Pathogenic bacteria are responsible for severe diseases worldwide. RNA stability is a major player controlling the expression of virulence factors. Ribonucleases (RNases) are the enzymes responsible for the maturation and degradation of RNA molecules (Arraiano et al., 2010; Silva et al., 2011). Exoribonucleases have been implicated in virulence in an increasing number of pathogens such as Salmonella enterica, Helicobacter pylori, Shigella flexneri, and Aeromonas hydrophila (see Andrade et al., 2009; Matos et al., 2011 and references below). However, the mechanisms underlying virulence are still mostly elusive (Arraiano et al., 2010; Lawal et al., 2011).
The recently published paper by Haddad et al. (2012) adds to this list Campylobacter jejuni, one of the most important human foodborne pathogens. Campylobacter is recognized as the leading bacterial cause of gastroenteritis and even more severe clinical manifestations can arise. The present work shows that C. jejuni bacteria lacking an 3′–5′ exoribonuclease called polynucleotide phosphorylase (PNPase) is significantly less virulent than the wild-type strain (Haddad et al., 2012).
Different steps have been identified in the ability of different pathogenic bacteria to promote infection, namely motility, adherence, invasion, intracellular replication, or spreading to the neighboring cells. Inactivation of the C. jejuni PNPase is shown to affect many of these steps, with pnp mutants showing distinct phenotypes such as limitations in swimming, substantial delay in the colonization of the chicken gut and a decreased ability to adhere and invade cells. Defects in motility are suggested to be responsible for many of the attenuation of the virulent traits of C. jejuni in the mutant pnp strain. Interestingly, the authors suggest that PNPase may be able to affect flagella-dependent motility by modulation of the NANA synthetase (neuB), involved in the post-translational modification of the flagellin subunit. Furthermore, proteomic studies also showed that PNPase affects the synthesis of proteins involved in virulence, such as LuxS and PEB3. This work confirms the importance of exoribonucleases, namely PNPase, in cell biology, and virulence (Haddad et al., 2012).
Bacterial pathogens rapidly adapt to environmental challenges. Adaptation requires a rapid adjustment in RNA levels, requiring not only transcriptional regulation, but also fine-tuning control of RNA stability. Stress-resistance plays an essential role in the capacity of many pathogenic bacteria to establish and maintain long-term intracellular residence in host cells. Many ribonucleases are regulated by stress conditions, being critical enzymes involved in the adaptation of bacteria to new environmental conditions. In particular, PNPase is a cold-shock protein in Escherichia coli being essential for growth at low temperatures (Zangrossi et al., 2000). Haddad et al. (2009) had previously shown that PNPase was also crucial for C. jejuni growth under cold-shock conditions. This was a relevant discovery especially when considering that this pathogen can persist and grow at refrigerated temperatures. PNPase also seems to be involved in C. jejuni resistance to acidic and oxidative stresses, as the pnp strain shows variations in the levels of the stress-response proteins KatA, DnaK, and Hsp90 (Haddad et al., 2012). In S. enterica, PNPase was shown to be important for acute infection and lethality in a murine model as result of increasing expression of the pathogenicity islands (Clements et al., 2002). In Yersinia pseudotuberculosis and Y. pestis PNPase was shown to be essential for the function of the Yersinia type tree secretion system (TTSS), an organelle that injects effector proteins directly into host cells (Rosenzweig et al., 2007). Interestingly, PNPase has been involved in the post-transcriptional regulation of small non-coding RNAs (Andrade and Arraiano, 2008; De Lay and Gottesman, 2011; Andrade et al., 2012). In C. jejuni, not much is known about this class of regulatory RNAs but transcriptomic studies have identified five candidate regions for harboring sRNAs (Chaudhuri et al., 2011). It is an exciting hypothesis that PNPase is able to regulate small RNAs possibly involved in the virulence traits of C. jejuni although this lacks experimental evidence at the time.
Together with PNPase, RNase II, and RNase R are the major exoribonucleases involved in RNA degradation in E. coli (Figure 1). Orthologs have been described in all domains of life (Arraiano et al., 2010). RNase R, a hydrolytic exoribonuclease, is also known to be involved in the virulence of several microorganisms. Like PNPase, RNase R is a cold-shock protein essential for the survival at low temperatures of several microorganisms, such as E. coli, Pseudomonas putida, P. syringae, and A. hydrophila. In some microorganisms RNase R was shown to be necessary for the expression of several invasion factors and mutations on its gene resulted in the reduced expression of virulence phenotypes in S. flexneri and in enteroinvasive E. coli (Tobe et al., 1992). Legionella pneumophila is an intracellular parasite of free-living protozoa which inhabits man-made water distribution systems, and is the most frequent cause of human legionellosis, community-acquired, and nosocomial pneumonia in adults. In this microorganism, RNase R is the only hydrolytic exoribonuclease present. Its activity was shown to be essential for growth and viability at low temperatures and induces competence (Charpentier et al., 2008). Similarly to what was shown in E. coli (Cairrão et al., 2003), RNase R is also a cold-shock protein in A. hydrophila. In this highly toxic microorganism, which is resistant to multiple medications, chlorine, and cold temperatures, RNase R was shown to be essential for viability at lower temperatures and its absence leads to a reduction in motility. The infection of mouse cells with A. hydrophila rnr mutant strains showed that their virulence was attenuated in comparison to the wild-type, which confirms the role of RNase R in pathogenesis (Erova et al., 2008).
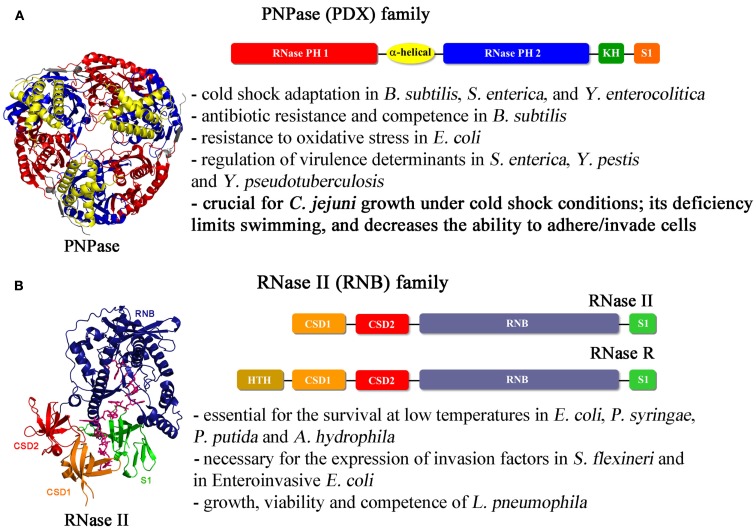
Figure 1.
Schematic representation of the domains found in the exoribonucleases from PNPase and RNase II families and structures of representative members. (A) Top: PNPase (PDX family) primary structure: two RNase PH catalytic domains followed by two RNA binding domains (KH and S1). PNPase is a trimer and assembles in a donut like shape (on the left; Shi et al., 2008). The S1 binding domain was shown to restore PNPase deletion effect in Yersinia (Rosenzweig et al., 2007). (B) Top: linear representation of RNase II and RNase R domains: the central catalytic RNB, and the CSD1, CSD1, and S1 RNA binding domains. Members of the family can have additional domains at the N-terminal region, namely Helix-Turn-Helix in RNase R. On the left is represented the E. coli RNase II–RNA complex crystal structure, showing the distinct domains of the enzyme, and the 13-mer bound RNA (Frazão et al., 2006). The RNB domain (in blue), is the responsible for the catalytic activity of the protein which, for example, is important for the development of competence in L. pneumophila (Charpentier et al., 2008).
Considering the important functions that these proteins have in the establishment of virulence, ribonucleases (namely RNase II, RNase R, and PNPase) offer a new perspective for developing efficient compounds in clinical treatments: they can be potential targets to design compounds able to kill specific microorganisms or to reduce their virulence ability. The further study of the function of exoribonucleases in the control of pathogenesis will certainly help in the comprehension of RNA-related processes involved in infection.
Acknowledgments
To FCT for funding, namely through grant PEst-OE/EQB/LA0004/2011.
References
- Andrade J. M., Arraiano C. M. (2008). PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA 14, 543–551 10.1261/rna.683308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade J. M., Pobre V., Matos A. M., Arraiano C. M. (2012). The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA 18, 844–855 10.1261/rna.029413.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade J. M., Pobre V., Silva I. J., Domingues S., Arraiano C. M. (2009). The role of 3’-5’ exoribonucleases in RNA degradation. Prog. Mol. Biol. Transl. Sci. 85, 187–229 10.1016/S0079-6603(08)00805-2 [DOI] [PubMed] [Google Scholar]
- Arraiano C. M., Andrade J. M., Domingues S., Guinote I. B., Malecki M., Matos R. G., Moreira R. N., Pobre V., Reis F. P., Saramago M., Silva I. J., Viegas S. C. (2010). The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 34, 883–923 10.1111/j.1574-6976.2010.00242.x [DOI] [PubMed] [Google Scholar]
- Cairrão F., Cruz A., Mori H., Arraiano C. M. (2003). Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 50, 1349–1360 10.1046/j.1365-2958.2003.03766.x [DOI] [PubMed] [Google Scholar]
- Charpentier X., Faucher S. P., Kalachikov S., Shuman H. A. (2008). Loss of RNase R induces competence development in Legionella pneumophila. J. Bacteriol. 190, 8126–8136 10.1128/JB.01035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R. R., Yu L., Kanji A., Perkins T. T., Gardner P. P., Choudhary J., Maskell D. J., Grant A. J. (2011). Quantitative RNA-seq analysis of the Campylobacter jejuni transcriptome. Microbiology 157, 2922–2932 10.1099/mic.0.050278-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements M. O., Eriksson S., Thompson A., Lucchini S., Hinton J. C., Normark S., Rhen M. (2002). Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. U.S.A. 99, 8784–8789 10.1073/pnas.132047099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N., Gottesman S. (2011). Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA 17, 1172–1189 10.1261/rna.2531211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erova T. E., Kosykh V. G., Fadl A. A., Sha J., Horneman A. J., Chopra A. K. (2008). Cold shock exoribonuclease R (VacB) is involved in Aeromonas hydrophila pathogenesis. J. Bacteriol. 190, 3467–3474 10.1128/JB.00075-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazão C., Mcvey C. E., Amblar M., Barbas A., Vonrhein C., Arraiano C. M., Carrondo M. A. (2006). Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature 443, 110–114 10.1038/nature05080 [DOI] [PubMed] [Google Scholar]
- Haddad N., Burns C. M., Bolla J. M., Prevost H., Federighi M., Drider D., Cappelier J. M. (2009). Long-term survival of Campylobacter jejuni at low temperatures is dependent on polynucleotide phosphorylase activity. Appl. Environ. Microbiol. 75, 7310–7318 10.1128/AEM.01366-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad N., Tresse O., Rivoal K., Chevret D., Nonglaton Q., Burns C. M., Prévost H., Cappelier J. M. (2012). Polynucleotide phosphorylase has an impact on cell biology of Campylobacter jejuni. Front. Cell. Infect. Microbiol. 2:30. 10.3389/fcimb.2012.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawal A., Jejelowo O., Chopra A. K., Rosenzweig J. A. (2011). Ribonucleases and bacterial virulence. Microb. Biotechnol. 4, 558–571 10.1111/j.1751-7915.2010.00212.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos R. G., Pobre V., Reis F. P., Malecki M., Andrade J. M., Arraiano C. M. (2011). “Structure and degradation mechanisms of 3’ to 5’ exoribonucleases,” in Ribonucleases (Nucleic Acids and Molecular Biology), (Heidelberg: Springer-Verlag; ), 26,193–222 [Google Scholar]
- Rosenzweig J. A., Chromy B., Echeverry A., Yang J., Adkins B., Plano G. V., Mccutchen-Maloney S., Schesser K. (2007). Polynucleotide phosphorylase independently controls virulence factor expression levels and export in Yersinia spp. FEMS Microbiol. Lett. 270, 255–264 10.1111/j.1574-6968.2007.00689.x [DOI] [PubMed] [Google Scholar]
- Shi Z., Yang W. Z., Lin-Chao S., Chak K. F., Yuan H. S. (2008). Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation. RNA 14, 2361–2371 10.1261/rna.1244308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva I. J., Saramago M., Dressaire C., Domingues S., Viegas S. C., Arraiano C. M. (2011). Importance and key events of prokaryotic RNA decay: the ultimate fate of an RNA molecule. Wiley Interdiscip. Rev. RNA. 2, 818–836 10.1002/wrna.94 [DOI] [PubMed] [Google Scholar]
- Tobe T., Sasakawa C., Okada N., Honma Y., Yoshikawa M. (1992). vacB, a novel chromosomal gene required for expression of virulence genes on the large plasmid of Shigella flexneri. J. Bacteriol. 174, 6359–6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangrossi S., Briani F., Ghisotti D., Regonesi M. E., Tortora P., Dehò G. (2000). Transcriptional and post-transcriptional control of polynucleotide phosphorylase during cold acclimation in Escherichia coli. Mol. Microbiol. 36, 1470–1480 10.1046/j.1365-2958.2000.01971.x [DOI] [PubMed] [Google Scholar]