sodium fluoride for plasma glucose and tubes containing EDTA for plasma insulin and NEFA. Plasma from the EDTA tubes was separated immediately by centrifugation at 2850 RPM for 10 min at 5°C and stored at −20°C. Plasma NEFA concentrations were measured by enzymatic reaction (Wako Diagnostics, Richmond, VA) and insulin concentrations by ELISA (Millipore Corporation, Billerica, MA) within 2d of the study. The specimens in sodium fluoride tubes taken during the IVGTT were maintained on ice after centrifugation and analyzed with a YSI 2300 Stat Plus analyzer within 1h of collection.
Glucose minimal modeling and calculations of RaNEFA
Glucose and insulin responses during an IVGTT were analyzed using the minimal model technique and the MINMOD Millenium software22. Four indices were reported from the fitting of the glucose data. Glucose effectiveness is the capacity of glucose to mediate its own disposal, acute insulin response to glucose (AIRg), the insulin release during the first 10 min after dextrose bolus, insulin sensitivity (SI), the capacity of insulin to promote glucose disposal, and disposition index the product of AIRg and SI which represents the interaction between insulin secretion and action. For the calculations of time-varying RaNEFA, the fatty acid infusate composition, palmitate enrichments in infusates, and plasma NEFA compositions were analyzed by gas chromatography and gas chromatography/mass spectrometry, as described by Barrows et al23. Plasma concentrations of long-chain acylcarnitines (LCAC) were measured as described previously24.
Systems biology and NEFA modeling, calculations and statistics
Model development and testing were carried out as previously described25, 26. Briefly, models were formulated as systems of nonlinear ordinary differential equations using standard principles of chemical kinetics, enzyme kinetics, and transport kinetics. The resulting models were subjected to simulated protocols corresponding to the experimental clinical protocols described above. Simulations of the combined models-of-experiments were carried out using standard techniques of numerical integration, and simulated time courses were compared to the experimental data as a means of testing the hypothesis represented by the model. To give each tested model its best chance to account for a given experimental data set, model parameters such as rate constants, maximal velocities, substrate constants, and inhibition constants were adjusted using standard weighted least squares optimization. As in nearly all published models of insulin-regulated metabolism, the measured time course of plasma insulin concentration is used as a known input, or forcing function. These methods were implemented in the ProcessDB© software (Integrative Bioinformatics Inc., www.integrativebioinformatics.com/processdb.html). Models were combined with appropriate experimental protocols (infusion rates, meal contents, bolus glucose amounts, etc) in ProcessDB© and exported to the Berkeley Madonna© solver (www.berkeleymadonna.com) for numerical integration and parameter optimization. Post-simulation calculations were performed using Microsoft Excel© (2000, Seattle, WA) and statistical analyses on Statview© for Windows (Version 5.0.1, SAS Institute Inc., Berkeley, CA). A P-value < 0.05 was considered statistically significant.
There are many approaches to kinetic modeling of biological systems. Some investigators focus on parameters, some on predictions, and some on hypothesis testing. A focus on parameters results in minimal models for which each parameter value is formally identifiable with coefficients of variation, given that the model is correct, often less than 30%27. A focus on prediction28 is often desirable because it helps guide experimental design. The approach taken in the present paper falls into the third category. The model presented here is a mechanistic working hypothesis whose goal is to account successfully for complex data sets. It is not intended as a minimal model and the optimization process does not yield fully-identified parameters. Instead, the optimizer’s search of parameter space is used to give each hypothesis its best chance to account for the experimental data. Hence, the principal benefits of the hypothesis testing mode of modeling are quantitative rejection of inadequate hypotheses and quantitative corroboration of successful hypotheses26, 29.
RESULTS
Subjects studied were of Hispanic (2 male and 7 females) and African-American (1 male and 5 females) ethnicity. Clinical and laboratory values for the subjects (46 ± 8 yrs of age) revealed characteristics common of obesity. Mean BMI was 35.8 ± 7.1 kg/m2, plasma TG 1.24 ± 0.42 mmol/L, HDLc 1.30 ± 0.33 mmol/L, LDLc 3.15 ± 3.12 mmol/L, glucose 5.3 ± 0.8 mmol/L, and waist circumference 114 ± 27 cm for men and 108 ± 14 cm for women. The subjects’ homeostasis model assessment of insulin resistance (HOMA-IR) was 2.3 ± 1.2 (range 0.9–4.4)30, 31 and HbA1c was 5.9 ± 0.3%. The data above were not different between the two ethnic groups, nor did ethnicity influence any of the kinetic data presented below. Thus, the results are presented for the group as a whole.
NEFA dynamics
We first tested the null hypothesis that changes in NEFA release from adipose were able to account for the observed reductions in plasma NEFA concentrations after a MTT. Figures 1A and 1B show the concentrations of glucose and insulin before and after the mixed meal. Total RaNEFA was measured using continuous IV infusion of 13C4-palmitate (fig. 1C). If inhibition of adipose fatty acid release is sufficient to explain the observed decreases in plasma NEFA concentration then a model consisting of a constant NEFA FCR and a time-varying NEFA input into plasma measured by the RaNEFA would be capable of accounting for the observed changes. As shown in figure 1D, this was not the case (dotted line). Plasma NEFA concentrations declined faster and further than could be accounted for by the measured decreases in RaNEFA. We therefore tested a more complex hypothesis by augmenting the simple mass action (constant FCR) uptake flux with an insulin-facilitated uptake flux as follows:
where Nplasma = plasma NEFA, kmaxFacUptake is the maximal insulin-sensitive rate constant, hFacUptake is the Hill coefficient and KmFacUptake is the Michaelis constant, all for insulin-activated, facilitated uptake (FacUptake) into tissues. All parameters for facilitated uptake characterize the net effect of insulin on proteins such as CD36, fatty acid transfer protein, di- and mono-acylglycerol transferase, and other events combined that would increase the transport of fatty acids into the cells. The term, similar to that used in4 Iplasma(t - DelayFacUptake) represents plasma insulin delayed by a time denoted DelayFacUptake, corresponding to the time required for insulin distribution, binding, and intracellular signaling to affect these proteins. This insulin-sensitive uptake process is the defining characteristic of the model presented in this paper; the model is denoted the INEFA model. In contrast to the inability of the classical model to fit the MTT data (dotted line, fig. 1D), adding an insulin-facilitated process to the constant FCR (mass action) term (solid line, fig. 1D) allowed the model to account for the observed changes in NEFA concentrations during the MTT given the directly measured RaNEFA. The first term in the Puptake rate law thus represents the classical insulin-insensitive passive net diffusive component of NEFA uptake with kdiff equivalent to the constant FCR. The second term adds a fatty acid transport system whose maximal rate is a saturable function of plasma insulin concentration. As assessed by comparing the largest value of calculated fatty acid uptake flux to its control value, facilitated uptake is increased by a factor of 2.2 while adipose release rate, as measured by RaNEFA, is inhibited by a factor of 1.9. Measurement of total RaNEFA not only provides the opportunity to quantify insulin-sensitive fatty acid uptake, but also permits direct quantitative characterization of insulin sensitive fatty acid release into plasma. This was accomplished by least squares optimization of parameters in the following rate law for adipose tissue fatty acid release:
Here, PAdipRel is the flux (μmol min−1kg−1) of fatty acids released into plasma as measured by the total RaNEFA technique. This includes fatty acids released from both intra-adipocyte lipolysis (mediated by TG-lipase and hormone sensitive lipase) and extracellular lipolysis, mediated by the spillover of fatty acids from LPL-mediated lipoprotein hydrolysis, primarily occurring at the adipose32, 33. The term VmAdipRel (μmol min−1kg−1) represents the maximal flux of insulin-sensitive adipose fatty acid release. The term Iplasma (t-DelayAdipRel) represents the time course of plasma insulin concentration delayed by time, DelayAdipRel accounts for delays encountered in propagating the insulin signal from the plasma into the interstitial space, receptor binding, and intracellular signaling cascades. KiInsulin represents the sensitivity of fatty acid release to insulin (half-maximally affected). Finally, hAdipRel is the Hill cooperativity coefficient for insulin sensitivity of fatty acid release; increasing values of the Hill coefficient correspond to more and more on-or-off, switch-like behavior. Both nonlinear rate laws were derived based on parallels to the classical rapid equilibrium methods of enzyme kinetics25.
The equations for PAdipRel and Puptake represent the input and output processes for the INEFA model. For each individual subject these equations were fitted simultaneously to the non-steady state plasma NEFA concentrations and total RaNEFA data. The individual fits for three subjects are shown in figure 2A (NEFA concentrations) and 2B (RaNEFA) and the fits for each of the 15 subjects are shown in supplementary figure I for NEFA concentrations and supplementary figure II, for the RaNEFA models. This procedure yielded quantitative parameter estimates for all the INEFA model parameters, which are summarized in table 1.
Table 1.
Parameter values obtained for the insulin sensitive processes of plasma NEFA metabolism derived from the meal tolerance test (MTT) and the IVGTT
| Process | Parameter description | Parameter name in model | Units | MTT | IVGTT | P-value† | ||
|---|---|---|---|---|---|---|---|---|
| Mean* | SD | Mean* | SD | |||||
| Fatty Acid Release | Insulin signaling delay | DelayAdipRel | Min | 47.4 | 27.0 | 36.7 | 16.4 | 0.101 |
| Vmax adipose release | VmAdipRel | μmol min−1 kgBW−1 | 5.61 | 2.6 | 5.9 | 3.5 | 0.407 | |
| Insulin inhibition constant | KiInsulin | pmol/kg BW | 11.0 | 9.5 | 9.7 | 9.7 | 0.360 | |
| Hill cooperativity coefficient | hAdipRel | Unitless | 1.59 | 1.6 | 1.2 | 0.6 | 0.211 | |
|
| ||||||||
| Fatty Acid Uptake | Diffusive fatty acid uptake | kdiff | min−1 | 0.08 | 0.04 | 0.09 | 0.06 | 0.307 |
| Insulin signaling delay | DelayFacUptake | Min | 24.1 | 25.9 | 29.4 | 26.7 | 0.293 | |
| kmax of fatty acid uptake | kmaxFacUptake | min−1 | 0.12 | 0.12 | 0.10 | 0.03 | 0.348 | |
| Km for insulin | KmFacUptake | pmol/kg BW | 12.3 | 9.2 | 13.0 | 10.2 | 0.421 | |
| Hill cooperativity coefficient | hFacUptake | Unitless | 4.4 | 3.8 | 3.8 | 2.8 | 0.312 | |
N=15 subjects for all parameters.
Abbreviations: BW, body weight; Kdiff, constant for non-protein-mediated (diffusive) fatty acid uptake
Paired t-test for parameter values generated from the MTT and IVGTT.
One goal of this study was to establish the feasibility of quantifying the insulin sensitivity of fatty acid release and fatty acid uptake in individual subjects based on a single protocol involving one tracer infusion and one non-steady state physiological perturbation. Since the resulting data provide one time course (plasma NEFA concentration) containing all the information on the four parameters of fatty acid uptake and another time course (total RaNEFA) containing all the information on the three parameters characterizing fatty acid release, the method appears well suited to future work targeting detailed parameter identifiability and characterization of additional subject populations. The INEFA model is diagrammed in figure 3 and the complete INEFA model equations are as follows:
where Nplasma is plasma NEFA concentration, PAdipRel is the fatty acid flux delivered to plasma via adipose lipolysis and release (from adipose lipoprotein lipase, triacylglycerol lipase, hormone sensitive and monoglyceride lipase), Puptake is the flux of fatty acids taken up by peripheral tissues, Iplasma is plasma insulin concentration, Vm is maximal velocities or maximal tissue capacities, Ki and Km are inhibition and Michaelis constants, kmaxFacUptake is the maximum insulin-stimulated rate constant for fatty acid uptake, DelayAdipRel and DelayFacUptake are delays associated with insulin signaling, and hi are Hill coefficients permitting cooperativity in regulatory mechanisms.
FIGURE 3. INEFA model diagram.
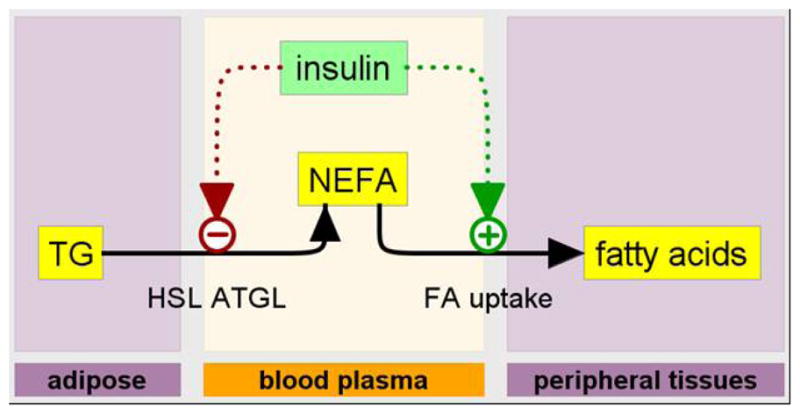
The INEFA model of insulin-regulated plasma NEFA dynamics including two insulin-regulated processes: 1) fatty acid release from adipocytes and 2) fatty acid uptake by peripheral tissues (adipose, heart, skeletal muscle). Model state variables are molecules (indicated by molecule names in rectangles) in particular physiological places (adipose, blood plasma, and peripheral tissues) indicated by the labeled “swim lanes.” Dotted arrows represent the physiological distribution and intracellular signaling delays associated with the two insulin-mediated regulatory control systems.
Model extension to analysis of standard IVGTT
Finally, we tested the hypothesis that the INEFA model, including the insulin-sensitive fatty acid uptake required to model the MTT, could predict the changes in NEFA dynamics during an insulin-modified IVGTT. Presented in figure 4 are the mean concentrations of glucose, insulin, and NEFA during the IVGTT. The nadir for NEFA concentration occurred at 70 min and was 0.21 ± 0.11 mmol/L, representing a 72.7 ± 14.9% suppression of NEFA from fasting. For the glucose/insulin system, calculated metabolic parameters using the MINMOD Millennium program revealed that the acute insulin response to glucose was 603 ± 396 μU/mL · min, the disposition index was 1238 ± 644, while the glucose effectiveness was 0.010 ± 0.007 min−1. Mean whole-body insulin sensitivity was 2.51 ± 1.46 × 10−4 min−1 · μU−1 · mL−1 (range 0.89–6.25), which was slightly greater than the value of 2.5 × 10−4 min−1 · μU−1 · mL−1, a commonly-used used cut-off, below which subjects are characterized as insulin resistant34.
Using measured IVGTT plasma insulin concentrations (fig. 4B) as a known input, the INEFA model was capable of accounting for the full range of plasma NEFA concentration time courses recorded during IVGTT. Data and model fits for the three representative subjects are shown in figure 2C. Model fits for all 15 subjects are shown in supplementary figure III. Importantly, the parameter values for these fits were constrained by knowledge of the parameters extracted from the MTT data in the same subjects. Standard least squares fitting of the IVGTT NEFA data was undertaken with upper and lower bounds on adipose fatty acid release parameters (VmAdipRel, KiInsulin, DelayAdipRel, and hAdipRel) and tissue fatty acid uptake parameters (kdiff, KmaxFacUptake, KmFacUptake, DelayFacUptake, and hFacUptake) set to ± 50% of the values obtained for each individual subject in the MTT analysis above. Since the INEFA model uses plasma insulin as a known input, it was occasionally also useful to allow the optimizer to adjust the basal value of Iplasma that was used as the model input before and after the period when insulin measurements were actually taken. It should be emphasized that resolution of two insulin sensitive processes depends on measuring RaNEFA. The time course of plasma NEFA concentrations alone, in either MTT or IVGTT, can neither demonstrate nor quantify insulin-sensitive fatty acid uptake.
Thus, a single model (INEFA) with four parameters characterizing adipose fatty acid release and five parameters characterizing tissue fatty acid uptake provided a mechanistic accounting for observed NEFA dynamics in both MTT and IVGTT. We validated the model derived from the meal studies by showing that it could also account for the NEFA response to an IVGTT. As shown in table 1, a paired t-test on each parameter (comparing meal-derived and IVGTT-derived values) reveals no significant difference. Development of the INEFA model thus uncovered the presence of a physiological control mechanism that activates tissue fatty acid uptake at the same time adipose release is inhibited. By testing against MTT and IVGTT data in 15 overweight subjects, we then corroborated the hypothesis that insulin is the required regulator of human fatty acid uptake.
Correlations between MINMOD and INEFA parameters
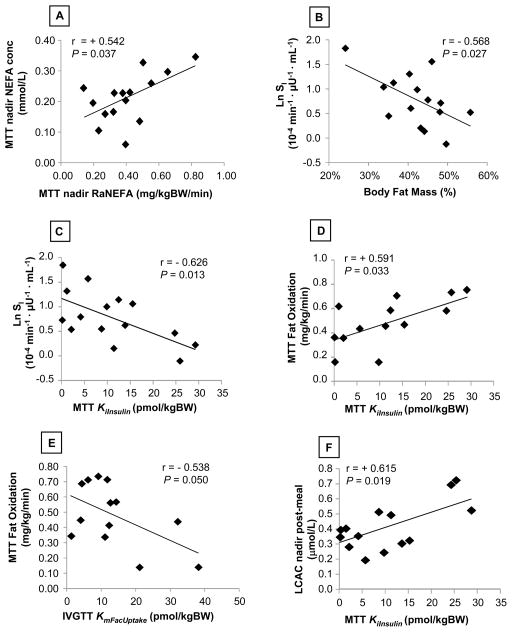
Given the wide use of the IVGTT in clinical research to generate measures of glucose metabolism, we determined whether any relationships existed between the standard MINMOD parameters, subject characteristics, and parameters of fatty acid metabolism obtained from the INEFA model. As expected, NEFA concentrations after the meal were a function of the release rates (fig. 5A) and the greater the body fat mass of the subjects the lower the insulin sensitivity (fig. 5B). However, a better relationship was observed between the Ki for insulin’s effect on adipose fatty acid release after the meal (KiInsulin) and the natural log of the IVGTT Si supporting the concept that one mediator of the negative association between body fat and insulin sensitivity is a lack of suppression of fatty acid release (fig. 5C, r = −0.626, P = 0.013). Further, whole body oxidation of fatty acids after the meal correlated positively with the MTT KiInsulin (fig. 5D) and negatively with the IVGTT KmFacUptake (fig. 5E). These data support the effect of insulin on both fatty acid release rates and peripheral uptake rates to influence the oxidation of lipid in tissues. Lastly, plasma long-chain acylcarnitines (LCAC) represent the products of incomplete fatty acid oxidation in the mitochondria23. The present results connect higher KiInsulin to reduce fatty acid release to overload of mitochondrial β-oxidation (fig. 5F).
FIGURE 5. Relationships between INEFA model parameters, body fat, and metabolic variables.
The figures demonstrate the relationships between parameters obtained from the INEFA model (KiInsulin for insulin suppression of fatty acid release and KmFacUptake for insulin stimulation of fatty acid uptake), those obtained during the IVGTT (SI), and during the MTT (RaNEFA, body composition, and plasma concentrations of NEFA and long-chain acylcarnitines, LCAC). Abbreviations: MTT, meal-tolerance test; FatOx, whole-body fat oxidation assessed by indirect calorimetry, and LCAC represent the saturated, mono-, and poly-unsaturated acylcarnitines with chain lengths from 12–18.
Discussion
Modeling and systems biology are particularly well suited to the complexities of metabolism and metabolic disease, explaining perhaps, the extensive literature linking these two disciplines - reviewed in3, 4, 13, 22, 26, 35–38. In this paper we have applied these techniques to human fatty acid metabolism and have demonstrated unequivocally that the classical (constant FCR) model of peripheral fatty acid uptake is incomplete. The constant FCR model fails to account for plasma NEFA dynamics during a MTT when total RaNEFA is known. Some additional mechanism is inescapably required.
Because there is evidence, based on studies in cultured cells39, 40 that insulin promotes fatty acid uptake, we tested the hypothesis that fatty acid uptake is, in part, regulated by insulin in a group of subjects with a wide range of insulin sensitivities. This hypothesis was corroborated using measured plasma insulin as the stimulus. The results thus make a strong case for insulin activation of human fatty acid uptake from plasma. However, not all published works favor a significant contribution of insulin-stimulated NEFA uptake in tissues. NEFA uptake in perfused rodent skeletal muscle was shown to be independent of insulin41 and NEFA concentration itself appears to play a role in its clearance rate7. Insulin increases cellular uptake of glucose which through glycolysis could potentially increase fatty acid re-esterification and reduce lipolysis by providing alpha-glycerol phosphate to form the glycerol backbone for TG synthesis. However, Caruso et al found no insulin-independent effect of hyperglycemia to reduce adipose lipolysis in humans in vivo42.
Since it has been proposed that insulin increases both skeletal muscle and adipose blood flow43, 44, an alternative hypothesis might include insulin-mediated control of vascular fatty acid delivery. This mechanism would require that fatty acid uptake is flow-limited, but the literature as a whole appears to support diffusion/transport as the limiting determinant of fatty acid uptake18. This position, combined with the increasingly prominent cell biological evidence for insulin activated plasma membrane fatty acid transport15, 39, 40, led us to focus on the parenchymal plasma membrane as the site of insulin action.
Clinical research is most practical and least expensive when only blood samples are required, but just as with glucose, plasma NEFA data alone are insufficient to calculate the absolute magnitude of fatty acid fluxes entering and leaving the plasma compartment. Indeed, concentration changes can, at most, yield the integrated difference between input and output fluxes. This inescapable fact requires that we find additional constraints to support data analysis. Frequently, as in the present work, those constraints have come from tracer kinetics. Our use of tracer-based RaNEFA flux measurements can be conceived as a two-step process. First, the RaNEFA flux data were used as known inputs to the incompletely known fatty acid uptake system. This permits analysis of the factors regulating fatty acid uptake by tissues. Second, the RaNEFA data themselves were fitted to a rate law representing fatty acid release which permitted characterization of adipose insulin sensitivity.
Other modeling approaches to this need for constraints have included carefully-selected priors during parameter optimization3 and an explicit reliance on estimates from the literature4. Remarkably, despite very different approaches to this issue, our range of maximal adipose release rates (2.04 – 10.38 μmol min−1 kgBW−1) includes the value calculated from table 4 of Periwal et al. as (l0 + l2)(0.045 L/kgBW) = 2.04 μmol/min/kg4. The INEFA mean value, 5.61 μmol/min/kg is greater, but this general agreement suggests that different methods are converging on the same answer. Useful constraints come in many forms. In the early literature on insulin and glucose kinetic modeling, Sherwin et al identified the utility of extravascular insulin concentration as the direct regulator of glucose uptake38. Since these investigators suggested that the same interstitial insulin concentration is obtained in both skeletal muscle and adipose tissue, we tested the hypothesis that extravascular insulin (calculated by applying our measured plasma insulin to Model A of Sherwin et al38) would provide the requisite delays and dynamics to account for the observed NEFA data. This would have been a considerable simplification since insulin kinetics have been shown to vary little in human subjects over a large range of BMI45. Unfortunately, if extravascular insulin dynamics were modeled as rate-determining for adipocyte release rate and/or myocyte fatty acid uptake, then the FCR of plasma NEFA was forced to be 10-fold slower than FCRs measured in our subjects by the 13C-palmitate RaNEFA technique or reported by others6, 10, 11, 13. This inconsistency suggests that the rate-determining steps in controlling plasma NEFA reside in the cellular insulin signaling pathways. This is desirable, because it offers the possibility of inferring changes in insulin signaling from plasma NEFA measurements during IVGTT and MTT. We conclude that the dynamics of extravascular insulin, by themselves, are insufficient to account for the required delays in insulin signaling. For this reason, we do not include an explicit extravascular insulin compartment; to do so would needlessly increase the number of parameters to be estimated from the insulin and NEFA data.
The INEFA model builds on the foundation laid by pioneering models of NEFA metabolism. Consequently, it has many structural features in common with those models. All published models include insulin inhibition of adipose lipolysis3, 4. This is well known as a major physiological determinant of the plasma NEFA time course. Another feature shared by all models is some method of delaying the action of insulin. One useful approach is the “remote insulin compartment”3. We adopted the closely-related first order delay from Jelic4. The model presented by Jelic4 included insulin activation of LPL. Unfortunately we were unable to resolve an LPL contribution and this would be a goal for future studies. We adopted the concept of first order delays, not only for insulin inhibition of adipose lipases4 but also for insulin activation of fatty acid uptake. Other models3, 37 treat this delay by including a remote insulin compartment, driven by plasma insulin and turning over at a some slower rate to be determined by fitting the NEFA data. Each method has strengths; the first order delay is arguably more physiological and the parameters of a delayed or “remote” insulin compartment may be more readily identifiable.
Some published models control all insulin responsive processes with the same insulin-derived signal, as in the model XH in Periwal3, while Jelic4 permitted different signaling and response times for different processes, an aspect also highlighted in the Model YH of Periwal3. We found different response times to be both necessary and reasonable in terms of the underlying molecular cell biology. An innovative approach by one group5, 46 extends the minimal model concept even further by treating plasma glucose, rather than plasma insulin, as the driver of the surrogate control signal, obviating the need for insulin measurements altogether. Thus, the spectrum of models ranges from those with a focus on parameter estimation, to those endeavoring to represent explicitly the relevant physiological and molecular processes as they are currently understood. Various rate laws have been used to quantify the lipolytic flux of fatty acids from adipose tissue. Periwal et al have done a careful evaluation of at least 11 such rate laws and favor one that is the sum of an insulin-insensitive basal flux and a standard Hill term for the insulin-sensitive component3. This same rate law, without a Hill coefficient, was also used by Jelic et al4. For our subjects, the constant term was consistently driven to zero during parameter optimization so that the rate law is identical to the H(l0 = 0) model type in Periwal3, except that we also include the first order delay for insulin signaling as proposed in4 Periwal’s value of X2, which corresponds to our KiInsulin, is 3.1-fold smaller3. Hill coefficients are quite close; our MTT mean is 1.59 and theirs is 1.88. Differences in parameter values may well follow from our conclusion that an insulin-insensitive component of adipose lipolysis is unnecessary. Our values for the hormone sensitive lipase signaling delay (mean 55 min, range: 3–162 min) surround the value of 30 min reported for normal subjects4, but again reflect a wide range of signaling speeds in our subjects with metabolic syndrome.
Remarkably, mean insulin Ki for adipose release (KiInsulin) and the Km values for insulin activation of fatty acid uptake in the INEFA model were found to be very similar numbers (11.0 and 12.3 pmol/kgBW). If these were distributed in plasma (0.045 L/kgBW) they would correspond to approximately 290 pM or 42 μU/mL. If distributed in the extravascular-extracellular fluid (0.097 L/kgBW), then 140 pM or 20 μU/mL. For either volume of distribution, our Ki for release is substantially greater than the values in either Jelic (7.2 μU/mL) or Periwal (11.3 μU/mL). This is consistent with reduced insulin sensitivity in our metabolic syndrome subjects. Importantly, however, Ki and Km values in a given subject were sometimes very different. This suggests that in some subjects adipose NEFA release is more sensitive to insulin than is fatty acid uptake - while in other subjects the opposite is true. It may be interesting to determine whether this ratio of insulin sensitivities correlates with any of the phenotype variables. If the Ki/Km ratio is high (>2), then this would indicate more insulin resistance at the adipose, while if this ratio if closer to 1, it would indicate good suppression of adipose fatty acid release rates after meal, and relatively poor activation of fatty acid uptake after a meal.
From a clinical perspective, we found that the sensitivity of the glucose system to insulin is also reflected in the ability of insulin to reduce fatty acid release into plasma (fig. 5C). Further, plasma long-chain acylcarnitines (LCAC) represent the products of incomplete fatty acid oxidation in the mitochondria and we have shown previously in these subjects that the greater the fatty acid flux after a meal, the higher the plasma concentration of LCAC occurring at the same time24. That result is extended here by showing that in the setting of poor, insulin-stimulated glucose disposal (fig. 5C), higher quantities of insulin were also needed to suppress adipose fatty acid release (KiInsulin). In these same subjects, higher post-meal fat oxidation (fig. 5D) was observed along with higher concentrations of products reflecting incomplete fatty acid oxidation (LCAC, fig. 5F). The data are internally consistent and offer an integrated view of the impact of adipose insulin resistance on plasma NEFA flux and concentration. The sum of these relationships reveals that the insulin sensitivity values calculated from an IVGTT with MINMOD mirror the more physiologic inter-relationships observed between glucose and fatty acid metabolism occurring after consumption of a mixed meal in obese subjects.
In summary, we have developed and tested a new model (INEFA) of human fatty acid metabolism whose structure is, in part, informed by recent molecular and cell biological findings. By doing so we have provided evidence, in a direct parallel to insulin regulation of glucose metabolism, that insulin is a significant activator of fatty acid uptake from plasma to tissues in humans with metabolic syndrome. Importantly, the methods described here permit quantification of the separate insulin sensitivities of both adipose fatty acid release and peripheral fatty acid uptake in a single clinical protocol. It will, of course, be important to confirm the generality of this control mechanism in other groups of subjects and in different disease states.
Supplementary Material
Acknowledgments
The authors would like to express their appreciation to the research subjects for their time and enthusiasm, to the staff of the Clinical Translational Research Center and particularly to Nurse Dora Bradford at UT Southwestern Medical Center for their excellent care of the research subjects.
SOURCES OF FUNDING
This study was funded with support from NIH grants 5RL1DK081187 (PI E.J. Parks), 5PL1DK081183 (PI E. Livingston), UL1DE019584 (PI J. Horton), and CTSA NIH Grant UL1-RR024982. This study is registered with ClinicalTrials.gov (NCT01371396).
Abbreviations
- AIR
acute insulin response to glucose
- BW
body weight
- CTRC
clinical translational research center
- DI
disposition index
- FCR
fractional catabolic rate
- INEFA
insulin activation of plasma non-esterified fatty acid uptake
- IVGTT
insulin-modified, frequently-sampled intravenous glucose tolerance test
- LCAC
long-chain acylcarnitines
- LPL
lipoprotein lipase
- MTT
meal-tolerance test
- NEFA
non-esterified fatty acids
- RaNEFA
rate of appearance of NEFA into plasma
- Sg
glucose effectiveness
- SI
insulin sensitivity
- TG
triacylglycerols
Footnotes
Portions of this work were presented at the American Diabetes Association National Meeting in Orlando, FL, June 2010.
DISCLOSURE
R.D. Phair and S.A. Lapidot are employed by Integrative Bioinformatics, Inc, developer of the ProcessDB© modeling software used in this study. The authors had no conflicts of interest except for R.D. Phair, who is Chief Science Officer of Integrative Bioinformatics, Inc.
References
- 1.Bergman RN, Ider YZ, Bowden CR, Cobelli C. Quantitative estimation of insulin sensitivity. Am J Physiol Endocrinol Metab. 1979;236:E667–E677. doi: 10.1152/ajpendo.1979.236.6.E667. [DOI] [PubMed] [Google Scholar]
- 2.Bergman RN. Orchestration of glucose homeostasis: From a small acorn to the california oak. Diabetes. 2007;56:1489–1501. doi: 10.2337/db07-9903. [DOI] [PubMed] [Google Scholar]
- 3.Periwal V, Chow CC, Bergman RN, Ricks M, Vega GL, Sumner AE. Evaluation of quantitative models of the effect of insulin on lipolysis and glucose disposal. Am J Physiol Regul Integr Comp Physiol. 2008;295:R1089–R1096. doi: 10.1152/ajpregu.90426.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jelic K, Hallgreen CE, Colding-Jorgensen M. A model of NEFA dynamics with focus on the postprandial state. Ann Biomed Eng. 2009;37:1897–1909. doi: 10.1007/s10439-009-9738-6. [DOI] [PubMed] [Google Scholar]
- 5.Boston RC, Moate PJ. A novel minimal model to describe NEFA kinetics following an intravenous glucose challenge. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1140–R1147. doi: 10.1152/ajpregu.00749.2007. [DOI] [PubMed] [Google Scholar]
- 6.Chow CC, Periwal V, Csako G, Ricks M, Courville AB, Miller BV, Vega GL, Sumner AE. Higher acute insulin response to glucose may determine greater free fatty acid clearance in African-American women. J Clin Endocrin Metab. 2011;96:2456–2463. doi: 10.1210/jc.2011-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpentier AC, Frisch F, Brassard P, Lavoie F, Bourbonnais A, Cyr D, Giguère R, Baillargeon JP. Mechanism of insulin-stimulated clearance of plasma nonesterified fatty acids in humans. Am J Physiol Endocrinol Metab. 2007;292:E696–E701. doi: 10.1152/ajpendo.00423.2006. [DOI] [PubMed] [Google Scholar]
- 8.Frayn KN. Metabolic Regulation: A Human Perspective. 2. Oxford-Blackwell; 2003. pp. 339–340. [Google Scholar]
- 9.Stump DD, Fan X, Berk PD. Oleic acid uptake and binding by rat adipocytes define dual pathways for cellular fatty acid uptake. J Lipid Res. 2001;42:509–520. [PubMed] [Google Scholar]
- 10.Nestel PJ, Ishikawa T, Goldrick RB. Diminished plasma free fatty acid clearance in obese subjects. Metabolism. 1978;27:589–597. doi: 10.1016/0026-0495(78)90025-2. [DOI] [PubMed] [Google Scholar]
- 11.Shadid S, Kanaley JA, Sheehan MT, Jensen MD. Basal and insulin-regulated free fatty acid and glucose metabolism in humans. Am J Physiol Endocrinol Metab. 2007;292:E1770–E1774. doi: 10.1152/ajpendo.00655.2006. [DOI] [PubMed] [Google Scholar]
- 12.Mittendorfer B, Liem O, Patterson BW, Miles JM, Klein S. What does the measurement of whole-body fatty acid rate of appearance in plasma by using a fatty acid tracer really mean? Diabetes. 2003;52:1641–1648. doi: 10.2337/diabetes.52.7.1641. [DOI] [PubMed] [Google Scholar]
- 13.Eaton RP, Berman M, Steinberg D. Kinetic studies of plasma free fatty acid and triglyceride metabolism in man. J Clin Invest. 1969;48:1560–1579. doi: 10.1172/JCI106122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taskinen MR, Bogardus C, Kennedy A, Howard BV. Multiple disturbances of free fatty acid metabolism in noninsulin-dependent diabetes. J Clin Invest. 1985;76:637–644. doi: 10.1172/JCI112016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stahl A, Evans JG, Pattel S, Hirsch D, Lodish HF. Insulin causes fatty acid transport protein translocation and enhanced fatty acid uptake in adipocytes. Dev Cell. 2002;2:477–488. doi: 10.1016/s1534-5807(02)00143-0. [DOI] [PubMed] [Google Scholar]
- 16.Jain SS, Chabowski A, Snook LA, Schwenk RW, Glatz JF, Luiken JJ, Bonen A. Additive effects of insulin and muscle contraction on fatty acid transport and fatty acid transporters, FAT/CD36, FABPpm, FATP1, 4 and 6. FEBS Lett. 2009;583:2294–2300. doi: 10.1016/j.febslet.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 17.Bonen A, Luiken JJ, Liu S, Dyck DJ, Kiens B, Kristiansen S, Turcotte LP, Van Der Vusse GJ, Glatz JF. Palmitate transport and fatty acid transporters in red and white muscles. Am J Physiol Endocrinol Metab. 1998;275:E471–E478. doi: 10.1152/ajpendo.1998.275.3.E471. [DOI] [PubMed] [Google Scholar]
- 18.Glatz JF, Luiken JJ, Bonen A. Membrane fatty acid transporters as regulators of lipid metabolism: Implications for metabolic disease. Physiol Rev. 2010;90:367–417. doi: 10.1152/physrev.00003.2009. [DOI] [PubMed] [Google Scholar]
- 19.Sunny NE, Parks EJ, Browning JD, Burgess SC. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metabolism. 2011;14:804–811. doi: 10.1016/j.cmet.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, Franklin BA, Gordon DJ, Krauss RM, Savage PJ, Smith SC, Spertus JA, Costa F. Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute scientific statement. Curr Opin Cardiol. 2006;21:1–6. doi: 10.1097/01.hco.0000200416.65370.a0. [DOI] [PubMed] [Google Scholar]
- 21.Parks EJ, Krauss RM, Christiansen MP, Neese RA, Hellerstein MK. Effects of a low-fat, high-carbohydrate diet on VLDL-triglyceride assembly, production and clearance. J Clin Invest. 1999;104:1087–1096. doi: 10.1172/JCI6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boston RC, Stefanovski D, Moate PJ, Sumner AE, Watanabe RM, Bergman RN. MINMOD Millennium: a computer program to calculate glucose effectiveness and insulin sensitivity from the frequently sampled intravenous glucose tolerance test. Diabetes Technol Ther. 2003;5:1003–1015. doi: 10.1089/152091503322641060. [DOI] [PubMed] [Google Scholar]
- 23.Barrows BR, Parks EJ. Contributions of different fatty acid sources to very low-density lipoprotein-triacylglycerol in the fasted and fed states. J Clin Endo Metab. 2006;91:1446–1452. doi: 10.1210/jc.2005-1709. [DOI] [PubMed] [Google Scholar]
- 24.Ramos-Roman MA, Sweetman L, Valdez MJ, Parks EJ. Postprandial changes in plasma acylcarnitine concentrations as markers of fatty acid flux in metabolic syndrome. Metabolism. 2012;61:202–212. doi: 10.1016/j.metabol.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phair RD. Development of kinetic models in the nonlinear world of molecular cell biology. Metabolism. 1997;46:1489–1495. doi: 10.1016/s0026-0495(97)90154-2. [DOI] [PubMed] [Google Scholar]
- 26.Phair RD, Misteli T. Kinetic modelling approaches to in vivo imaging. Nat Rev Mol Cell Biol. 2001;2:898–907. doi: 10.1038/35103000. [DOI] [PubMed] [Google Scholar]
- 27.Cobelli C, Foster DM. Compartmental models: Theory and practice using the SAAM II software system. Adv Exp Med Biol. 1998;445:79–101. doi: 10.1007/978-1-4899-1959-5_5. [DOI] [PubMed] [Google Scholar]
- 28.Cirit M, Haugh JM. Quantitative models of signal transduction networks: How detailed should they be? Commun Integr Biol. 2011;4:353–356. doi: 10.4161/cib.4.3.15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beard DA, Kushmerick MJ. Strong inference for systems biology. PLoS Comput Biol. 2009;5:e1000459. doi: 10.1371/journal.pcbi.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 31.Salgado AL, Carvalho L, Oliveira AC, Santos VN, Vieira JG, Parise ER. Insulin resistance index (HOMA-IR) in the differentiation of patients with non-alcoholic fatty liver disease and healthy individuals. Arq Gastroenterol. 2010;47:165–169. doi: 10.1590/s0004-28032010000200009. [DOI] [PubMed] [Google Scholar]
- 32.Coppack SW, Fisher RM, Gibbons GF, Humphreys SM, McDonough MJ, Potts JL, Frayn KN. Postprandial substrate deposition in human forearm and adipose tissues in vivo. Clin Sci (Lond) 1990;79:339–348. doi: 10.1042/cs0790339. [DOI] [PubMed] [Google Scholar]
- 33.Evans K, Clark ML, Frayn KN. Effects of an oral and intravenous fat load on adipose tissue and forearm lipid metabolism. Am J Physiol. 1999;276:E241–E248. doi: 10.1152/ajpendo.1999.276.2.E241. [DOI] [PubMed] [Google Scholar]
- 34.Haffner SM, Howard G, Mayer E, Bergman RN, Savage PJ, Rewers M, Mykkänen L, Karter AJ, Hamman R, Saad MF. Insulin sensitivity and acute insulin response in African-Americans, non-Hispanic whites, and Hispanics with NIDDM: the Insulin Resistance Atherosclerosis Study. Diabetes. 1997;46:63–69. doi: 10.2337/diab.46.1.63. [DOI] [PubMed] [Google Scholar]
- 35.Barrett PH, Chan DC, Watts GF. Thematic review series: patient-oriented research. Design and analysis of lipoprotein tracer kinetics studies in humans. J Lipid Res. 2006;47:1607–1619. doi: 10.1194/jlr.R600017-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.Patterson BW, Mittendorfer B, Elias N, Satyanarayana R, Klein S. Use of stable isotopically labeled tracers to measure very low density lipoprotein-triglyceride turnover. J Lipid Res. 2002;43:223–233. [PubMed] [Google Scholar]
- 37.Roy A, Parker RS. Dynamic modeling of free fatty acid, glucose, and insulin: an extended “minimal model”. Diabetes Technol Ther. 2006;8:617–626. doi: 10.1089/dia.2006.8.617. [DOI] [PubMed] [Google Scholar]
- 38.Sherwin RS, Kramer KJ, Tobin JD, Insel PA, Liljenquist JE, Berman M, Andres R. A model of the kinetics of insulin in man. J Clin Invest. 1974;53:1481–1492. doi: 10.1172/JCI107697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldberg IR, Eckel RH, Abumrad NA. Regulation of fatty acid uptake into tissues: lipoprotein lipase- and CD36-mediated pathways. J Lipid Res. 2009;50:S86–S90. doi: 10.1194/jlr.R800085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell JA, Reed MA, Consitt LA, Martin OJ, Haynie KR, Hulver MW, Muoio DM, Dohm GL. Lipid partitioning, incomplete fatty acid oxidation, and insulin signal transduction in primary human muscle cells: Effects of severe obesity, fatty acid incubation, and fatty acid translocase/cd36 overexpression. J Clin Endocrinol Metab. 2010;95:3400–3410. doi: 10.1210/jc.2009-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yee AJ, Turcotte LP. Insulin fails to alter plasma LCFA metabolism in muscle perfused at similar glucose uptake. Am J Physiol Endocrinol Metab. 2002;283:E73–E77. doi: 10.1152/ajpendo.00553.2001. [DOI] [PubMed] [Google Scholar]
- 42.Caruso M, Divertie GD, Jensen MD, Miles JM. Lack of effect of hyperglycemia on lipolysis in humans. Am J Physiol Endocrinol Metab. 1990;259:E542–E547. doi: 10.1152/ajpendo.1990.259.4.E542. [DOI] [PubMed] [Google Scholar]
- 43.Summers LK, Samra JS, Frayn KN. Impaired postprandial tissue regulation of blood flow in insulin resistance: A determinant of cardiovascular risk? Atherosclerosis. 1999;147:11–15. doi: 10.1016/s0021-9150(99)00172-0. [DOI] [PubMed] [Google Scholar]
- 44.Labbé SM, Croteau E, Grenier-Larouche T, Frisch F, Ouellet R, Langlois R, Guérin B, Turcotte EE, Carpentier AC. Normal postprandial nonesterified fatty acid uptake in muscles despite increased circulating fatty acids in type 2 diabetes. Diabetes. 2011;60:408–415. doi: 10.2337/db10-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prager R, Wallace P, Olefsky JM. In vivo kinetics of insulin action on peripheral glucose disposal and hepatic glucose output in normal and obese subjects. J Clin Invest. 1986;78:472–481. doi: 10.1172/JCI112599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boston RC, Moate PJ. NEFA minimal model parameters estimated from the oral glucose tolerance test and the meal tolerance test. Am J Physiol Regul Integr Comp Physiol. 2008;295:R395–R403. doi: 10.1152/ajpregu.90317.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.