Abstract
Benzo[a]pyrene 7,8-diol 9,10-epoxide adducts in DNA are implicated in mutagenesis, and their formation from the diol epoxides and subsequent incorrect replication by human DNA polymerases provide an attractive mechanism for the induction of cancer by this highly carcinogenic hydrocarbon and its diol epoxide metabolites. Here, we describe the crystal structure of such an adduct at the exocyclic amino group of a purine nucleoside. The present adduct derives from trans opening at C10 of the (-)-(7S,8R)-diol (9R,10S)-epoxide enantiomer by the exocyclic N2-amino group of deoxyguanosine. In the crystal, the pyrene rings of adjacent molecules stack with each other, but the guanine bases do not stack either intermolecularly with each other or intramolecularly with the pyrene. The most notable features of the molecular structure are (i) independent and unambiguous proof of the absolute configuration of the adduct based on the spatial relationship between the known chiral carbon atoms of the deoxyribose and the four asymmetric centers in the hydrocarbon moiety; (ii) visualization of the relative orientations of the pyrene and guanine ring systems as well as the conformation of the partially saturated hydrocarbon ring (comprising carbon atoms 7, 8, 9, and 10), both of which conformational features in the crystal are in good agreement with deductions from NMR and CD measurements in solution; and (iii) the presence in the crystal of a syn glycosidic torsion angle, a conformation that is unusual in B-DNA but that may be involved in error-prone replication of these benzo[a]pyrene 7,8-diol 9,10-epoxide deoxyguanosine adducts by DNA polymerases.
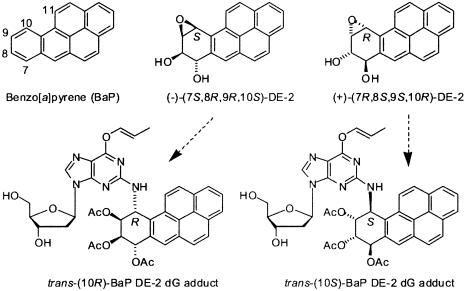
The polycyclic aromatic hydrocarbon benzo[a]pyrene (BaP), first isolated from coal tar and characterized by Cook et al. in 1933 (1), is one of the most prevalent environmental carcinogens to which humans are exposed (2). It is metabolized in mammals (3) by oxidation on the angular benzo-ring (carbon atoms 7, 8, 9, and 10; Fig. 1) to give two enantiomeric pairs of diastereomeric 7,8-diol 9,10-epoxides (DEs), designated as DE-1 (benzylic 7-hydroxyl group and epoxide oxygen cis; also referred to as the syn diastereomer) and DE-2 (benzylic 7-hydroxyl group and epoxide oxygen trans; also referred to as the anti diastereomer). Of these four DE isomers, (+)-(7R,8S,9S,10R)-DE-2 predominates on metabolism of the hydrocarbon in mammals (3) and is also by far the most tumorigenic isomer in animal models (4). Reaction of BaP DEs with nucleic acids in cells or in vitro occurs mainly by trans and to a lesser extent cis ring opening of their epoxide rings at C10 by the exocyclic 2- and 6-amino groups of the guanine and adenine bases, respectively, to give 16 possible covalent adducts (5-8), with the trans-(10S) deoxyguanosine (dG) adduct derived from the highly tumorigenic (+)-(7R,8S,9S,10R)-DE-2 predominating on reaction with DNA. Analogous ring opening of the nontumorigenic (-)-(7S,8R,9R,10S)-DE-2 enantiomer gives the trans-(10R)-BaP DE dG adduct. Because of the large difference in tumorigenicity between (+)- and (-)-BaP DE-2, comparison of the structures of nucleoside adducts derived from the two enantiomers is of considerable interest. Molecular features such as the preferred orientation of the hydrocarbon and guanine ring systems relative to each other, the preferred conformation of the glycosidic bond between the sugar and the purine base, and the conformation of the partially saturated tetrahydro benzo-ring can potentially influence the fit of these adducts in DNA into the active site of DNA processing enzymes.
Fig. 1.
Structures of BaP, its two optically active DE-2 7,8-diol 9,10-epoxides, and the (10R)- and (10S)-BaP DE-2 dG adducts derived from trans opening of (-)-(7S,8R,9R,10S)-DE-2 and (+)-(7R,8S,9S,10R)-DE-2, respectively, as their O6-allyl-7,8,9-triacetates. Only the (10R) adduct isomer provided crystals that gave satisfactory diffraction data.
Crystal structures for both the racemic BaP DE-1 (9) and DE-2 (10) diastereomers have confirmed their relative stereochemistry as deduced by NMR (11, 12). Absolute configurations of their optically active precursor trans-7,8-dihydrodiols were assigned by exciton chirality CD spectroscopy (13) of a derivative, and the configurations of the DEs were established by chemical synthesis from the assigned dihydrodiols (13). Configurational assignments for the dihydrodiols were subsequently confirmed by x-ray crystallography (14).
In contrast to the parent BaP DEs, crystal structures for their adducts with either dG or deoxyadenosine (dA) have proved extremely elusive. Crystals of nucleoside or nucleotide adducts either free or with selective blocking groups had not been described. In the present study, we report the preparation of crystals of trans-opened N2-dG (designated as N2g in the crystal structure) adducts of both (+)- and (-)-DE-2. Key to our ability to obtain these crystals has been the serendipitous selection of the blocking groups shown in Fig. 1. Although both sets of crystals were of adequate size for diffraction studies, only the crystals that correspond to trans ring opening of the (-)-(7S,8R,9R,10S)-DE-2 isomer to produce the trans-(10R) dG adduct gave x-ray diffraction data suitable for structure determination. We here describe the molecular structure of this adduct, which has acetyl blocking groups on the 7-, 8-, and 9-hydroxyl groups of the trans-opened BaP DE moiety and an allyl blocking group on O6 of the dG (Fig. 1) (designated as Olg in the crystal structure). Our structure makes possible an unambiguous assignment of absolute configuration for the chiral centers in the hydrocarbon portion of the adduct by internal comparison with the known absolute configuration of deoxyribose. The relative orientation of the pyrene and guanine rings and the conformation of the tetrahydro benzo-ring of the BaP moiety in the crystal are similar to these conformational features of the molecule in solution as deduced from CD and NMR spectra. This crystal structure provides a basis for comparative interpretation of CD and NMR properties of the trans-opened dG adducts derived from (-)-DE-2 and its highly carcinogenic (+)-DE-2 enantiomer. Thus it constitutes a valuable starting point to analyze enzyme-substrate interactions critical to understanding error-prone replication, subsequent mutation, and the induction of cancer by BaP DE adducts in DNA.
Materials and Methods
The (10R)-BaP DE dG adduct, formed by trans-opening of the (-)-(7S,8R,9R,10S)-DE-2 isomer, as its O6-allyl 7,8,9-triacetate, N2-[10R-(7S,8R,9S-triacetoxy-7,8,9,10-tetrahydrobenzo[a]pyrenyl)]-O6-allyl-2′-dG, and the corresponding (10S)-diastereomer from the (+)-(7R,8S,9S,10R)-DE-2 enantiomer (Fig. 1), were prepared as described for the mixed (10R/10S) diastereomers (15, 16) in 95% yield from the corresponding 3′,5′-bis-tert-butyldimethylsilyl ether by treatment with 7% HF-pyridine at room temperature for 20 h (17). Crystallization of the trans-(10R)-BaP DE dG adduct from ethanol afforded colorless prisms: mp 185-187°C; [α]D +31° (c = 0.9, CH2Cl2); highresolution MS (HRMS) calculated for C39H37O10N5 (M + Cs+) 868.1595; found 868.1616. Crystallization of the trans-(10S)-BaP DE dG adduct from ethanol afforded colorless chunky needles: mp 184-185°C; [α]D -64.4° (c = 6.75, CH2Cl2); HRMS calculated for C39H37O10N5 (M + Cs+) 868.1595; found 868.1592. Listings and traces of the NMR spectra for the trans-(10S)- and trans-(10R)-BaPDE dG adducts are given in Fig. 6, which is published as supporting information on the PNAS web site.
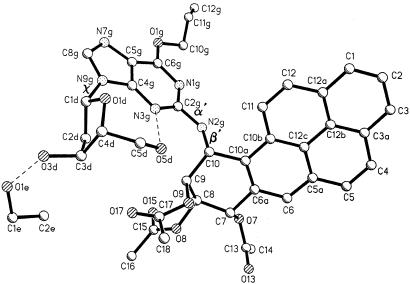
3D x-ray diffraction data on a crystal (0.40 × 0.32 × 0.13 mm) of the (10R) adduct obtained by slow evaporation of the ethanol solvent were measured with CuKα radiation with an automated four-circle diffractometer at -75°C. The θ-2θ scan technique was used to measure data up to 2θmax = 116°. Of 4,405 reflections measured, 3,991 were considered observed with |Fo| > 2σ(I). The structure was solved by direct phase determination and refined by full-matrix least-squares refinement on F2 data. Resolution of 0.90 Å was achieved. The labeling of the atoms, including a cocrystallized ethanol molecule, is shown in Fig. 2. All hydrogen atoms were located in difference maps and, except for the deoxyribose protons H(O3d) and H(O5d) (Fig. 2), were adjusted to idealized positions and allowed to ride on the C or N atom to which they were bonded. The coordinates for H(O3d) and H(O5d) were refined by least squares with those of all of the nonhydrogen atoms. The final reliability factor was R1 = 0.045 for 3,991 observed data and 515 parameters. Crystal data were as follows: C39H37N5O10.C2H5OH, formula weight 735.80 + 46.1, orthorhombic space group C2221, a = 10.233(1) Å, b = 19.636(1) Å, c = 38.277(3) Å, V = 7,691.2 Å3, Z = 8, Dcalc = 1.350 Mg/m3. Coordinates, bond lengths, and angles for the trans-(10R)-BaP DE dG adduct have been deposited with the Cambridge Crystallographic Data Centre.
Fig. 2.
Absolute configuration of N2-[10R-(7S,8R,9S-triacetoxy-7,8,9,10-tetrahydrobenzo[a]pyrenyl)]-O6-allyl-2′-dG [protected trans-(10R)-BaP DE dG adduct]. The labeling of the atoms in the molecule, the cocrystallized ethanol molecule, and the major torsional angles that define the conformation of the adduct, α′ = N1g-C2g-N2g-C10, β′ = C2g-N2g-C10-C9, and χ = O1d-C1d-N9g-C4g, are shown. The dashed lines indicate hydrogen bonds.
Crystals of the trans-(10S)-BaP DE dG adduct were examined as above. However, x-ray diffraction data from several of these crystals, one for example of size 0.56 × 0.13 × 0.14 mm, showed intensity falloff at relatively small scattering angles, possibly because of disorder in part of the molecule, thus obviating molecular structure determination. The cell constants for this crystal are: a = 20.39 Å, b = 22.82 Å, c = 57.63 Å, α = β = γ = 90°, space group most probably P212121.
Results and Discussion
Absolute Configuration. The present x-ray structure permits the unambiguous assignment of absolute configuration to the chiral atoms of the trans-opened DE moiety of the (10R)-BaP DE dG adduct. In previous studies, BaP DE adducts of guanosine (5, 6) and adenosine (6, 18) were prepared from the optically active (+)- and (-)-enantiomers of their parent DEs and were characterized by their CD spectra. Measurement of the CD spectra of the corresponding deoxyribonucleoside adducts (6, 7, 18) showed them to be virtually identical to those obtained for the ribonucleoside adducts formed from the optically active DEs of known absolute configuration. However, correlation of the CD spectra of these adducts with their absolute configurations depended not only on the known chirality of the parent DEs but also on NMR assignments of cis versus trans ring opening, because cis- and trans-opened adducts from a single optically active DE exhibited major CD bands that were opposite in sign. Our direct crystallographic determination of the absolute configuration of the trans-(10R)-BaP DE dG adduct is thus of considerable importance, since the unequivocal assignment of trans as opposed to cis opening and the known absolute configuration of the sugar unambiguously prove the absolute configuration of the crystalline adduct. In addition, this crystal structure confirms our previous assignments of absolute configuration (13, 14) to the DEs and their precursor 7,8-dihydrodiols.
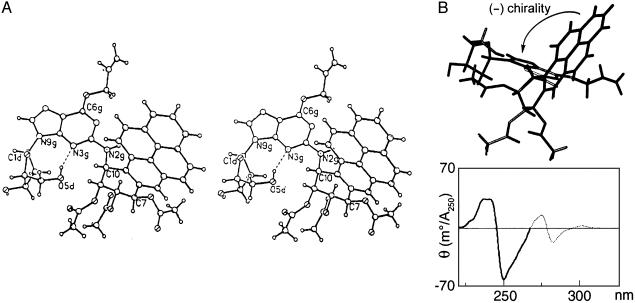
We had previously recognized (6) that the bands near 250 nm in the CD spectra of DE-nucleoside adducts arose from an exciton interaction between the aromatic hydrocarbon and the purine base. However, the apparent flexibility of these adducts around the two bonds connecting the purine and the hydrocarbon (C2g-N2g and N2g-C10) precluded reliable predictions of the relative orientations of the interacting chromophores, and analysis (19) of the CD spectra of these adducts in terms of exciton chirality was not attempted. The availability of a crystallographic picture of the relative spatial orientation of the pyrene and guanine chromophores (Fig. 3A) now permits interpretation of the split Cotton effects (centered at ≈245 nm) caused by this exciton interaction (5, 6). A schematic representation of the relative orientations of the pyrene and guanine rings (Fig. 3B) based on the structure shown in Fig. 3A indicates that the screw sense of the two chromophores in the crystal is left-handed (negative). The CD spectrum of the present O6-allyl derivative (15) does not show an interpretable exciton interaction with strong and comparable positive and negative Cotton effects, presumably because O-allyl substitution alters the guanine chromophore. However, the CD spectrum (15) (Fig. 3B) of the same diastereomer lacking the O-allyl substituent shows a negative first and a positive second Cotton effect, in agreement with their predicted directions (19) for the negative screw sense observed in the crystal. Virtually identical CD spectra are observed in the absence of any hydroxyl-blocking groups on either the sugar or hydrocarbon moiety (5, 7). Thus, the relative orientations of the purine and hydrocarbon chromophores are likely very similar in the crystal and solution and are independent of the blocking groups.
Fig. 3.
(A) Stereo diagram of a single molecule of the trans-(10R)-BaP DE dG adduct showing its absolute configuration. (B) A view of the crystalline adduct, looking down a line between C10a of the pyrene and C2g of the guanine chromophore, that depicts the negative screw sense of their transition moments near 250 nm. For both guanine (20, 21) and pyrene (22) these transition moments are parallel to their long axes. The relevant portion of the CD spectrum in methanol of a derivative lacking the O6-allyl group and with tert-butyldimethylsilyl protecting groups on the deoxyribose (15) shows a negative first Cotton effect and a positive second Cotton effect (heavy line), as predicted from the negative chirality observed in the crystal.
Molecular Conformation. The conformation of the present (10R)-BaP DE dG adduct can be characterized by three torsion angles between its three relatively rigid components, namely the polycyclic hydrocarbon, the base, and the sugar. These three flexible torsion angles are α′ (N1g-C2g-N2g-C10), β′ (C2g-N2g-C10-C9), and χ (O1d-C1d-N9g-C4g) (Fig. 2). The relative orientations of the pyrene and guanine rings as described in the preceding section are determined by the two torsion angles α′ and β′, which are 172.5° and 80.8°, respectively, in the crystal structure. Computations by Xie et al. (23) identified low-energy conformations of BaP DE dG adducts as the unblocked nucleosides by uniformly sampling the possible rotamers about the α′, β′, and χ angles at 5° intervals and determination of each of their energies with amber 4.0. In contrast to the diffraction data for the crystal, these computations assume the molecules to be in a “gas” state without near neighbors. Four low-energy domains were identified for the trans-(10R)-BaP DE dG adduct [referred to as (-)-trans anti]. Domain III, found by the calculations to be most favored (23), has α′ centered at 180° and β′ at 90°, both corresponding well to the crystal structure values of α′ = 172° and β′ = 81°. In trans-BaP DE dG adducted duplex DNA, the β′ torsion angle largely determines the direction in which the hydrocarbon moiety lies in the minor groove relative to the ends of the DNA strand (23). Thus, as shown by solution 2D NMR (24), the present trans-(10R) BaP dG adduct in duplex DNA exhibits angles of α′ = 152° and β′ = 78°, similar to those in our crystal structure, and orients toward the 3′ end of the adducted strand. In contrast, the trans-(10S)-BaP dG adduct derived from (+)-(R,S,S,R)-DE-2 has torsion angles of α′ = 137° and β′ = -102° and orients toward the 5′ end of the adducted strand (25).
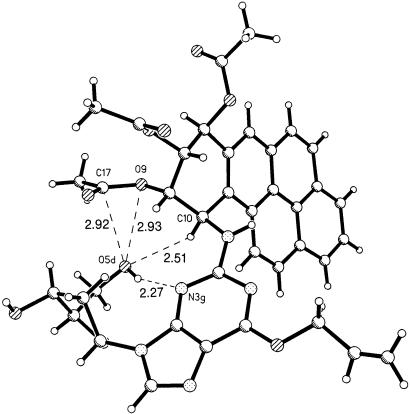
The experimentally determined value of the torsion angle χ (O1d-C1d-N9g-C4g) in the crystal is 73.3° and corresponds to an unusual syn conformation around the glycosidic bond. This finding contrasts with the calculated lowest-energy nucleoside structure (23) for which χ = 210° (anti glycosidic conformation) and with the normal anti glycosidic torsion in B-DNA (26) (see below). In the crystal, an intramolecular hydrogen bond (3.09 Å) between O5dH and N3g (Table 1) restricts χ to the syn domain, a conformation that provides for a compact arrangement of atoms with the sugar and the hydrocarbon moieties of the adduct approaching each other (see Fig. 4). The acetoxy group on C9 of the BaP overlays the -CH2OH group of the sugar. The shortest distances are between atoms O9···O5d (2.93 Å), C17···O5d (2.92 Å), H10a···O5d (2.51 Å), and the hydrogen bond H5Od···N3g (2.27 Å). If the acetyl group on O9 were replaced by an H atom, there would be a good possibility for forming an O9H···O5d hydrogen bond.
Table 1. H bonds at R = 4.5%.
| Type | Donor | Acceptor | D-A, Å | D-H, Å |
|---|---|---|---|---|
| Intermolecular | N2g | O3d* | 2.865‡ | 1.97 |
| Intermolecular | O3d | O1e | 2.681§ | 1.78 |
| O1e | N7g† | 2.815§ | 1.91 | |
| Intramolecular | O5d | N3g | 3.094¶ | 2.27 |
Symmetry equivalent -1+x, y, z
Symmetry equivalent 1/2+x, 3/2-y, 1-z
Direct NH···O hydrogen bond
Hydrogen bonds mediated by ethanol
Intramolecular
Fig. 4.
Near approaches of atoms within a single molecule to the back side of the sugar O5d atom. The hydrogen atom on O5d is a donor in the hydrogen bond to N3g.
The syn conformation is not uncommon in crystal structures containing the guanosine moiety. Thus, intramolecular hydrogen bonding in the crystal and possibly also crystal packing interactions likely play a role in stabilizing this conformation. Similar syn conformations, with somewhat shorter O5d···N3g hydrogen bonds than in our present structure (3.09 Å), have been observed in crystals of 7-methyl-8-oxo-7,8-dihydroguanosine monohydrate (O5d···N3g = 2.94 Å, χ = 65°) (27) and guanylyl-2′,5′-cytidine dihydrate (O5d···N3g = 2.79 Å, χ = 54°) (28). An N6 adduct of dA to the 7-methyl group of 7,12-dimethylbenzo[a]anthracene (29) also exhibits a syn conformation and an internal O5d···N3g hydrogen bond in the crystal state.
In both the trans-(10R)- and trans-(10S)-BaP DE dG adducted DNA duplexes (24, 25), as in normal B-DNA (26), the glycosidic torsion angle is anti (≈260°), in contrast with the syn conformation (73.3°) in the present crystal. However, the syn conformation may have a role as an intermediate in the replication of dG adducts in DNA. A BaP-DE dG adduct with the syn conformation was observed by NMR in a partial duplex, corresponding to a template-primer for DNA replication, containing the adduct at the fourth position from the 5′ end of a 13-mer oligonucleotide bound to a complementary 9-mer that terminated one base before the adduct (30).
The deoxyribose in our crystal is in the C2′-endo conformation, as in typical B-DNA (26), with a pseudorotation angle P (31) of 161°. The tetrahydro benzo-ring of the BaP DE moiety assumes a distorted half chair conformation with the guanine base bonded to C10. Its neighboring trans-acetoxy group at C9 is pseudoaxial and the C8 and C7 acetoxy groups are pseudoequatorial (Fig. 3A). Comparison of the measured dihedral angles between the vicinal methine hydrogens in the crystal and their values calculated from their NMR coupling constants in CDCl3 solution is given in Table 2. The agreement between these values indicates that the conformation of this ring in the crystal is comparable to that in solution. The N2 amino group of the guanine has a larger angle (72°) with the plane of the pyrene ring system than does H10 (44°), presumably caused by greater steric hindrance between H11 and the substituted amino group.
Table 2. Comparison of dihedral angles (in degrees) between the methine hydrogen atoms on the tetrahydro benzo-ring of N2-[10R-(7S,8R,9S-triacetoxy-7,8,9,10-tetrahydrobenzo[a]pyrenyl)]-O6-allyl-2′-deoxyguanosine in the crystal structure with calculated values from its solution NMR spectrum.
| Dihedral angle | Crystal | NMR* |
|---|---|---|
| H7-C7-C8-H8 | 153.0 | 142 |
| H8-C8-C9-H9 | 56.9 | 77 |
| H9-C9-C10-H10 | 72.7 | 64 |
At 300 MHz (CDCl3); this work.
Calculated by use of the Karplus equation, J = A + B cos ϕ + C cos 2ϕ, with A = 7, B = -1, and C = 5 Hz (32) from the observed coupling constants: J7,8 = 9.0; J8,9 = 2.3; and J9,10 = 3.5 Hz
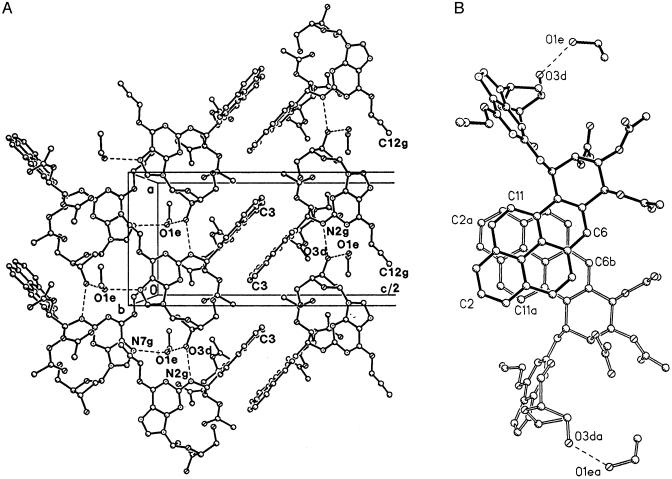
Crystal Packing and Stacking. The adduct molecules are arranged in antiparallel columns with their pyrene moieties partially stacked and interdigitated (Fig. 5A) on one side. On the other side (Fig. 5A), the base and sugar moieties of adjacent columns are contiguous, with N7g and O3d of neighboring molecules in adjacent columns linked by bridging hydrogen bonds to the solvent ethanol retained in the crystal.
Fig. 5.
(A) Crystal packing showing edge-on view of pyrene stacking. Molecules repeat along the a direction (vertical) with hydrogen bond links, N2gH···O3d. Pairs of hydrogen bonds to ethanol solvent molecules, O3dH···O1e and O1eH···N7g, hold together adjacent columns of the adduct as seen on the left. (B) Stacking of the pyrene moieties in pairs of molecules with a separation of ≈3.55 Å.
Both the aromatic pyrene moiety and the guanine base have planar pi-systems that are conducive to stacking in crystals and may also be involved in base-hydrocarbon interactions in adducted DNA molecules. There is no stacking of the guanine bases in this crystal either with each other or with the pyrene ring systems. In contrast, the pyrene moieties of neighboring pairs of antiparallel molecules almost completely overlap each other (Fig. 5B), with a separation of ≈3.55 Å between the planes. However, each pair of molecules exhibits little stacking with the next pair of molecules above it along the a axis in Fig. 5A, and intercalation of the pyrenes in the left-hand array of Fig. 5A between those in the oppositely oriented, right-hand array is incomplete.
Hydrogen bonds play several strategic roles in the structure. The intramolecular hydrogen bond O5dH···N3g, already mentioned, stabilizes the molecule in the syn conformation. The one direct intermolecular hydrogen bond, N2gH···O3d, links the adduct molecules into columns by translation along the a axis (Fig. 5). An ethanol solvent molecule mediates hydrogen bonding between columns by a pair of hydrogen bonds, O3dH···O1e and O1eH···N7g in Fig. 5. The effect is that two columns of adduct molecules, related by a 2-fold screw axis, are linked along the c axis direction with the aid of the ethanol OH group.
Summary and Conclusions
The present study describes the x-ray crystal structure of a nucleoside adduct from a polycyclic aromatic hydrocarbon DE. As such it permits direct assignment of absolute configuration to such an adduct and provides unambiguous proof of our previous configurational assignments to the four optically active BaP DEs and their eight dG adducts that arise by cis and trans opening of the DEs at C10. Similarity between the values of the J9,10 coupling constants of the dG adducts and their corresponding dA adducts supports the eight dA adduct structures as well.
Resemblances between the crystal structure and conformational features deduced from physical measurements on the dG adduct in solution and DNA suggest that, despite their apparent flexibility around the C2g-N2g bond of guanine and the N2g-C10 bond between guanine and the hydrocarbon, these adducts have strong conformational preferences that may well influence their interactions with enzymes such as DNA polymerases. (i) The relative orientation of the pyrene and guanine chromophores is consistent with solution CD spectra for both the 7,8,9-triacetate (see Fig. 3B) and the unacetylated nucleoside adducts (5-7). In addition, a similar orientation of these two aromatic moieties is observed in the present structure and an NMR structure (24) of this adduct in duplex DNA. (ii) The conformation of the tetrahydro benzoring in the crystal structure closely resembles that of the molecule in solution as deduced from the NMR coupling constants for the ring protons (Table 2).
The present adduct exhibits a syn glycosidic torsion angle in the crystal, even though the same trans-(10R)-BaP DE dG adduct prefers the normal anti conformation in duplex DNA (24). We speculate that syn-anti interconversion may be relatively facile and/or more strongly influenced by the adduct's environment than other conformational features of this molecule as described above. Consequently, factors such as hydrogen bond formation, crystal packing, stacking interactions within DNA (such as with the base pair 3′ to the adduct in a template-primer duplex, cf. ref. 30) or interactions with an enzyme's active site may influence the syn-anti conformational preference. Indeed, a possible role for the syn conformation, which can more effectively form hydrogen bonds with an incorrect incoming purine base than with a correct cytosine, has been proposed in erroneous replication of BaP DE dG adducts in DNA (33, 34).
Supplementary Material
Abbreviations: BaP, benzo[a]pyrene; DE, diol epoxide; dG, deoxyguanosine; dA, deoxyadenosine.
Data deposition: The atomic coordinates have been deposited in the Cambridge Structural Database, Cambridge Crystallographic Data Centre, Cambridge CB2 1EZ, United Kingdom (CSD reference no. 213703).
References
- 1.Cook, J. W., Hewett, C. L. & Hieger, I. (1933) J. Chem. Soc. 396-398.
- 2.Committee on the Biologic Effects of Atmospheric Pollutants (1972) Particulate Polycyclic Organic Matter (Natl. Acad. Sci., Washington, DC).
- 3.Thakker, D. R., Yagi, H., Akagi, H., Koreeda, M., Lu, A. Y. H., Levin, W., Wood, A. W., Conney, A. H. & Jerina, D. M. (1977) Chem. Biol. Interact. 16, 281-300. [DOI] [PubMed] [Google Scholar]
- 4.Buening, M. K., Wislocki, P. G., Levin, W., Yagi, H., Thakker, D. R., Akagi, H., Koreeda, M., Jerina, D. M. & Conney, A. H. (1978) Proc. Natl. Acad. Sci. USA 75, 5358-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore, P. D., Koreeda, M., Wislocki, P. G., Levin, W., Conney, A. H., Yagi, H. & Jerina, D. M. (1977) in Drug Metabolism Concepts, ACS Symposium Series no. 44, ed. Jerina, D. M. (Am. Chem. Soc., Washington, DC), pp. 127-154.
- 6.Sayer, J. M., Chadha, A., Agarwal, S. K., Yeh, H. J. C., Yagi, H. & Jerina, D. M. (1991) J. Org. Chem. 56, 20-29. [Google Scholar]
- 7.Cheng. S. C., Hilton, B. D., Roman, J. M. & Dipple, A. (1989) Chem. Res. Toxicol. 2, 334-340. [DOI] [PubMed] [Google Scholar]
- 8.Koreeda, M., Moore, P. D., Wislocki, P. G., Levin, W., Conney, A. H., Yagi, H. & Jerina, D. M. (1978) Science 199, 778-781. [DOI] [PubMed] [Google Scholar]
- 9.Neidle, S. & Cutbush, S. D. (1983) Carcinogenesis 4, 415-418. [DOI] [PubMed] [Google Scholar]
- 10.Neidle, S., Subbiah, A., Cooper, C. S. & Riberio, O. (1980) Carcinogenesis 1, 249-254. [DOI] [PubMed] [Google Scholar]
- 11.Yagi, H., Hernandez, O. & Jerina, D. M. (1975) J. Am. Chem. Soc. 97, 6881-6883. [DOI] [PubMed] [Google Scholar]
- 12.Yagi, H., Thakker, D. R., Hernandez, O., Koreeda, M. & Jerina, D. M. (1977) J. Am. Chem. Soc. 99, 1604-1611. [DOI] [PubMed] [Google Scholar]
- 13.Yagi, H., Akagi, H., Thakker, D. R., Mah, H. D., Koreeda, M. & Jerina, D. M. (1977) J. Am. Chem. Soc. 99, 2358-2359. [DOI] [PubMed] [Google Scholar]
- 14.Boyd, D. R., Gadaginamath, G. S., Kher, A., Malone, J. F., Yagi, H. & Jerina, D. M. (1980) J. Chem. Soc. Perkin Trans. 1, 2112-2116. [Google Scholar]
- 15.Kroth, H., Yagi, H., Seidel, A. & Jerina, D. M. (2000) J. Org. Chem. 65, 5558-5564. [DOI] [PubMed] [Google Scholar]
- 16.Kroth, H., Yagi, H., Sayer, J. M., Kumar, S. & Jerina, D. M. (2001) Chem. Res. Toxicol. 14, 708-719. [DOI] [PubMed] [Google Scholar]
- 17.Yagi, H., Ramesha, A. R., Kalena, G., Sayer, J. M., Kumar, S. & Jerina, D. M. (2002) J. Org. Chem. 67, 6678-6689. [DOI] [PubMed] [Google Scholar]
- 18.Jeffrey, A. M., Grzeskowiak, K., Weinstein, I. B., Nakanishi, K., Roller, P. & Harvey, R. G. (1979) Science 206, 1309-1311. [DOI] [PubMed] [Google Scholar]
- 19.Harada, N. & Nakanishi, K. (1983) Circular Dichroic Spectroscopy: Exciton Coupling in Organic Stereochemistry (University Science Books, New York).
- 20.Matsuoka, Y. & Nordén, B. (1982) J. Phys. Chem. 86, 1378-1386. [Google Scholar]
- 21.Clark, L. B. (1994) J. Am. Chem. Soc. 116, 5265-5270. [Google Scholar]
- 22.Thulstrup, E. W., Michl, J. & Eggers, J. H. (1970) J. Phys. Chem. 74, 3868-3878. [Google Scholar]
- 23.Xie, X.-M., Geacintov, N. E. & Broyde, S. (1999) Biochemistry 38, 2956-2968. [DOI] [PubMed] [Google Scholar]
- 24.de los Santos, C., Cosman, M., Hingerty, B. E., Ibanez, V., Margulis, L. A., Geacintov, N. E., Broyde, S. & Patel, D. J. (1992) Biochemistry 31, 5245-5252. [DOI] [PubMed] [Google Scholar]
- 25.Cosman, M., de los Santos, C., Fiala, R., Hingerty, B. E., Singh, S. B., Ibanez, V., Margulis, L. A., Live, D., Geacintov, N. E., Broyde, S. & Patel, D. J. (1992) Proc. Natl. Acad. Sci. USA 89, 1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnott, S. (1998) in Oxford Handbook of Nucleic Acid Structure, ed. Neidle, S. (Oxford Univ. Press, Oxford), pp. 1-36.
- 27.Larson, S. B., Cottam, H. B. & Robins, R. K. (1989) Acta Crystallogr. C 45, 1979-1983. [DOI] [PubMed] [Google Scholar]
- 28.Krishnan, R. & Seshadri, T. P. (1994) Biopolymers 34, 1637-1646. [Google Scholar]
- 29.Carrell, H. L., Glusker, J. P., Moschel, R. C., Hudgins, W. R. & Dipple, A. (1981) Cancer Res. 41, 2230-2234. [PubMed] [Google Scholar]
- 30.Cosman, M., Hingerty, B. E., Geacintov, N. E., Broyde, S. & Patel, D. J. (1995) Biochemistry 34, 15334-15350. [DOI] [PubMed] [Google Scholar]
- 31.Altona, C. & Sundaralingam, M. (1972) J. Am. Chem. Soc. 94, 8205-8212. [DOI] [PubMed] [Google Scholar]
- 32.Bothner-By, A. A. (1965) Adv. Magn. Res. 1, 195-316. [Google Scholar]
- 33.Chiapperino, D., Kroth, H., Kramarczuk, I. H., Sayer, J. M., Masutani, C., Hanaoka, F., Jerina, D. M. & Cheh, A. M. (2002) J. Biol. Chem. 277, 11765-11771. [DOI] [PubMed] [Google Scholar]
- 34.Perlow, R. A. & Broyde, S. (2003) J. Mol. Biol. 327, 797-818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.