Abstract
NSCLC is the leading cause of cancer-related death in the US. Patients with NSCLC are mostly treated with platinum-based chemotherapy, often in combination with radiation therapy. However, the development of chemoresistance is a major hurdle limiting treatment success. In this review, we summarize the current understanding of the genetic factors modulating chemoresistance to platinum chemotherapeutics and their association with clinical outcomes for NSCLC patients. We focus on candidate pathways responsible for drug influx and efflux, metabolism and detoxification, DNA damage repair, and other downstream cellular processes that modulate the effect of platinum-based therapy. We also discuss the application of pathway-based polygenic and genome-wide approaches in identifying genetic factors involved in NSCLC clinical outcomes. Overall, current studies have shown that the effects of each individual polymorphism on clinical outcomes are modest suggesting that a more comprehensive approach that incorporates polygenetic, phenotypic, epidemiologic and clinical variables will be necessary to predict prognosis for NSCLC patients receiving platinum-based chemotherapeutics.
Keywords: carboplatin, chemotherapy, cisplatin, clinical outcomes, NSCLC
1. Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide. In 2002, > 1.2 million individuals were diagnosed with lung cancer [1]. In the US alone, it is estimated that there were > 140,000 newly diagnosed lung cancer patients and > 160,000 resulting deaths in 2008 [2]. NSCLC comprises > 75% of lung cancer cases making it the leading cause of cancer-related death. The prognosis for NSCLC patients is highly dependent on the stage at diagnosis, and despite efforts to develop early screening tools, a majority of tumors are detected at an advanced stage [3]. Surgery is the standard of care for patients with resectable tumors, often with the addition of adjuvant chemotherapy with platinum-based agents such as cisplatin and carboplatin. Patients with advanced NSCLC have a much dimmer prognosis [4]. A recent meta-analysis of > 2700 patients has shown that the addition of chemotherapy to supportive care increases overall survival > 1 year for advanced NSCLC to 29% from 20% [5]. Unfortunately, although these agents have shown success in treating NSCLC, the use of platinum-based chemotherapeutics is limited by the development of chemoresistance and toxicity.
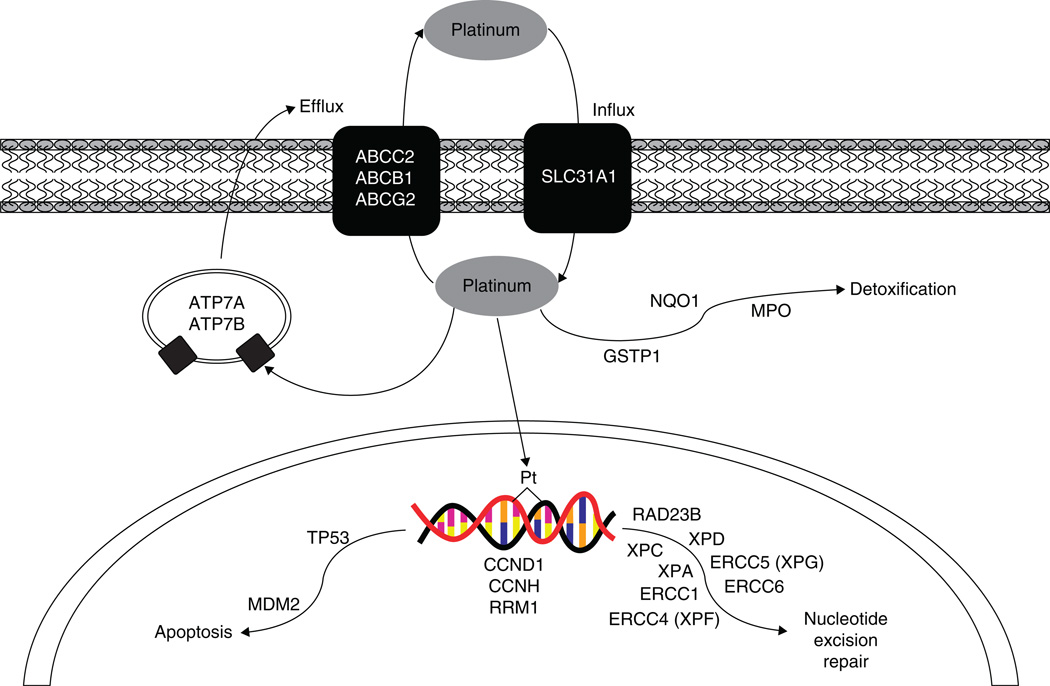
This development of chemoresistance can be intrinsic owing to germ-line genetic variation or acquired through altered mRNA or protein expression in key pharmacokinetic or pharmacodynamic pathways. Several excellent reviews have detailed the molecular mechanisms of resistance to platinum agents [6–8]. These include alterations in drug influx or efflux, detoxification through glutathione conjugation, DNA repair capacity and other cellular pathways required for proper response to DNA damage (Figure 1). Pharmacogenomic analysis of genetic variation in these candidate pathways has shown that an individual’s genetic background plays a role in determining response to platinum-based chemotherapy. More recently, unbiased, genome-wide approaches have been taken to identify unknown genetic modulators and these studies will surely become more valuable.
Figure 1.
Platinum drug action pathway.
At present, only clinical variables are used to guide treatment decisions for NSCLC patients such as stage and performance scores [9]. Through the identification of polymorphisms associated with response to therapy and overall survival, the physician may have more information to better select the appropriate treatment regimen. In this review, we focus on the association between genetic variation and clinical outcomes in advanced NSCLC.
2. Drug influx and efflux
A common characteristic of cancer cells that have become resistant to platinum agents is reduced intracellular accumulation of drug [10], which may be caused by altered influx or efflux of the compounds in or out of the target cell. Although the exact processes by which platinum compounds enter the cell are not fully understood, it is believed that the primary mechanism is passive diffusion [6]. Another route is through the copper transporter CTR1 (SLC31A1) [11]. This protein has been show to be downregulated in ovarian cancer cell lines following cisplatin treatment and homozygous knockouts of the gene increase resistance to both cisplatin and carboplatin [12]. SLC31A1 contains one coding region polymorphism that results in a proline to alanine change at codon 199 (rs2233915). This variant was genotyped as part of the HapMap Project and only found in individuals from Africa. Because of the lack of candidate single nucleotide polymorphisms (SNPs), there are at present no association studies of genetic variation in this gene and clinical outcomes in any cancer. However, transfection of CTR1 protein in resistant cell lines with low expression does not fully convert the cell to a sensitive phenotype, suggesting that another, unknown membrane transporter may also be involved in platinum uptake [13]. If identified, genetic variation with this candidate will be of interest.
Active drug efflux will also result in decreased levels of platinum within the cell. Genetic mutations in ATP7A and ATP7B result in severe copper deficiencies that cause Menkes and Wilson disease, respectively. Interestingly, these two copper transporters have been shown to be involved in the development of chemoresistance to platinum agents [14]. Overexpression of either gene resulted in increased resistance and ovarian cancer patients with high expression had a poorer prognosis [15,16]. As with CTR1, studies of the association between genetic variation in these two copper transporters and clinical outcomes have been limited due to lack of functional candidate SNPs.
Copper transporters are not the only mechanism for cisplatin efflux. Several drug transporters have also been show to be involved in pumping cisplatin out of the cell resulting in chemoresistance. The strongest evidence is for the multi-drug resistance protein (MRP2, or cMOAT), encoded for by the gene ABCC2. Expression levels of MRP2 were associated with chemoresistance in cancer cell lines [17]. Genetic variation of ABCC2 has been studied for effect on NSCLC clinical outcomes in patients treated with both cisplatin and irinotecan [18]. Homozygous variant genotypes of two SNPs in ABCC2, −24C>T (rs717620) and 3972C>T (rs3740066), were found to be associated with increased response rate and progression-free survival. However, as these patients were treated with cisplatin–irinotecan combination therapy, and ABCC2 is a known efflux transporter for irinotecan, it is difficult to assess whether the observed effect is owing to decreased cisplatin efflux.
The ABC transporter ABCB1 (MDR1 or p-glycoprotein) is responsible for the efflux of many commonly used chemotherapeutics and has been widely studied for involvement in the development of chemoresistance. It is thought to play a role in platinum efflux, although to a much lesser extent than the copper transporters or MRP2. Two common variants in ABCB1 have been studied in lung cancer, rs2032582 (Ser893Ala) and rs1045642 (Ile1145Ile). A small study of only 54 NSCLC patients treated with docetaxel and cisplatin, the Ser893Ala variant, was found to be associated with better response [19]. However, in our study of 229 advanced NSCLC patients treated with platinum-based chemotherapy, neither of these SNPs was associated with overall survival [20]. Other efflux transporters in the ABC family such as ABCG2 (breast cancer resistance protein or BCRP) and ABCC1 (MRP1) may also play a role in the development of chemoresistance to platinum agents, although genetic variation in these genes has not been analyzed.
3. Metabolism and detoxification
Once platinum compounds enter the cell, they become aquated creating a more reactive species that can bind to cellular targets, including DNA. However, this reactive form is readily inactivated by conjugation with glutathione, potentially resulting in increased chemoresistance. Reaction with glutathione is dependent on the glutathione levels within the cell and studies in NSCLC cell lines have shown that glutathione levels are correlated with resistance or sensitivity to cisplatin [21].
The platinum-glutathione conjugates can form spontaneously when glutathione levels are high or are aided by the enzyme glutathione S-transferase (GST). There are several members of the GST family, but much of the attention has been on the π subfamily, specifically GSTP1. This enzyme was shown to increase the rate of adduct formation and cisplatin sensitivity increased with transfection of antisense GSTP1 in colon cancer cell lines [22]. Two common nonsynonymous SNPs (nsSNPs) at codons 105 (Ile to Val, rs1695) and 114 (Ala to Val, rs1138272) have been well studied. In in vitro studies, the Val105 variant resulted in an 80% reduction in GSTP1 enzyme activity, whereas Val114 resulted in a much smaller decrease in activity of only 20% [23]. However, when these variants were expressed in cell lines, there were only minor differences in the formation rate of platinum-glutathione adducts [24].
Several association studies between the GSTP1 variants and response to platinum-based chemotherapy have been performed but the results have been inconsistent. In a study by Lu et al., of 425 advanced NSCLC patients, no association was observed between the Ala114Val polymorphism and survival, but patients with at least one variant Val105 allele had better survival compared to patients with wild-type genotype [25]. Although not significant, this trend was also present in a subset of the patients who received chemotherapy. A similar finding was reported in 108 NSCLC patients treated with platinum-based chemotherapy [26]. In this study, a trend was also found between Val105 and poor response to therapy. In our recent analysis of 229 NSCLC patients, those carrying two of the Val105 alleles who were treated with cisplatin-based chemotherapy had a significant nearly twofold increased risk of dying [20]. The discrepancy of results may be owing to patient, tumor and treatment heterogeneity, and insufficient sample size. Further studies of large sample size with homogeneous patient population (in terms of tumor stage and treatment regimen) are warranted to clarify the effect of GSTP1 SNPs on NSCLC outcomes.
Genetic variants in other enzymes involved in cellular detoxification, such as NAD(P)H dehydrogenase, quinone (NQO1) and myeloperoxidase (MPO), have also been studied [20]. These enzymes control the levels of reactive oxygen species in the cell, and thus contribute to the level of DNA damage present following cellular stress owing to chemotherapy treatment. NQO1 and MPO have opposing effects with NQO1 acting as a detoxifier and MPO as an activator [27]. NQO1*2 (rs1800566) is a proline to serine change that results in reduced enzyme activity [28]. The MPO variant rs2243828 is located within the 5′-flanking region of the gene and causes decreased MPO expression [29,30]. Although studies have linked these variants to NSCLC risk [27], there is no clear evidence that demonstrates association between these polymorphisms with clinical outcomes following platinum-based chemotherapy.
4. DNA damage repair
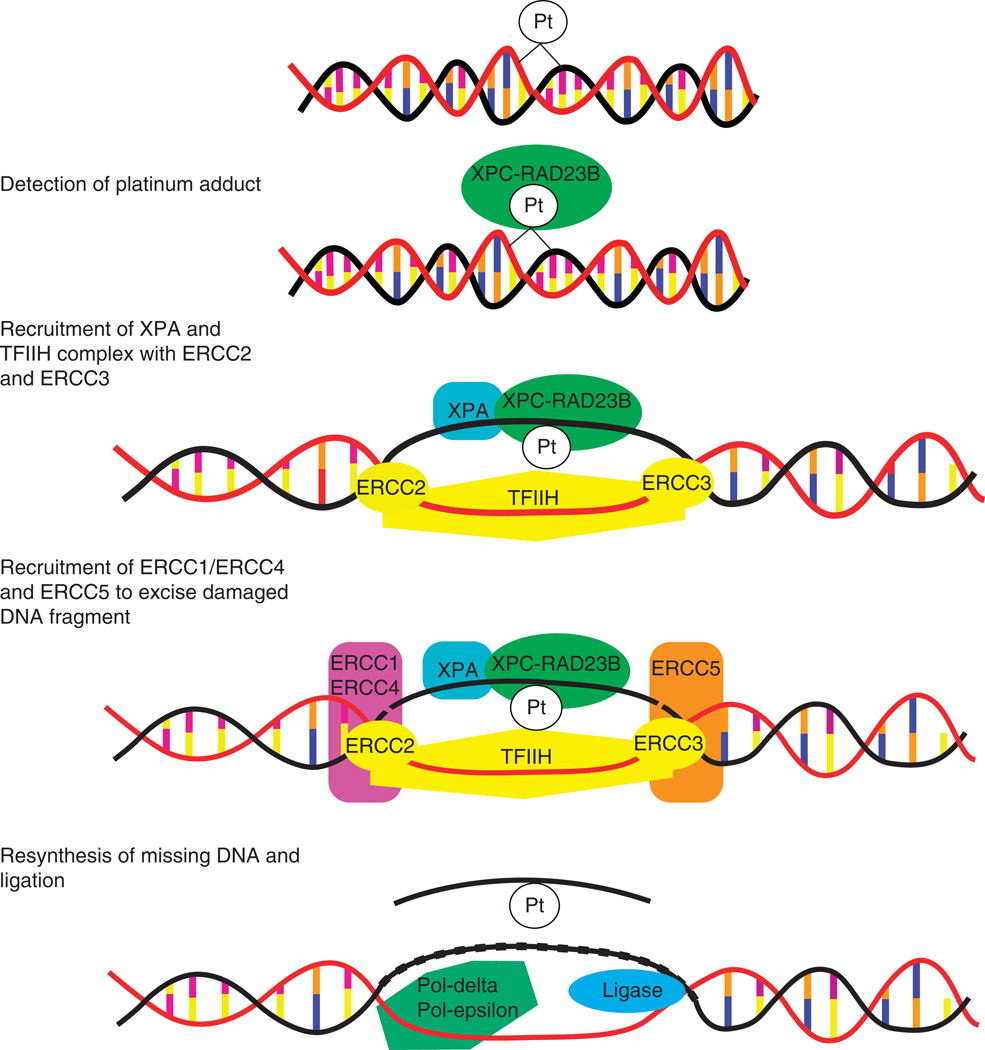
Platinum causes DNA damage through the creation of platinum-DNA adducts. The cell has mechanisms in place to remove these DNA adducts and repair the damage through nucleotide excision repair (NER; Figure 2). The first step involves recognition of the Pt-DNA adduct by the RAD23B/XPC complex, which then recruits the TFIIH complex to the site. This transcription factor complex includes the two helicases, ERCC3 (also known as XPB) and ERCC2 (XPD). These helicases unwind the DNA around the site, which then allows for the binding of XPA and RPA to stabilize the complex. ERCC5 (XPG) is the 3′-endonuclease and ERCC1 and ERCC4 (XPF) form the 5′-endonuclease. These proteins excise the lesion along with some nucleotides surrounding the damage site. The missing nucleotides are then replaced by re-synthesis and ligation (reviewed by Simon et al. [31] and Christmann et al. [32]). The ability of the cell to successfully undergo this process and remove the Pt-DNA lesions will result in either resistance or sensitivity to platinum-based chemotherapy. In NSCLC cell lines, elevated DNA repair capacity was associated with chemoresistance [33]. Furthermore, there is a large variation among individuals in their DNA repair capacity and it has been shown that this translates into variation in how NSCLC patients respond to platinum-based chemotherapy [34]. Because of the importance of this process in determining sensitivity and resistance, components of the NER and other DNA damage pathways have been well studied with regard to genetic variation and response to therapy.
Figure 2.
Nucleotide excision repair (NER) of platinum-DNA adducts.
There is a common nsSNP (Ala249Val, rs1805329) in the gene encoding RAD23B. This SNP was shown to be associated with variation in NER capacity as measured by the mutagen sensitivity assay [35]. In NSCLC patients, two copies of this variant were modestly associated with overall survival following cisplatin-based chemotherapy [20]. However, this finding was not supported in a large study by Matakidou et al. [36]. This difference may be due to the heterogeneity in patients analyzed: the former study analyzed advanced NSCLC cases treated with cisplatin whereas the latter one included both NSCLC and small cell lung cancer patients at all stages of disease.
RAD23B and XPC form a complex that recognizes Pt-DNA adducts, and similarly to RAD23B genetic variation, two variants in XPC were also found to be significantly associated with NER capacity [35]. One of these polymorphisms is an nsSNP (Gln940Lys, rs2228001), whereas the other is an insertion/deletion within intron 9 (often referred to as XPC-PAT). However, association studies have not identified a link of these polymorphisms, or two other XPC SNPs rs2228000 and rs3731062, with overall survival or response to platinum-based chemotherapy in NSCLC [20,36].
The association between two common nsSNPs in XPD and response to platinum-based chemotherapy in NSCLC is also not clear. In 39 advanced NSCLC patients treated with combination gemcitabine and cisplatin, neither the Asp312Asn (rs1799793) nor the Lys751Gln (rs13181) variants were significantly associated with overall survival, time to progression and response [37]. Similar null results were reported by several other studies [36,38–41], including a large study of 229 advanced patients who received first-line cisplatin chemotherapy [20]. However, two studies reported positive results. Gurubhagavatula et al. found that patients with two Asn312 alleles were at an increased risk of dying compared to those with wild-type genotypes [42] in 103 advanced NSCLC patients. Booton et al. observed in 108 advanced stage patients that a haplotype containing both Asn312 and Gln751 was associated with decreased response to chemotherapy and a corresponding increase in progressive disease rates resulting in decreased overall survival [43]. Because of these inconsistencies, it is unknown whether these two variants are truly associated with response to platinum-based chemotherapy.
For XPA genetic variation, Mellon et al. studied the Arg228Gln (rs1805160) and Val234Leu (rs3176749) polymorphisms in an in vitro system and observed that these nsSNP variants did not alter DNA repair capacity or cell survival [44]. However, a 5-UTR variant of XPA (rs1800975) was shown to be associated with increased lung cancer risk and resulted in impaired NER capacity following tobacco carcinogen exposure [35,45], but not overall survival [20]. It is possible that this variant is more involved in repair of smoking-related DNA damage than the removal of Pt-DNA adducts.
Several polymorphisms in ERCC5 (XPG) have been studied with the His1104Asp variant (rs17655) being associated with a poor response in a lung cancer population comprised of all stages and both NSCLC and small cell lung cancer [36]. This finding was not supported in two studies of only advanced NSCLC patients treated with platinum-based chemotherapy [20,46]. In the study by Sun et al., a synonymous SNP, His46His (rs1047768), was associated with clinical response [46].
Variation in ERCC1 has been extensively studied for association with platinum-based chemotherapy response and clinical outcomes for NSCLC. At the protein level, absence of ERCC1 expression is associated with better response to cisplatin chemotherapy and increased overall survival in the adjuvant setting [47]. A Phase III trial customizing cisplatin treatment of NSCLC patients based on quantitative ERCC1 mRNA expression has been carried out and the results suggest that determination of ERCC1 expression may be a useful tool in identifying patients who will respond to platinum-based chemotherapy [48]. There are two common ERCC1 genetic variants, one located in the 3′-UTR of ERCC1 (8092C>A, rs3212986) and another at codon 118 (Asn118Asn or 118T>C, rs11615). In 229 advanced NSCLC patients, Wu et al. demonstrated that patients with at least one variant rs3212986 allele had a significant increase in overall survival following cisplatin-based chemotherapy [20]. This is in contrast to a study of 128 patients in whom this variant was associated with decreased overall survival [49]. There is yet another study showing no effect on overall survival by 8092C>A [50]. As for the other common polymorphism, 118C>T, the results are relatively more consistent with the variant T allele being associated with a poor outcome and response to therapy in several studies [39,40,50,51]. But, there are studies of 135 and 65 advanced NSCLC patients that did not find any association with either polymorphism [41,52].
SNPs in other NER genes, such as ERCC4 (XPF) and ERCC6, have been analyzed for association with clinical outcomes in NSCLC, but no significant findings have been reported [20,36].
Taken together, individual polymorphisms have no or only modest effect on response to platinum-based chemotherapy. However, the cumulative effects of these SNPs on overall survival in the NER pathway are striking [20]. Advanced stage NSCLC patients with less than two unfavorable genotypes had a significant lengthening of their median survival time to 26.9 months compared to 10.9 months for those patients with five or more unfavorable genotypes (p < 0.001). These results demonstrate that combined analyses may be more powerful in detecting the true effect of these polymorphisms on clinical outcomes.
5. Other cellular pathways
5.1 Cell cycle control
Once the target cell recognizes the DNA damage and begins the repair process, the cell cycle must be arrested to allow sufficient time for DNA repair and to avoid replication of this damage. Several genes responsible for cell cycle control have been analyzed for association with response to platinum-based chemotherapy. Cyclin D1 is responsible for the G1/S transition, and because of this function it plays an integral role in maintaining genome integrity. A common genetic polymorphism (rs9344) in CCND1 results in the production of an alternatively spliced transcript and a poor prognosis for NSCLC patients and has been associated with response to platinum-based chemotherapy [53,54]. However, variation in expression for several cell cycle regulators, including cyclin D1, cyclin D3, cyclin E, p27Kip1, p16INK4A and Ki-67, was analyzed in 778 NSCLC patients treated with cisplatin-based therapy [55]. In this study, cyclin D1 expression was not predictive of receiving benefit from cisplatin-based therapy, but patients with p27Kip1-negative tumors did show a benefit resulting in longer survival. Another cyclin, cyclin H, is part of the TFIIH complex and links NER with cell cycle control. A polymorphism at codon 270 results in a valine to alanine substitution (rs2266690). Two large studies of NSCLC patients did not find an association between this variant and overall survival [20,36].
5.2 DNA synthesis
Once the Pt-DNA adduct is excised from the DNA strand, the missing nucleotides are replaced requiring the synthesis of deoxyribonucleotides. Several enzymes are responsible for this process, and several have been candidate genes for variation in clinical outcomes of NSCLC patients, including ribonucleotide reductase M1 (RRM1). RRM1 gene expression has been shown to be a prognositic marker for NSCLC [56,57], but the effect of genetic variation in RRM1 is less known. Bepler et al. resequenced the RRM1 promoter and identified two functional polymorphisms that altered promoter activity. These variants showed a modest association with overall and disease-free survival in NSCLC [58]. However, in another study of 135 advanced NSCLC cases, one of these SNPs (rs12806698) was not found to be associated with overall survival, except for patients with a performance status of 0 [52]. It remains to be seen if there is a genetic basis for the observed variation in gene expression that can be used as a direct prognostic marker.
5.3 Apoptosis
If the Pt-DNA lesions are not repaired, the damaged DNA triggers activation of the apoptosis pathway. This is an essential step for the effectiveness of platinum-based chemotherapeutics in killing tumor cells and deficiencies in this process can contribute to the development of chemoresistance. Several polymorphisms have been analyzed for association with response to therapy and overall survival. TP53 (p53) is considered the gatekeeper for cell survival or death. There are several TP53 polymorphisms, but the most commonly studied is the arginine to proline change at codon 72 (rs1042522). The Arg72 variant was shown to be better at inducing apoptosis compared to the Pro allele [59,60]. Han et al. reported that patients carrying two copies of Pro72 were more likely to be resistant to cisplatin–irinotecan chemotherapy in advanced NSCLC [61]. However, this result was not duplicated in a population of 148 advanced NSCLC patients also treated with cisplatin-based chemotherapy [20]. TP53 is regulated by the oncogene MDM2. Genetic variation in MDM2 has also been shown to be associated with clinical outcomes in NSCLC [61]. For this intronic SNP (rs2279744), those with the TT genotype had increased p53 expression and increased overall survival.
6. Genome-wide approaches
With advances in genotyping and gene expression technologies, several studies have taken an unbiased approach towards understanding the genetic factors influencing response to platinum-based chemotherapy. These studies include those analyzing lymphoblastoid cell lines generated as part of the international HapMap Project, lung cancer cell lines from the NCI-60 panel and tumor tissue from NSCLC patients. In lymphoblastoid cell lines obtained from pedigrees of Centre d’Etudes du Polymorphisme Humain individuals of European background, it was estimated that ~ 30 – 40% of the variation in cisplatin-induced cytotoxicity was owing to heritable factors [62,63]. This has been followed by several other studies analyzing genetic factors contributing to cisplatin and carboplatin cytotoxicity in HapMap cell lines from Centre d’Etudes du Polymorphisme Humain and African individuals [63–66]. The advantage of utilizing these cell lines is that the HapMap provides genome-wide SNP data and, thus, are useful tools for performing linkage and association studies. This approach has identified several loci and, interestingly, none of the significant genes have previously been associated with cisplatin response. However, the results of these studies have not been replicated in any cancer patients treated with platinum-based chemotherapeutics. There is a possibility that these results are specific for cytotoxicity in lymphoblastoid cell lines or, owing to the genome-wide nature of the studies, false-positives.
Another useful tool is the NCI-60 tumor cell line panel [67]. This panel is comprised of tumor cell lines from several sites, including nine NSCLCs. These cells have been analyzed for sensitivity/resistance to many chemotherapeutic agents and, with the availability of genome-wide gene expression data, have been used to identify variation in gene expression associated with cell survival [68]. For cisplatin resistance, interestingly, the candidate pathways identified were what would be expected based on what is known about the pharmacokinetics and pharmacodynamics of this drug [69]. In another study, hypermethylation of TP73, a homologue of p53, was shown to increase sensitivity to cisplatin and other alkylating agents [70]. However, because of the limited number of NSCLC cell lines included in the NCI-60 panel, it is difficult to study individual variation in response to chemotherapy. Therefore, the main use of these cell lines is to identify candidate pathways for analysis in a larger patient population.
Expression studies utilizing tumor tissue from NSCLC patients have shown to be successful in identifying prognostic signatures. Chen et al. published a five-gene signature based on tissue samples from 125 stage I–III NSCLC patients [71]. In a replication set of 60 samples, these five genes were able to successfully identify high and low risk groups for overall and relapse-free survival. Similarly, a signature containing 133 genes was shown to be valuable in predicting recurrence in 89 patients with early-stage NSCLC [72]. Unfortunately, the results of these studies and others do not overlap, most likely owing to tumor heterogeneity, lack of statistical power and data analysis differences [73]. Another strategy is to query microRNA expression differences. A microRNA profile was shown to be associated with overall survival and relapse in tumor samples from 112 NSCLC [74]. This signature containing five microRNAs was validated and shown to be an independent predictor of clinical outcome for early stage NSCLC.
Although tissue expression-based approaches show promise, the disadvantage of these studies is that they rely on tumor tissue. Advanced NSCLC patients who receive platinum-based chemotherapy are often not candidates for surgery and a portion of patients with locally advanced or even local disease are also not surgical candidates owing to other underlying medical conditions. This limitation would make expression-based profiles impossible for a large subset of NSCLC patients. Therefore, it would be advantageous to link gene expression profiles to underlying genetic variation. This would allow for an assessment of prognosis through DNA easily obtained from a blood sample and not dependent on tumor tissue acquisition.
With the emergence the of genome-wide SNP arrays, genome-wide association studies (GWAS) in large patient populations are now possible. These GWAS have been shown useful in identifying previously unknown risk loci for lung cancer [75–77]. The case component of these studies, with the inclusion of well characterized treatment and outcome information, can be used for clinical outcome study. The available GWAS data would be invaluable in identifying predictors of clinical outcomes. One of the first genome-wide pharmacogenomic studies was recently published in the New England Journal of Medicine [78], and surely more will follow. The danger of these studies is the high probability of false-positive findings. However, these studies will be immensely useful in uncovering new susceptibility loci.
7. Expert opinion
Platinum-based chemotherapeutics are the mainstay of treatment for advanced NSCLC. Unfortunately, survival rates for patients after treatment are still poor at only 29% [5]. The primary obstacle to successful treatment with cisplatin, carboplatin and other platinum agents is the development of chemoresistance. Although much work has been on genetic variation influencing risk of NSCLC, as this review has shown, there is a growing body of evidence that genetic variation also modulates how a patient responds to platinum chemotherapy.
These studies are difficult to design, complete and interpret owing to several issues related to how NSCLC patients are treated. Advanced NSCLC is commonly treated with cisplatin (or carboplatin) combination therapy, often with the addition of radiation. This can make it difficult for sorting out the influence of genetic variation on sensitivity or resistance to platinum agents when the effect may be owing to the other chemotherapeutics used or radiotherapy. This has resulted in several reports of single SNP effects on overall survival or response that have been inconclusive and inconsistent. Further studies with larger and well-characterized patient populations are necessary to explain the effects of genetic variation on each aspect of an NSCLC patient’s treatment regimen. These studies will also need to be anchored by functional genomic studies that clearly demonstrate how the genetic variant is able to alter response to platinum agents.
Much of the focus on genetic variation influencing clinical outcomes following platinum-based therapy has been on SNPs within the DNA damage and repair pathway. Studies have shown that the effects of each individual SNP are modest and suggest that a single genetic variant alone is not strongly associated with resistance or sensitivity to chemotherapy. However, when the effect of several SNPs within the same pathway is analyzed, the effect is much more dramatic as shown by Wu et al. [20]. These results also highlight the need to use advanced statistics to better identify patients at risk for a poor prognosis. Statistical tools such as survival tree analysis and classification and regression tree analysis are methods that allow for the identification of gene–gene and gene–environment interactions and have been extensively used for identifying higher-order interactions modulating cancer risk [79–83] and more recently for NSCLC and esophageal overall survival [20,84,85].
A further hurdle to overcome is the relationship between chemoresistance and toxicity. If we are able to identify patients who respond well to specific drugs and, therefore, decreased chemoresistance, how do we balance the expected increased risk of toxicity? Studies need to be developed that not only identify appropriate chemotherapeutics, but also the best dose and duration for each patient to avoid the common dose-limiting side effects of platinum-based therapy. Studies have shown that many of the same genes involved in chemoresistance are involved in the development of toxicity, including genetic variation in ERCC1 [86]. Obviously, this is a much bigger issue and one that needs to be addressed in the future to individualize therapy for each patient.
The success of GWAS in identifying novel susceptibility loci for several cancers has opened the door for similar studies of clinical outcomes following platinum-based chemotherapy for NSCLC. GWAS may uncover the hidden genetic variants that influence response to therapy. GWAS are not fool-proof although and are limited by the great number of false-positive findings. This requires large sample populations with several replications and validations to identify the true causative variant. This will only be possible through collaborative efforts to cross-validate and pool information.
We propose that the best mechanism to accurately predict clinical outcome is the creation of a risk prediction model that incorporates genetic, phenotypic, epidemiologic and clinical variables calibrated specifically for NSCLC patients receiving platinum-based chemotherapeutics. These comprehensive risk prediction models have been developed for cancer risk prediction [87–90]. This same approach can be implemented for clinical outcomes to supply the physician with the information necessary to select the most appropriate treatment regimen.
Finally, before any of this information can truly make a difference in the clinic, prospective studies are needed to validate and confirm that knowledge of the patient’s underlying genetic variation makes a significant difference in clinical outcome for advanced NSCLC patients. This is not an easy task, but with well-designed studies utilizing current knowledge as the foundation, pharmacogenomics can go from the bench to the bedside.
Acknowledgments
This work was supported by National Cancer Institute grants R01 CA111646 and P50 CA070907.
Footnotes
Declaration of interest
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350(4):379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 4.Rapp E, Pater JL, Willan A, et al. Chemotherapy can prolong survival in patients with advanced non-small-cell lung cancer–report of a Canadian multicenter randomized trial. J Clin Oncol. 1988;6(4):633–641. doi: 10.1200/JCO.1988.6.4.633. [DOI] [PubMed] [Google Scholar]
- 5. Chemotherapy in addition to supportive care improves survival in advanced non-small-cell lung cancer: a systematic review and meta-analysis of individual patient data from 16 randomized controlled trials. J Clin Oncol. 2008;26(28):4617–4625. doi: 10.1200/JCO.2008.17.7162. •• Systematic study of supporting the advantage of including chemotherapy in the treatment of NSCLC.
- 6. Kartalou M, Essigmann JM. Mechanisms of resistance to cisplatin. Mutat Res. 2001;478(1–2):23–43. doi: 10.1016/s0027-5107(01)00141-5. •• Comprehensive review of the mechanisms underlying platinum resistance.
- 7. Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33(1):9–23. doi: 10.1016/j.ctrv.2006.09.006. • Good review of the mechanisms underlying platinum resistance.
- 8.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22(47):7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 9.Alberts WM. Diagnosis and management of lung cancer executive summary: ACCP evidence-based clinical practice guidelines. Chest. (2nd Edition) 2007;132(3 Suppl):1S–19S. doi: 10.1378/chest.07-1860. [DOI] [PubMed] [Google Scholar]
- 10.Kelland LR, Abel G, McKeage MJ, et al. Preclinical antitumor evaluation of bis-acetato-ammine-dichlorocyclohexylamine platinum(IV): an orally active platinum drug. Cancer Res. 1993;53(11):2581–2586. [PubMed] [Google Scholar]
- 11.Ishida S, Lee J, Thiele DJ, Herskowitz I. Uptake of the anticancer drug cisplatin mediated by the copper transporter Ctr1 in yeast and mammals. Proc Natl Acad Sci USA. 2002;99(22):14298–14302. doi: 10.1073/pnas.162491399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holzer AK, Manorek GH, Howell SB. Contribution of the major copper influx transporter CTR1 to the cellular accumulation of cisplatin, carboplatin, and oxaliplatin. Mol Pharmacol. 2006;70(4):1390–1394. doi: 10.1124/mol.106.022624. [DOI] [PubMed] [Google Scholar]
- 13.Song IS, Savaraj N, Siddik ZH, et al. Role of human copper transporter Ctr1 in the transport of platinum-based antitumor agents in cisplatin-sensitive and cisplatin-resistant cells. Mol Cancer Ther. 2004;3(12):1543–1549. [PubMed] [Google Scholar]
- 14.Komatsu M, Sumizawa T, Mutoh M, et al. Copper-transporting P-type adenosine triphosphatase (ATP7B) is associated with cisplatin resistance. Cancer Res. 2000;60(5):1312–1316. [PubMed] [Google Scholar]
- 15.Nakayama K, Kanzaki A, Terada K, et al. Prognostic value of the Cu-transporting ATPase in ovarian carcinoma patients receiving cisplatin-based chemotherapy. Clin Cancer Res. 2004;10(8):2804–2811. doi: 10.1158/1078-0432.ccr-03-0454. [DOI] [PubMed] [Google Scholar]
- 16.Samimi G, Varki NM, Wilczynski S, et al. Increase in expression of the copper transporter ATP7A during platinum drug-based treatment is associated with poor survival in ovarian cancer patients. Clin Cancer Res. 2003;9(16 Pt 1):5853–5859. [PubMed] [Google Scholar]
- 17.Liedert B, Materna V, Schadendorf D, et al. Overexpression of cMOAT (MRP2/ABCC2) is associated with decreased formation of platinum-DNA adducts and decreased G2-arrest in melanoma cells resistant to cisplatin. J Invest Dermatol. 2003;121(1):172–176. doi: 10.1046/j.1523-1747.2003.12313.x. [DOI] [PubMed] [Google Scholar]
- 18.Han JY, Lim HS, Yoo YK, et al. Associations of ABCB1, ABCC2, and ABCG2 polymorphisms with irinotecan-pharmacokinetics and clinical outcome in patients with advanced non-small cell lung cancer. Cancer. 2007;110(1):138–147. doi: 10.1002/cncr.22760. [DOI] [PubMed] [Google Scholar]
- 19.Sohn JW, Lee SY, Lee SJ, et al. MDR1 polymorphisms predict the response to etoposide-cisplatin combination chemotherapy in small cell lung cancer. Jpn J Clin Oncol. 2006;36(3):137–141. doi: 10.1093/jjco/hyi231. [DOI] [PubMed] [Google Scholar]
- 20. Wu X, Lu C, Ye Y, et al. Germline genetic variations in drug action pathways predict clinical outcomes in advanced lung cancer treated with platinum-based chemotherapy. Pharmacogenet Genomics. 2008;18(11):955–965. doi: 10.1097/FPC.0b013e32830efdd4. •• Comprehensive study of the association between genetic variations in candidate genes and response to platinum-based chemotherapeutics in NSCLC.
- 21.Lai SL, Hwang J, Perng RP, Whang-Peng J. Modulation of cisplatin resistance in acquired-resistant nonsmall cell lung cancer cells. Oncol Res. 1995;7(1):31–38. [PubMed] [Google Scholar]
- 22.Goto S, Iida T, Cho S, et al. Overexpression of glutathione S-transferase pi enhances the adduct formation of cisplatin with glutathione in human cancer cells. Free Radic Res. 1999;31(6):549–558. doi: 10.1080/10715769900301121. [DOI] [PubMed] [Google Scholar]
- 23.Moyer AM, Salavaggione OE, Wu TY, et al. Glutathione s-transferase p1: gene sequence variation and functional genomic studies. Cancer Res. 2008;68(12):4791–4801. doi: 10.1158/0008-5472.CAN-07-6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peklak-Scott C, Smitherman PK, Townsend AJ, Morrow CS. Role of glutathione S-transferase P1-1 in the cellular detoxification of cisplatin. Mol Cancer Ther. 2008;7(10):3247–3255. doi: 10.1158/1535-7163.MCT-08-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu C, Spitz MR, Zhao H, et al. Association between glutathione S-transferase pi polymorphisms and survival in patients with advanced nonsmall cell lung carcinoma. Cancer. 2006;106(2):441–447. doi: 10.1002/cncr.21619. [DOI] [PubMed] [Google Scholar]
- 26.Booton R, Ward T, Heighway J, et al. Glutathione-S-transferase P1 isoenzyme polymorphisms, platinum-based chemotherapy, and non-small cell lung cancer. J Thorac Oncol. 2006;1(7):679–683. [PubMed] [Google Scholar]
- 27.Kiyohara C, Yoshimasu K, Takayama K, Nakanishi Y. NQO1, MPO, and the risk of lung cancer: a HuGE review. Genet Med. 2005;7(7):463–478. doi: 10.1097/01.gim.0000177530.55043.c1. [DOI] [PubMed] [Google Scholar]
- 28.Siegel D, McGuinness SM, Winski SL, Ross D. Genotype-phenotype relationships in studies of a polymorphism in NAD(P) H:quinone oxidoreductase 1. Pharmacogenetics. 1999;9(1):113–121. doi: 10.1097/00008571-199902000-00015. [DOI] [PubMed] [Google Scholar]
- 29.Austin GE, Lam L, Zaki SR, et al. Sequence comparison of putative regulatory DNA of the 5′ flanking region of the myeloperoxidase gene in normal and leukemic bone marrow cells. Leukemia. 1993;7(9):1445–1450. [PubMed] [Google Scholar]
- 30.Piedrafita FJ, Molander RB, Vansant G, et al. An Alu element in the myeloperoxidase promoter contains a composite SP1-thyroid hormone-retinoic acid response element. J Biol Chem. 1996;271(24):14412–14420. doi: 10.1074/jbc.271.24.14412. [DOI] [PubMed] [Google Scholar]
- 31.Simon GR, Ismail-Khan R, Bepler G. Nuclear excision repair-based personalized therapy for non-small cell lung cancer: from hypothesis to reality. Int J Biochem Cell Biol. 2007;39(7–8):1318–1328. doi: 10.1016/j.biocel.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193(1–2):3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 33.Zeng-Rong N, Paterson J, Alpert L, et al. Elevated DNA repair capacity is associated with intrinsic resistance of lung cancer to chemotherapy. Cancer Res. 1995;55(21):4760–4764. [PubMed] [Google Scholar]
- 34.Bosken CH, Wei Q, Amos CI, Spitz MR. An analysis of DNA repair as a determinant of survival in patients with non-small-cell lung cancer. J Natl Cancer I. 2002;94(14):1091–1099. doi: 10.1093/jnci/94.14.1091. [DOI] [PubMed] [Google Scholar]
- 35.Lin J, Swan GE, Shields PG, et al. Mutagen sensitivity and genetic variants in nucleotide excision repair pathway: genotype-phenotype correlation. Cancer Epidemiol Biomarkers Prev. 2007;16(10):2065–2071. doi: 10.1158/1055-9965.EPI-06-1041. [DOI] [PubMed] [Google Scholar]
- 36. Matakidou A, el Galta R, Webb EL, et al. Genetic variation in the DNA repair genes is predictive of outcome in lung cancer. Hum Mol Genet. 2007;16(19):2333–2340. doi: 10.1093/hmg/ddm190. • Comprehensive study of genetic variation and clinical outcomes for lung cancer.
- 37.Camps C, Sarries C, Roig B, et al. Assessment of nucleotide excision repair XPD polymorphisms in the peripheral blood of gemcitabine/cisplatin-treated advanced non-small-cell lung cancer patients. Clin Lung Cancer. 2003;4(4):237–241. doi: 10.3816/clc.2003.n.004. [DOI] [PubMed] [Google Scholar]
- 38.Giachino DF, Ghio P, Regazzoni S, et al. Prospective assessment of XPD Lys751Gln and XRCC1 Arg399Gln single nucleotide polymorphisms in lung cancer. Clin Cancer Res. 2007;13(10):2876–2881. doi: 10.1158/1078-0432.CCR-06-2543. [DOI] [PubMed] [Google Scholar]
- 39.Isla D, Sarries C, Rosell R, et al. Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15(8):1194–1203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 40.Ryu JS, Hong YC, Han HS, et al. Association between polymorphisms of ERCC1 and XPD and survival in non-small-cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer. 2004;44(3):311–316. doi: 10.1016/j.lungcan.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 41.Tibaldi C, Giovannetti E, Vasile E, et al. Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2008;14(6):1797–1803. doi: 10.1158/1078-0432.CCR-07-1364. [DOI] [PubMed] [Google Scholar]
- 42.Gurubhagavatula S, Liu G, Park S, et al. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22(13):2594–2601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 43.Booton R, Ward T, Heighway J, et al. Xeroderma pigmentosum group D haplotype predicts for response, survival, and toxicity after platinum-based chemotherapy in advanced nonsmall cell lung cancer. Cancer. 2006;106(11):2421–2427. doi: 10.1002/cncr.21885. [DOI] [PubMed] [Google Scholar]
- 44.Mellon I, Hock T, Reid R, et al. Polymorphisms in the human xeroderma pigmentosum group A gene and their impact on cell survival and nucleotide excision repair. DNA Repair. 2002;1(7):531–546. doi: 10.1016/s1568-7864(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 45.Wu X, Zhao H, Wei Q, et al. XPA polymorphism associated with reduced lung cancer risk and a modulating effect on nucleotide excision repair capacity. Carcinogenesis. 2003;24(3):505–509. doi: 10.1093/carcin/24.3.505. [DOI] [PubMed] [Google Scholar]
- 46.Sun X, Li F, Sun N, et al. Polymorphisms in XRCC1 and XPG and response to platinum-based chemotherapy in advanced non-small cell lung cancer patients. Lung Cancer. 2009 doi: 10.1016/j.lungcan.2008.11.014. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 47.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355(10):983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 48.Cobo M, Isla D, Massuti B, et al. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol. 2007;25(19):2747–2754. doi: 10.1200/JCO.2006.09.7915. [DOI] [PubMed] [Google Scholar]
- 49.Zhou W, Gurubhagavatula S, Liu G, et al. Excision repair cross-complementation group 1 polymorphism predicts overall survival in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res. 2004;10(15):4939–4943. doi: 10.1158/1078-0432.CCR-04-0247. [DOI] [PubMed] [Google Scholar]
- 50.Park SY, Hong YC, Kim JH, et al. Effect of ERCC1 polymorphisms and the modification by smoking on the survival of non-small cell lung cancer patients. Med Oncol. 2006;23(4):489–498. doi: 10.1385/MO:23:4:489. [DOI] [PubMed] [Google Scholar]
- 51.Su D, Ma S, Liu P, et al. Genetic polymorphisms and treatment response in advanced non-small cell lung cancer. Lung Cancer. 2007;56(2):281–288. doi: 10.1016/j.lungcan.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 52.de las Penas R, Sanchez-Ronco M, Alberola V, et al. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann Oncol. 2006;17(4):668–675. doi: 10.1093/annonc/mdj135. [DOI] [PubMed] [Google Scholar]
- 53.Betticher DC, Thatcher N, Altermatt HJ, et al. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11(5):1005–1011. [PubMed] [Google Scholar]
- 54.Gautschi O, Hugli B, Ziegler A, et al. Cyclin D1 (CCND1) A870G gene polymorphism modulates smoking-induced lung cancer risk and response to platinum-based chemotherapy in non-small cell lung cancer (NSCLC) patients. Lung Cancer. 2006;51(3):303–311. doi: 10.1016/j.lungcan.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 55.Filipits M, Pirker R, Dunant A, et al. Cell cycle regulators and outcome of adjuvant cisplatin-based chemotherapy in completely resected non-small-cell lung cancer: the International Adjuvant Lung Cancer Trial Biologic Program. J Clin Oncol. 2007;25(19):2735–2740. doi: 10.1200/JCO.2006.08.2867. [DOI] [PubMed] [Google Scholar]
- 56.Bepler G, Sharma S, Cantor A, et al. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22(10):1878–1885. doi: 10.1200/JCO.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 57.Zheng Z, Chen T, Li X, et al. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356(8):800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 58.Bepler G, Zheng Z, Gautam A, et al. Ribonucleotide reductase M1 gene promoter activity, polymorphisms, population frequencies, and clinical relevance. Lung Cancer. 2005;47(2):183–192. doi: 10.1016/j.lungcan.2004.07.043. [DOI] [PubMed] [Google Scholar]
- 59.Dumont P, Leu JI, Della Pietra AC, 3rd, et al. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33(3):357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 60.Thomas M, Kalita A, Labrecque S, et al. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19(2):1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han JY, Lee GK, Jang DH, et al. Association of p53 codon 72 polymorphism and MDM2 SNP309 with clinical outcome of advanced nonsmall cell lung cancer. Cancer. 2008;113(4):799–807. doi: 10.1002/cncr.23668. [DOI] [PubMed] [Google Scholar]
- 62.Dolan ME, Newbold KG, Nagasubramanian R, et al. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64(12):4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 63.Shukla SJ, Duan S, Badner JA, et al. Susceptibility loci involved in cisplatin-induced cytotoxicity and apoptosis. Pharmacogenet Genomics. 2008;18(3):253–262. doi: 10.1097/FPC.0b013e3282f5e605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang RS, Duan S, Shukla SJ, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am J Hum Genet. 2007;81(3):427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang RS, Duan S, Kistner EO, et al. Genetic variants associated with carboplatin-induced cytotoxicity in cell lines derived from Africans. Mol Cancer Ther. 2008;7(9):3038–3046. doi: 10.1158/1535-7163.MCT-08-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shukla SJ, Duan S, Wu X, et al. Whole-genome approach implicates CD44 in cellular resistance to carboplatin. Hum Genomics. 2009;3(2):128–142. doi: 10.1186/1479-7364-3-2-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6(10):813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 68.Weinstein JN, Pommier Y. Transcriptomic analysis of the NCI-60 cancer cell lines. C R Biol. 2003;326(10–11):909–920. doi: 10.1016/j.crvi.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 69.Riedel RF, Porrello A, Pontzer E, et al. A genomic approach to identify molecular pathways associated with chemotherapy resistance. Mol Cancer Ther. 2008;7(10):3141–3149. doi: 10.1158/1535-7163.MCT-08-0642. [DOI] [PubMed] [Google Scholar]
- 70.Shen L, Kondo Y, Ahmed S, et al. Drug sensitivity prediction by CpG island methylation profile in the NCI-60 cancer cell line panel. Cancer Res. 2007;67(23):11335–11343. doi: 10.1158/0008-5472.CAN-07-1502. [DOI] [PubMed] [Google Scholar]
- 71. Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356(1):11–20. doi: 10.1056/NEJMoa060096. • One of the major papers to use an expression-based signature for prognosis.
- 72.Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355(6):570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 73.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. P Natl Acad Sci USA. 2006;103(15):5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57. doi: 10.1016/j.ccr.2007.12.008. • First microRNA-based signature for clinical outcomes in lung cancer.
- 75.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 77.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Link E, Parish S, Armitage J, et al. SLCO1B1 variants and statin-induced myopathy–a genomewide study. N Engl J Med. 2008;359(8):789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 79.Chen M, Kamat AM, Huang M, et al. High-order interactions among genetic polymorphisms in nucleotide excision repair pathway genes and smoking in modulating bladder cancer risk. Carcinogenesis. 2007;28(10):2160–2165. doi: 10.1093/carcin/bgm167. [DOI] [PubMed] [Google Scholar]
- 80.Huang M, Dinney CP, Lin X, et al. High-order interactions among genetic variants in DNA base excision repair pathway genes and smoking in bladder cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2007;16(1):84–91. doi: 10.1158/1055-9965.EPI-06-0712. [DOI] [PubMed] [Google Scholar]
- 81.Wu X, Gu J, Grossman HB, et al. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006;78(3):464–479. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang H, Lippman SM, Huang M, et al. Genetic polymorphisms in double-strand break DNA repair genes associated with risk of oral premalignant lesions. Eur J Cancer. 2008;44(11):1603–1611. doi: 10.1016/j.ejca.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ye Y, Yang H, Grossman HB, et al. Genetic variants in cell cycle control pathway confer susceptibility to bladder cancer. Cancer. 2008;112(11):2467–2474. doi: 10.1002/cncr.23472. [DOI] [PubMed] [Google Scholar]
- 84.Hildebrandt MA, Yang H, Hung MC, et al. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J Clin Oncol. 2009;27(6):857–871. doi: 10.1200/JCO.2008.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu X, Gu J, Wu TT, et al. Genetic variations in radiation and chemotherapy drug action pathways predict clinical outcomes in esophageal cancer. J Clin Oncol. 2006;24(23):3789–3798. doi: 10.1200/JCO.2005.03.6640. [DOI] [PubMed] [Google Scholar]
- 86.Suk R, Gurubhagavatula S, Park S, et al. Polymorphisms in ERCC1 and grade 3 or 4 toxicity in non-small cell lung cancer patients. Clin Cancer Res. 2005;11(4):1534–1538. doi: 10.1158/1078-0432.CCR-04-1953. [DOI] [PubMed] [Google Scholar]
- 87.Etzel CJ, Kachroo S, Liu M, et al. Development and validation of a lung cancer risk prediction model for African-Americans. Cancer Prev Res. 2008;1(4):255–265. doi: 10.1158/1940-6207.CAPR-08-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spitz MR, Etzel CJ, Dong Q, et al. An expanded risk prediction model for lung cancer. Cancer Prev Res. 2008;1(4):250–254. doi: 10.1158/1940-6207.CAPR-08-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spitz MR, Hong WK, Amos CI, et al. A risk model for prediction of lung cancer. J Natl Cancer Inst. 2007;99(9):715–726. doi: 10.1093/jnci/djk153. [DOI] [PubMed] [Google Scholar]
- 90.Wu X, Lin J, Grossman HB, et al. Projecting individualized probabilities of developing bladder cancer in white individuals. J Clin Oncol. 2007;25(31):4974–4981. doi: 10.1200/JCO.2007.10.7557. [DOI] [PubMed] [Google Scholar]