Abstract
Defensins constitute a major class of cationic antimicrobial peptides in mammals and vertebrates, acting as effectors of innate immunity against infectious microorganisms. It is generally accepted that defensins are[d1] bactericidal by disrupting the anionic microbial membrane. Here, we provide evidence that membrane activity of human α-defensins does not correlate with antibacterial killing. We[d2] further show that the α-defensin Human Neutrophil Peptide 1 (HNP-1) binds to the cell wall precursor lipid II and that reduction of lipid II levels in the bacterial membrane significantly reduces bacterial killing. The interaction between defensins and Lipid II suggests the inhibition of cell wall synthesis as a novel antibacterial mechanism of this important class of host defense peptides.
Keywords: human neutrophil peptide 1, defensin, lipid II
Introduction
Defensins form a large subfamily of cationic antimicrobial peptides that kill a broad range of microorganisms (1–4). Human defensins are cysteine-rich, cationic peptides with molecular masses ranging from 3 to 5 kDa. Based on the connectivity of the six conserved cysteine residues and sequence homology, human defensins are classified into α and β families. Both families of defensins have similar three-dimensional structures as determined by X-ray crystallography and NMR studies (5–9), sharing a common fold of three-stranded anti-parallel β-sheets constrained by three intra-molecular disulfide bonds. Human[d1] defensins were discovered originally as natural peptide antibiotics in neutrophils. These defensins were named Human Neutrophil Peptides (HNP) 1–3 of the α-defensin family (10). Subsequently, a fourth α-defensin was discovered in neutrophils, termed HNP-4 (11–13). More recently, two additional α-defensins were described, termed Human Defensin 5 and 6 (14, 15). HD-5 and HD-6 are stored in[d2] the granules of Paneth cells, specialized epithelial cells in the small intestine (16).
Defensins are widely accepted to kill bacteria through pore formation in the microbial membrane, causing leakage of intracellular contents and cell lysis (17, 18). The specific disruption of the bacterial membrane by defensins is believed to be driven by electrostatic attractions between these cationic peptides and the negatively charged membrane. However, alternative mechanisms for bacterial killing have been proposed, including membrane-independent mechanisms and targeting of intra-cellular compounds by defensins (19–21).
Recent observations on the bacterial killing by human defensins could not fully be explained by the membrane-disruption model, suggesting a more nuanced mode of action. First, α-Defensins were shown to preferentially kill Gram-positive bacteria, whereas β-defensins kill Gram-negative strains more effectively (22, 23). However, human β-defensins carry more positive charges, indicating that cationicity of defensins alone does not explain this strain-specificity. Second, disruption of the membrane via stable pore formation is believed to require peptide structure. However, we and others have shown that bacterial killing by defensins can[d3] be structure independent (24, 25). Third, we recently observed that α-defensins composed entirely of D-amino acids show greatly reduced anti-bacterial activity against Staphylococcus aureus compared to the L-peptide, suggesting that the microbial membrane is not the sole target (26). Here, we further examine the bacterial killing by α-defensins and provide evidence for an[d4] interaction with the bacterial target Lipid II.
Materials and Methods
Materials
Chemicals used for solid phase peptide synthesis were obtained as described (27). Staphylococcus aureus ATCC 29213 was obtained from Microbiologics (St. Cloud, MN). The phospholipids palmitoyl-oleoyl-phosphatidylcholine (POPC), palmitoyl-oleoyl-phosphatidylglycerol (POPG) and dipalmitoyl-phosphatidyl choline (DPPC) were purchased from Avanti Polar Lipids (Alabaster, AL). 8[d5]-aminonaphthalene-1,3,6-trisulfonic acid sodium salt (ANTS) and p-xylenebis(pyridinium) bromide (DPX) were from Molecular Probes (Eugene, OR). Poly-L-lysine (MW=3800) was obtained from Sigma. Bacitracin, D-cycloserine and fosfomycine[d6] were purchased from Sigma, Calbiochem and LKT Laboratories respectively.
Solid phase peptide synthesis
Chemical synthesis and folding of defensins was carried out as described (27, 28). The molecular mass of the peptides was verified by electrospray ionization mass spectrometry (ESI-MS) as described (27). Peptide stock solutions prepared with water were quantified spectroscopically using molar extinction coefficients at 280nm calculated according to the algorithm of Pace et al (29).
LUVs preparation
LUVs with the low molecular weight fluorophore/quencher pair (ANTS/DPX) encapsulated were prepared using the standard extrusion method. Specifically, phospholipids were dissolved in chloroform at a desired molar ratio, dried as a film by solvent evaporation. After removal[d7] of residual solvent, the lipid film was hydrated in the fluorescent solution containing 5 mM HEPES, 12.5 mM ANTS, 45 mM DPX, and 20 mM NaCl, pH 7.0, freeze-thawed for 10 cycles and extruded 10 times through 0.4-µm polycarbonate membranes. LUVs were separated from unencapsulated materials by gel filtration chromatography using a Sepharose CL-4B column eluted with 5 mM HEPES, 100 mM NaCl, pH 7.4 (high-salt). For leakage assays in a low-salt buffer, purified vesicles were further diluted with 5 mM HEPES containing 10 mM NaCl, pH 7.4.
Leakage assay
Leakage of ANTS from LUVs, monitored on a LS-55 Perkin Elmer luminescence spectrometer, was characterized by an increase in fluorescence, which was quenched by DPX when encapsulated together inside liposomes (30). 270 µl ANTS/DPX-encapsulated LUVs (in either high-salt or low-salt buffers) were added to each well of a 96-well plate to a final lipid concentration of 600 µM. 30 µl H2O was added to the first well of each row as a blank, and 30 µl 2.5% (v/v) Triton X-100 to the last (twelfth) well as the control for 100% leakage. Upon addition of 30 µl of a twofold dilution series of defensin, the fluorescence signal was recorded at 515 nm with an excitation wavelength of 353 nm, 10 nm bandwidths and a 390 nm cut-off filter in the emission path. Percent leakage is expressed as:
where Ft is the fluorescence determined at different time points after addition of defensin, F0 is the background fluorescence of the “blank” cells, and F100 is the fluorescence of the control cells containing 0.25% Triton X-100.
Lipid II purification
Short-chain water-soluble Lipid II containing a lipid tail of three isoprene units[d8] was generated and purified essentially as described (31). Typically, M. flavus vesicles (120 µmol lipid-Pi) were incubated together with 500 µmol UDP-GlcNAc, 500 µmol UDP-MurNAC-pentapeptide and 400 µmol farnesyl phosphate in[d9] 100 mM Tris-HCl pH 8.0, 5 mM MgCl2. The incubation lasted two hours at room temperature for 3-P. The synthesis of 3-Lipid II was followed using RP-8 reversed phase TLC (Merck) developed in 75% methanol. For purification, the membranes were removed by centrifugation at 40,000 × g and the supernatant was collected and loaded on a C18 HPLC column and eluted with a linear gradient from 50 mM ammonium bicarbonate to 100 % methanol in 30 minutes. Farnesyl-Lipid II (3-Lipid II) eluted at approximately 60% methanol. Its identity was confirmed by mass spectroscopy.
Surface Plasmon Resonance
Surface Plasmon Resonance binding experiments were carried out on a BIAcore T100 system (BIAcore Inc., Piscataway, NY) at 25 °C. The assay buffer was 10 mM HEPES, 150 mM NaCl, 0.05% surfactant P20, pH 7.4 (± 3 mM EDTA). L-HNP1 (780 RUs) or D-HNP1 (790 RUs) were immobilized on CM5 sensor chips using the amine-coupling chemistry recommended by the manufacturer. Lipid II was introduced into the flow-cells at 30 µl/min in the running buffer. Association and dissociation were assessed for 300 and 600 second, respectively. Resonance signals were corrected for nonspecific binding by subtracting the background of the control flow-cell. After each analysis, the sensor chip surfaces were regenerated with 15 mM HCl for 30 s at a flow rate 100 µl/min, and equilibrated with the buffer prior to next injection. Binding isotherms were analyzed with manufacturer-supplied software for BIAcore T100 and/or GraphPad Prism 4.0.
Antibacterial activity assay
The antibacterial activity of HNP1 against Staphylococcus aureus ATCC 29213 was carried out in a 96-well turbidimetric assay essentially as described previously (22). Lipid II levels in S. aureus were manipulated by the addition of three different inhibitors of cell wall synthesis: bacitracin (250 µg/ml), D-cycloserine (64 µg/ml) and fosfomycine (250 µg/ml). Bacterial cultures were pretreated with these compounds for 30 min under shaking at 37 °C. Subsequently, cells were exposed to HNP1 peptide ranging from 256 to 1 µg/ml for 15 min, after which HNP1 activity was neutralized by the addition of Mueller Hinton broth. Bacterial growth was monitored for 12 hours and data were analyzed as described (22).
Results
Membrane lipid interaction of α-defensins
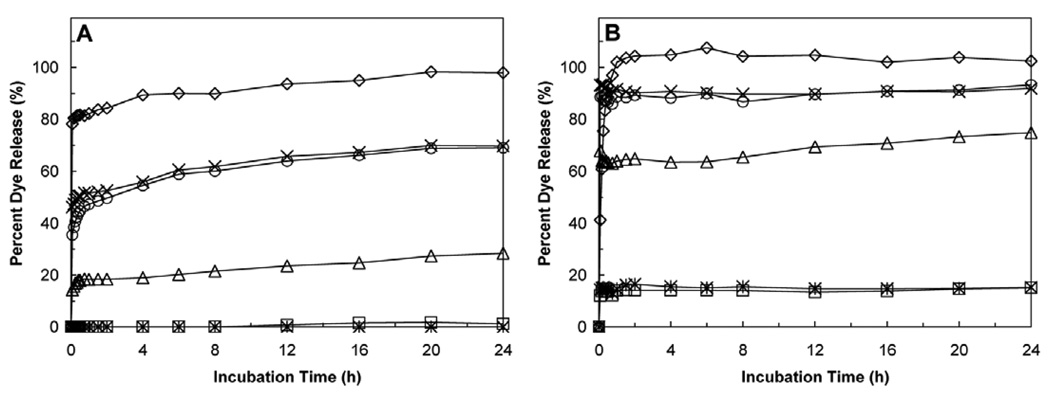
Defensins are believed to kill bacteria by permeabilizing the membrane, causing leakage of intracellular content and eventually cell lysis and death. We tested the ability of six human α-defensins to induce leakage of fluorophores encapsulated in LUVs (Fig. 1). Figure 1 shows typical time-dependent leakage curves for HNP1-4 and HD5-6 with POPG LUVs at high and low salt concentrations over a period of 24 h. All the six α-defensins tested at concentrations ranging from 0.19 to 100 µg/ml, whenever capable of inducing liposomal leakage, were fast acting as evidenced by a fluorescence plateau reached within the first hour. The[d10] plateau effect reflects the observation that at the start of the experiment the fractional fluorescence increases rapidly due to leakage of ANTS and DPV from the vesicles into the exterior solution, after which dilution removes the quenching effect of DPX. The observed differences between individual defensins over time likely reflect differences in the kinetics of induction of LUV leakage. Membrane activity of defensins invariably decreased at high salt concentrations and varied significantly between the defensins tested.
Figure 1.
Time-dependent percent release of ANTS-DPX from POPG LUVs induced by the six human α-defensins at 10 µg/ml over a period of 24 h (circle = HNP1, cross = HNP2, triangle = HNP3, diamond = HNP4, star = HD5, square = HD6). (A) At high salt concentration (5 mM HEPES, 100 mM NaCl, pH 7.4). (B) At low salt concentration (5 mM HEPES, 10 mM NaCl, pH 7.4). LUVs were 250 nm in diameter and 600 µM (phospholipids) in concentration.
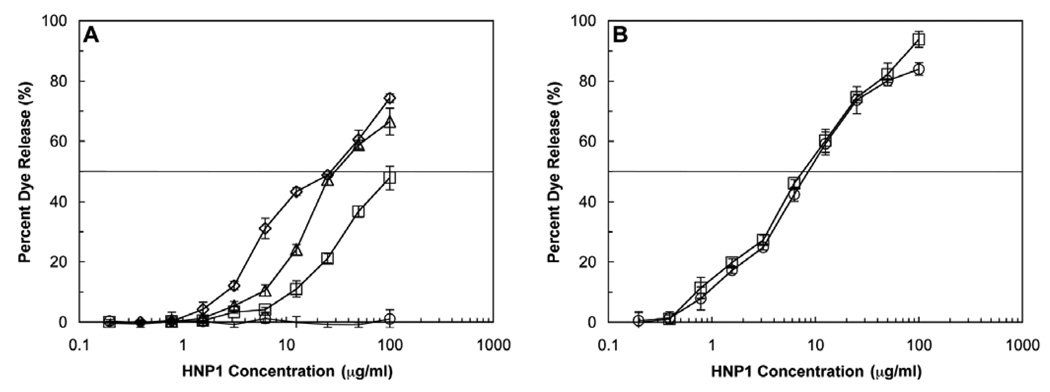
Next, we examined the effects of negative surface charge on defensin-induced membrane leakage. We used HNP1, extensively in our laboratory (23, 26, 32, 33), as a model for α-defensins in these experiments. To elucidate the role of electrostatic forces in defensin-induced membrane leakage, we prepared LUVs composed of the unsaturated lipid pair POPC (charge: 0) and POPG (charge: −1) at four different ratios, i.e., POPC:POPG = 1:0, 3:2, 2:3, and 0:1. As shown in Figure 2A, leakage from LUVs became increasingly pronounced across the entire HNP1 concentration range as the content of the negatively charged lipid POPG increased from 0, 40%, 60% to 100%, equivalent to a[d11] charge on the membrane surface of 0, −0.4, −0.6 and −1, respectively. LUVs composed solely of the neutral lipid POPC were resistant to the attack by HNP1 at all concentrations used, regardless of the incubation time. We recently reported that HNP1 composed entirely of D-amino acids (D-HNP1) was significantly less bactericidal than L-HNP1 against S. aureus (26). We compared the ability of both L- and D-HNP1 to induce leakage from LUVs (DPPC/POPG (1:1)) and no significant difference in activity between the two enantiomers was found (Figure 2B).
Figure 2.
(A) Effects of surface charge on HNP1-induced leakage from LUVs of four different compositions: POPC (circle, surface charge = 0), POPC/POPG = 3:2 (square, surface charge = −0.4), POPC/POPG = 2:3 (triangle, surface charge = −0.6), and POPG (diamond, surface charge = −1). (B) LUV (POPG:DPPC 1:1) leakage induced by L-HNP1 (circles) or D-HNP1 (squares). A two-fold dilution series of HNP1 peptides from 0.19 to 100 µg/ml was incubated with 600 µM LUVs for one hour before readings were taken. Error bars indicate the standard error in triplicate experiments.
Taken together, these findings suggest that defensin-induced permeabilization of lipid vesicles depends on electrostatic interaction, however varies greatly between different α-defensins. Most importantly, the ability of individual α-defensins to cause membrane leakage (Figure 1) correlates poorly with their ability to kill bacteria (22). For example, HNP4, the most membrane active defensin in the panel of six[d12] (Figure 1), is ineffective against Gram-positive bacteria (22). Vice versa, HNP1 and HD-5 are potently bactericidal, however display reduced, or in the case of HD-5 little membrane activity even at high concentrations. Finally[d13], our observation that D-HNP1 and L-HNP1 disrupt LUVs equally efficiently suggests that native HNP1 preferentially interacts with a bacterial membrane component, possibly of chiral nature.
HNP1 binds to Lipid II
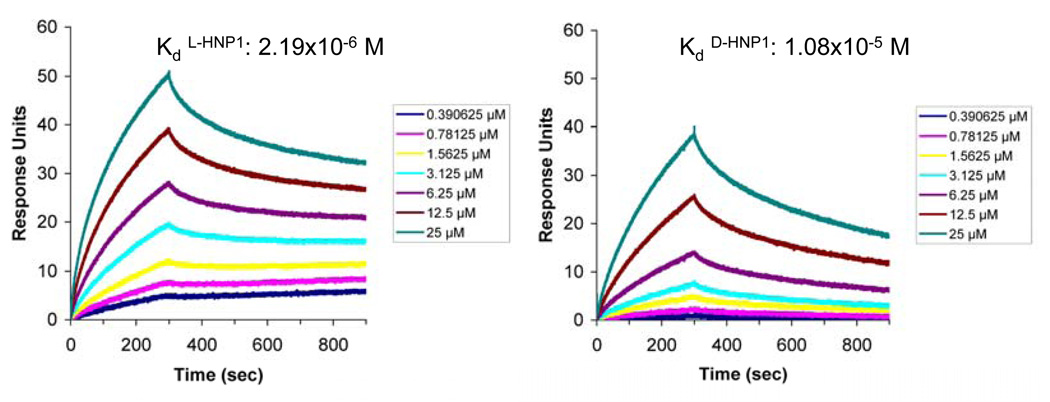
Recently, α-defensins were shown to bind with high affinity to glycosylated proteins (34) and carbohydrates (35). HNP1 kills Gram-positive bacteria very efficiently, however showed reduced[d14] membrane leakage, especially at high salt concentrations. Based on these studies, we reasoned that defensins could interact with components of the bacterial cell wall or cytoplasmic membrane. We studied the possibility of an interaction between defensins and Lipid II, a peptidoglycan precursor, for two reasons: (i) the Gram-positive cell wall consists of a thick layer of peptidoglycan and (ii) Lipid II is a known target for antibiotic peptides (36). D-HNP1 and a linear form of HNP1 were studied also, since both linear as well as enantiomeric α-defensin peptides appeared less bactericidal against S. aureus, but equally bactericidal against E. coli (24, 26). We used a Surface Plasmon Resonance (SPR) approach to determine the binding of HNP1 to Lipid II directly. Initial binding of L-HNP1, D-HNP1 and linear HNP1 to soluble Lipid II immobilized on the chip surface was determined. As shown in supplemental Figure 1, linear HNP1 showed little or no binding to Lipid II at 0.1, 1 or at 10 µM. Both L- and D-HNP1 bound Lipid II dose-dependently, however binding of wild-type HNP-1 was more efficient than that of the D-form. Conversely, the L- and D-HNP1 peptides were individually immobilized on a CM5 chip and binding of the purified, soluble form of Lipid II ranging in concentration from 25 to 0.78 µM to both peptides was determined. As shown in Figure 3, soluble Lipid II bound to both the L-form as well as the D-form of HNP1. Fitting of the kinetic data to a 1:1 binding model indicated that Lipid II binds the L-HNP1 peptide with an approximately five times higher affinity than the D-peptide (2.19×10−6 M vs. 1.08×10−5M).
Figure 3.
Binding kinetics of soluble Lipid II on immobilized HNP1 as determined by SPR at room temperature. Representative sensorgrams of one out of two separate experiments of soluble Lipid II (from 20 to 0.390625 µM) using a sensorchip with 780 RUs of L-HNP1 (left panel) or 790 RUs of D-HNP1 (right panel). Indicated[d20] Kd values represent the average of the two separate experiments (individual values: L-HNP-1: 1.79×10−6 and 2.59×10−6; D-HNP-1: 1.11×10−5 and 1.05×10−5 respectively).
HNP1 functionally interacts with lipid II
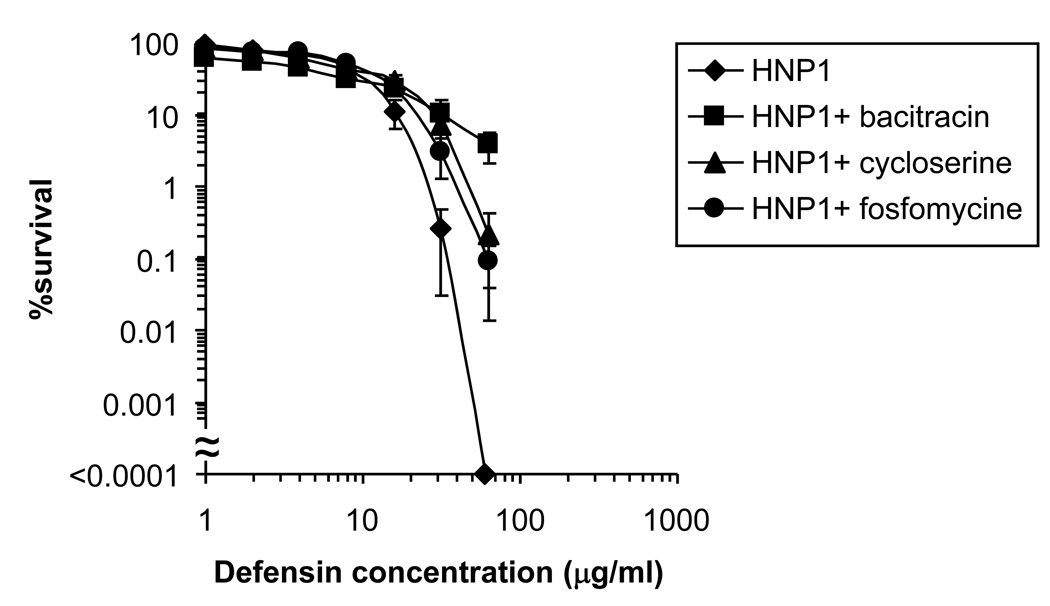
To examine whether the observed interaction between HNP1 and Lipid II is functionally relevant in the environment of the membrane, we determined the ability of HNP1 to kill S. aureus with altered levels of Lipid II. Three different inhibitors of cell wall synthesis, fosfomycine, D-cycloserine, and bacitracin, were used to reduce the lipid II levels in S. aureus cells. Bacitracin binds directly to undecaprenyl pyrophosphate, the portion of lipid II that remains in the membrane once GlcNAc-MurNAc is polymerized, and prevents its use in subsequent cycles of lipid II synthesis. Fosfomycine is an inhibitor of MurA, the enzyme responsible for the first step in peptidoglycan synthesis. D-Cycloserine inhibits both alanine racemase and D-Ala–D-Ala ligase, two enzymes required for the synthesis of the D-Ala–D-Ala dipeptide of lipid II. All three inhibitors thus block the synthesis of lipid II (37). S. aureus cells were exposed to each of the Lipid II synthesis inhibitors for 30 min and subsequently exposed to HNP1 at concentrations ranging from 256 to 1 µg/ml for 15 min (Figure 4). Following[d15] pre-treatment with Lipid II biosynthesis inhibitors and exposure of HNP-1 to the bacteria for 2 hours, according to our original protocol (22), we observed no difference in bacterial killing (data not shown). Most likely, the effects of any bacterial pre-treatments are negated by killing efficiency and kinetics of HNP1 during the prolonged, two hour exposure to the bacteria. As observed previously, after 15 min, HNP1 efficiently killed S. aureus (38). At[d16] peptide concentrations of 256 and 128 µg/ml, bacterial growth did not measurably recover after 12 h incubation and data points could not be plotted. Fosfomycine and D-cycloserine and in particular bacitracin treatment attenuated[d17] killing of S. aureus by HNP1 markedly. Taken together, these data indicate that efficient killing of S. aureus by HNP1 depends on membrane lipid II levels.
Figure 4.
Lipid II-dependent bacterial killing by HNP1. Survival curves of S. aureus ATCC 29213 exposed to HNP1 at concentrations varying two-fold from 1 to 256 µg/ml. Bacteria were pre-treated with bacitracin (250 µg/ml), D-cycloserine (64 µg/ml) and fosfomycine (250 µg/ml) for 30 min under shaking at 37 °C as indicated, followed by exposure to HNP1 for 15 min. Each curve is the mean of three separate experiments[d21] (±S.D.). Points scored as zero survival could not be plotted.
Discussion
Disruption of the functional integrity of the bacterial membrane is a common mode of action of many antibacterial compounds and is believed to be the primary mode of bacterial killing by defensins. An early study reported on the bactericidal activity of HNP1-3 against E. coli, suggesting a sequential permeabilization of the outer and inner membranes (18). More recent observations on the bactericidal activity of α-defensins have expanded and nuanced these findings. We and others reported that linear, unstructured defensins retained their antibacterial activity in a strain-selective manner (24, 39). The activity of HD-5 against E. coli appeared structure-independent, whereas the unstructured peptide showed greatly reduced activity against S. aureus (24). More recently, we observed that the D-forms of HNP1 and HD-5 were significantly less active than their native L-forms against S. aureus, but equally bactericidal against E. coli (26). Combined, these findings suggested different bactericidal mechanisms of α-defensins against[d18] E. coli or S. aureus. In addition, these findings suggested a possible interaction between defensins and an unidentified cellular component of S. aureus.
In this study, we find that HNP1 functionally[d19] interacts with lipid II, an essential precursor of cell wall synthesis (36). A number of antibacterial compounds target lipid II, thus affecting cell wall synthesis or membrane function (36). For example, the antibacterial action of nisin, an amphiphilic peptide produced by certain strains of Lactococcus lactis, is a result of its high affinity for lipid II as well as its ability to assemble into nisin-lipid II complexes (31). Such complexes have the ability to form pores in the membrane, explaining the high efficacy of nisin (40). Here, we show that bacterial killing by α-defensins depends on lipid II levels by blocking the synthesis of lipid II. All three lipid II synthesis inhibitors reduced bacterial killing, in particular when S. aureus cells were pretreated with bacitracin. A similar observation was made recently in the case of nisin (37). In this study, depolarization of the S. aureus membrane induced by nisin was suppressed by pretreatment of cells with lipid II inhibitors, especially by bacitracin. Interestingly, nisin and bacitracin share a common target in binding the lipid II molecule, both binding the pyrophosphate moiety of undecaprenyl-pyrophosphate (37, 40). Since the antibacterial activity of both nisin and defensin was reduced most strongly by treatment with bacitracin, it is tempting to speculate that defensins, like nisin, may use lipid II as an initial binding target and perhaps even similarly disrupt the membrane via complex pore formation.
Our observation that HNP1 binds to lipid II partly rationalizes our previous findings on the strain-selective and structure-dependent difference in bactericidal activity of human α-defensins. However, questions still remain why α-defensins preferentially kill Gram-positive bacteria. For example, we found that D-HNP1 binds to lipid II with a five-fold weaker affinity than the L-form. This difference could be explained by the fact that lipid II itself is a chiral molecule. However, D-HNP1 was found to be ~19 times weaker in S. aureus killing compared to L-HNP1 as judged by their respective vLD90 values, defined as the defensin concentration required to kill 90% of bacteria (26). Defensins therefore may interact with other membrane components in addition to lipid II. Other possible interactions at the bacterial membrane could include negatively charged molecules such as (lipo)teichoic acid in the case of Gram-positive bacteria or lipopolysaccharide or teichoic acid in the case of Gram-negative bacteria. Precursors of teichoic acid synthesis are, like Lipid II, undecaprenyl-linked (41), and may therefore constitute a possible binding target for HNP1 also. In addition, we observed that bacterial killing by α-defensins correlates poorly with their lipid membrane activity. Nevertheless, increase of negative charge of the phospholipid headgroup increased HNP1 membrane activity, suggesting that bactericidal activity may involve direct defensin-lipid interactions. In summary, our findings suggest the inhibition of peptidoglycan synthesis through binding of lipid II as a novel mechanism of bacterial killing for defensins.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health grants AI061482 and AI072732 to W.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 2.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE, Lehrer R. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 1998;16:82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 4.De Leeuw EaLW. Human Defensins: Turning Defense into Offense? Infect Disord Drug Targets. 2007;7:67–70. doi: 10.2174/187152607780090702. [DOI] [PubMed] [Google Scholar]
- 5.Hill CP, Yee J, Selsted ME, Eisenberg D. Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization. Science. 1991;251:1481–1485. doi: 10.1126/science.2006422. [DOI] [PubMed] [Google Scholar]
- 6.Hoover DM, Rajashankar KR, Blumenthal R, Puri A, Oppenheim JJ, Chertov O, Lubkowski J. The structure of human beta-defensin-2 shows evidence of higher order oligomerization. J Biol Chem. 2000;275:32911–32918. doi: 10.1074/jbc.M006098200. [DOI] [PubMed] [Google Scholar]
- 7.Hoover DM, Chertov O, Lubkowski J. The structure of human beta-defensin-1: new insights into structural properties of beta-defensins. J Biol Chem. 2001;276:39021–39026. doi: 10.1074/jbc.M103830200. [DOI] [PubMed] [Google Scholar]
- 8.Pardi A, Zhang XL, Selsted ME, Skalicky JJ, Yip PF. NMR studies of defensin antimicrobial peptides. 2. Three-dimensional structures of rabbit NP-2 and human HNP-1. Biochemistry. 1992;31:11357–11364. doi: 10.1021/bi00161a013. [DOI] [PubMed] [Google Scholar]
- 9.Szyk A, Wu Z, Tucker K, Yang D, Lu W, Lubkowski J. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci. 2006;15:2749–2760. doi: 10.1110/ps.062336606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganz T, Selsted ME, Szklarek D, Harwig SS, Daher K, Bainton DF, Lehrer RI. Defensins. Natural peptide antibiotics of human neutrophils. J Clin Invest. 1985;76:1427–1435. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilde CG, Griffith JE, Marra MN, Snable JL, Scott RW. Purification and characterization of human neutrophil peptide 4, a novel member of the defensin family. J Biol Chem. 1989;264:11200–11203. [PubMed] [Google Scholar]
- 12.Singh A, Bateman A, Zhu QZ, Shimasaki S, Esch F, Solomon S. Structure of a novel human granulocyte peptide with anti-ACTH activity. Biochem Biophys Res Commun. 1988;155:524–529. doi: 10.1016/s0006-291x(88)81118-5. [DOI] [PubMed] [Google Scholar]
- 13.Gabay JE, Scott RW, Campanelli D, Griffith J, Wilde C, Marra MN, Seeger M, Nathan CF. Antibiotic proteins of human polymorphonuclear leukocytes. Proc Natl Acad Sci U S A. 1989;86:5610–5614. doi: 10.1073/pnas.86.14.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones DE, Bevins CL. Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem. 1992;267:23216–23225. [PubMed] [Google Scholar]
- 15.Jones DE, Bevins CL. Defensin-6 mRNA in human Paneth cells: implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett. 1993;315:187–192. doi: 10.1016/0014-5793(93)81160-2. [DOI] [PubMed] [Google Scholar]
- 16.Porter EM, Liu L, Oren A, Anton PA, Ganz T. Localization of human intestinal defensin 5 in Paneth cell granules. Infect Immun. 1997;65:2389–2395. doi: 10.1128/iai.65.6.2389-2395.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kagan BL, Selsted ME, Ganz T, Lehrer RI. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc Natl Acad Sci U S A. 1990;87:210–214. doi: 10.1073/pnas.87.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, Selsted ME. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J Clin Invest. 1989;84:553–561. doi: 10.1172/JCI114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 20.Hancock RE, Rozek A. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol Lett. 2002;206:143–149. doi: 10.1111/j.1574-6968.2002.tb11000.x. [DOI] [PubMed] [Google Scholar]
- 21.Wu M, Maier E, Benz R, Hancock RE. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry. 1999;38:7235–7242. doi: 10.1021/bi9826299. [DOI] [PubMed] [Google Scholar]
- 22.Ericksen B, Wu Z, Lu W, Lehrer RI. Antibacterial activity and specificity of the six human {alpha}-defensins. Antimicrob Agents Chemother. 2005;49:269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou G, de Leeuw E, Li C, Pazgier M, Li C, Zeng P, Lu WY, Lubkowski J, Lu W. Toward understanding the cationicity of defensins: ARG and LYS versus their noncoded analogs. J Biol Chem. 2007 doi: 10.1074/jbc.M611003200. [DOI] [PubMed] [Google Scholar]
- 24.de Leeuw E, Burks SR, Li X, Kao JP, Lu W. Structure-dependent functional properties of human defensin 5. FEBS Lett. 2007;581:515–520. doi: 10.1016/j.febslet.2006.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maemoto A, Qu X, Rosengren KJ, Tanabe H, Henschen-Edman A, Craik DJ, Ouellette AJ. Functional analysis of the alpha-defensin disulfide array in mouse cryptdin-4. J Biol Chem. 2004;279:44188–44196. doi: 10.1074/jbc.M406154200. [DOI] [PubMed] [Google Scholar]
- 26.Wei G, de Leeuw E, Pazgier M, Yuan W, Zou G, Wang J, Ericksen B, Lu WY, Lehrer RI, Lu W. Through the looking glass, mechanistic insights from enantiomeric human defensins. J Biol Chem. 2009;284:29180–29192. doi: 10.1074/jbc.M109.018085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z, Ericksen B, Tucker K, Lubkowski J, Lu W. Synthesis and characterization of human alpha-defensins 4–6. J Pept Res. 2004;64:118–125. doi: 10.1111/j.1399-3011.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- 28.Wu Z, Powell R, Lu W. Productive folding of human neutrophil alpha-defensins in vitro without the pro-peptide. J Am Chem Soc. 2003;125:2402–2403. doi: 10.1021/ja0294257. [DOI] [PubMed] [Google Scholar]
- 29.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellens H, Bentz J, Szoka FC. H+ and Ca2+induced fusion and destabilization of liposomes. Biochemistry. 1985;24:3099–3106. doi: 10.1021/bi00334a005. [DOI] [PubMed] [Google Scholar]
- 31.Breukink E, van Heusden HE, Vollmerhaus PJ, Swiezewska E, Brunner L, Walker S, Heck AJ, de Kruijff B. Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J Biol Chem. 2003;278:19898–19903. doi: 10.1074/jbc.M301463200. [DOI] [PubMed] [Google Scholar]
- 32.Wu Z, Li X, Ericksen B, de Leeuw E, Zou G, Zeng P, Xie C, Li C, Lubkowski J, Lu WY, Lu W. Impact of pro segments on the folding and function of human neutrophil alpha-defensins. J Mol Biol. 2007;368:537–549. doi: 10.1016/j.jmb.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou G, de Leeuw E, Lubkowski J, Lu W. Molecular determinants for the interaction of human neutrophil alpha defensin 1 with its propeptide. J Mol Biol. 2008;381:1281–1291. doi: 10.1016/j.jmb.2008.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W, Owen SM, Rudolph DL, Cole AM, Hong T, Waring AJ, Lal RB, Lehrer RI. Activity of alpha- and theta-defensins against primary isolates of HIV-1. J Immunol. 2004;173:515–520. doi: 10.4049/jimmunol.173.1.515. [DOI] [PubMed] [Google Scholar]
- 35.Lehrer RI, Jung G, Ruchala P, Andre S, Gabius HJ, Lu W. Multivalent binding of carbohydrates by the human alpha-defensin, HD5. J Immunol. 2009;183:480–490. doi: 10.4049/jimmunol.0900244. [DOI] [PubMed] [Google Scholar]
- 36.Breukink E, de Kruijff B. Lipid II as a target for antibiotics. Nat Rev Drug Discov. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 37.Lunde CS, Hartouni SR, Janc JW, Mammen M, Humphrey PP, Benton BM. Telavancin disrupts the functional integrity of the bacterial membrane through targeted interaction with the cell wall precursor lipid II. Antimicrob Agents Chemother. 2009;53:3375–3383. doi: 10.1128/AAC.01710-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou G, de Leeuw E, Li C, Pazgier M, Zeng P, Lu WY, Lubkowski J, Lu W. Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs. J Biol Chem. 2007;282:19653–19665. doi: 10.1074/jbc.M611003200. [DOI] [PubMed] [Google Scholar]
- 39.Hadjicharalambous C, Sheynis T, Jelinek R, Shanahan MT, Ouellette AJ, Gizeli E. Mechanisms of alpha-defensin bactericidal action: comparative membrane disruption by Cryptdin-4 and its disulfide-null analogue. Biochemistry. 2008;47:12626–12634. doi: 10.1021/bi800335e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl H, de Kruijff B. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 41.Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.