Abstract
The majority of Helicobacter pylori reside on gastric epithelial cell surfaces and in the overlying mucus, but a small fraction of H. pylori enter host epithelial and immune cells. To explore the role of the nudA invasin in host cell entry, a ΔnudA deletion derivative of strain J99 was constructed and transformants were verified by PCR and by fluorescence in situ hybridization. AGS cells were inoculated with either wild type (WT) strain J99 or its ΔnudA mutant to determine the fraction of bacteria that were bound to the cells and were present inside these cells using the gentamicin protection assay. We observed no significant difference between either the density of H. pylori bound to AGS cell membranes or the density of intracellular H. pylori. To further explore this finding, separate chambers of each culture were fixed in glutaraldehyde for transmission electron microscopy (TEM) and immunogold TEM. This addition to the “classical” gentamicin assay demonstrated that there were significantly more intracellular, and fewer membrane-bound, H. pylori in WT-infected AGS cells than in ΔnudA allele infected cells. Thus, the sum of intracellular and membrane-bound H. pylori was similar in the two groups. Since no other similar TEM study has been performed, it is at present unknown whether our observations can be reproduced by others Taken together however, our observations suggest that the “classical” gentamicin protection assay is not sufficiently sensitive to analyze H. pylori cell entry and that the addition of TEM to the test demonstrates that nudA plays a role in H. pylori entry into AGS cells in vitro. In addition, deletion of the invasin gene appears to limit H. pylori to the AGS cell surface, where it may be partly protected against gentamicin. In contrast, this specific environment may render H. pylori more vulnerable to host defense and therapeutic intervention, and less prone to trigger normal immune, carcinogenic, and other developmental response pathways.
Keywords: H. pylori, intracellular, adhesion, adhesin, nudA, electron microscopy
Introduction
The pathogenicity of many bacteria colonizing the gastrointestinal tract often depends on their ability to gain access to cells that are normally non-phagocytic. Helicobacter pylori colonizes the stomach of over half the world population and is the main cause of peptic ulcer disease and gastric cancer. It often is considered to be a non-invasive pathogen present only in the lumen of the stomach and attached to gastric epithelial cells although a number of in vivo and in vitro studies have demonstrated that H. pylori is in fact invasive (Dubois and Berg, 1997; Engstrand et al., 1997; Amieva et al., 2002; Semino-Mora et al., 2003; Necchi et al., 2007). In addition, H. pylori can repopulate the extracellular environment after elimination of extracellular bacteria with gentamicin, suggesting it may be considered a facultative intracellular bacterium (Amieva et al., 2002). Finally, H. pylori may be present inside metaplastic, dysplastic, and neoplastic epithelial cells (Semino-Mora et al., 2003). The multiplicity of these observations and the fact that various methods were used to reach the same conclusions strongly support their validity.
Bacterial invasion of eukaryotic cells appears to be mediated by Nudix enzymes, initially called MutT because E. coli MutT was the first Nudix hydrolase to be described (Maki and Sekiguchi, 1992). The Bartonella bacilliformis Nudix hydrolase, encoded by the ialA gene, was shown to be associated with the ability to invade human erythrocytes using the gentamicin assay complemented by transmission electron microscopy (TEM; Mitchell and Minnick, 1995). Similarly, invasion of human brain microvascular endothelial cells by E. coli is accompanied by increased expression of the K1 ortholog ygdP and the early stages of infection of infection by Rickettsia prowazekii ortholog is temporarily accompanied by an increased transcription of the invA gene (Lundin et al., 2003). Finally, the invA gene was highly conserved in protein sequence and present in all tested members of the pathogenic Leptospira species (Luo et al., 2011).
Helicobacter pylori appears to be a suitable system to study the biological role of Nudix hydrolases since the NudA protein is the only dinucleoside polyphosphate hydrolase homolog present in the two first strains that were sequenced (Tomb et al., 1997; Alm et al., 1999). J99 H. pylori harbors one Nudix hydrolase ortholog, Nudix hydrolase A, nudA, with the gene numbers JHP1149 (Alm et al., 1999), 26,695 strain harbors HP1228 (named invA; Tomb et al., 1997), and a GenBank search showed that a nudA gene is present in 30 additional completely sequenced H. pylori strains. Due to the functional heterogeneity within this group of proteins, H. pylori NudA may be involved in (1) DNA repair, (2) oxidative stress and/or heat shock response, or (3) bacterial invasion of epithelial cells through degradation of toxic substances induced during invasion (Lundin et al., 2003). These authors describe the enzymatic function of the Nudix hydrolase NudA in H. pylori and they constructed a nudA insertion mutant to determine the biological role of this protein. Using the classical gentamicin protection assay (Kwok et al., 2002), the authors found no quantifiable differences in invasion frequency by the NudA H. pylori J99 strain mutant compared to WT but they did not show the data (Lundin et al., 2003). Their conclusion was that they found no supportive evidence for a role for the NudA protein in H. pylori invasion of AGS cells, although they warned that this could be due to the fact that complete eradication of extracellular bacteria is rarely obtained in the gentamicin protection assay (Amieva et al., 2002). They also concluded that the gentamicin assay of AGS cells invasion by H. pylori lacked the sensitivity needed to demonstrate differences in their experimental setup.
In the present study, we examined the role of H. pylori invasin J99 NudA in H. pylori entry into gastric epithelial cells. To this effect, we generated a ΔnudA allele of strain J99 in which nudA was replaced by a chloramphenicol resistance gene (CAM). Absence of the nudA gene was verified and the effect of this deletion on colonization was studied in vitro using the classical gentamicin assay aided by ultrastructural studies as used by others (Mitchell and Minnick, 1995).
Materials and Methods
Construction of a Δnuda allele mutant
A ΔnudA allele in which nudA was replaced by the chloramphenicol resistance cassette (CamR) was constructed from a low-pass isolate of the J99 strain (kindly provided by Dr. R. Peek) and using a PCR method as reported earlier (Chalker et al., 2001; Tan and Berg, 2004). Culture of single colony isolates in the presence of chloramphenicol was then used to select for ΔnudA alleles carrying CAM. Genomic DNA was extracted from fresh H. pylori isolates using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA) and this DNA was used for confirmation of the mutant. Presence of CAM and absence of nudA in single colony isolates was then verified using PCR (Figure 1) and fluorescence in situ hybridization (Figure 2). The CamR and nudA sequences were confirmed using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems Inc.).
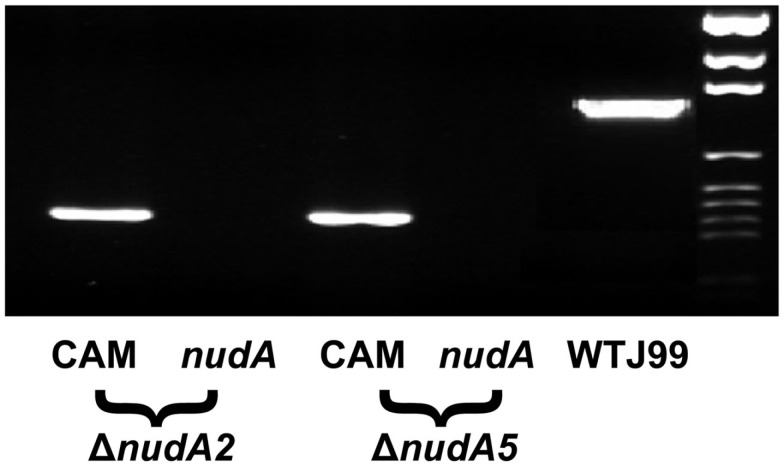
Figure 1.
Verification of post-transformation bacterial DNA in the ΔnudA allele PCR products on agarose gel. Two pairs of primers were designed for verification of post-transformation of bacterial transformed with ΔnudA allele PCR products. One pair was specific for nudA and the other pair was specific for the CamR gene. The wild type strain J99 PCR product was used as positive control. The figure illustrates that the nudA gene was present only in wild type strain J99 but absent in nudA mutant strains while the CamR gene was present in nudA mutant strains but not in the wild type.
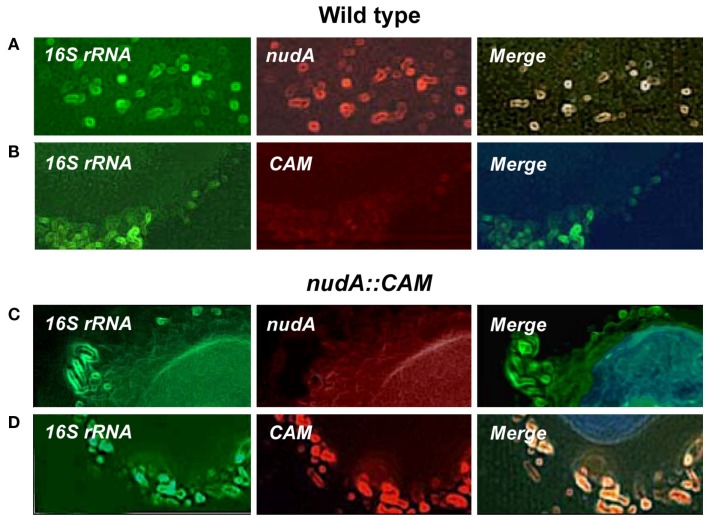
Figure 2.
Illustration of the expression of H. pylori 16S rRNA, nudA, and chloramphenicol resistance cassette (CAM) in AGS cells infected with H. pylori J99 WT (A,B) and ΔnudA allele (C,D). (A) 16S rRNA and nudA expression and complete merge of the two genes (green, red, and yellow, respectively). (B) 16S rRNA expression, no expression of CAM, and no merge. (C) 16S rRNA expression, no expression of nudA, and no merge. (D) 16S rRNA and CAM expression and complete merge of the two genes (green, red, and yellow, respectively). Original magnification: 1,000×.
Gentamicin protection viability assay experimental design
Human gastric adenocarcinoma cell line AGS was purchased from ATCC and maintained in 5% CO2 atmosphere in Dulbecco’s Modified Eagle Medium (DMEM, Gibco/BRL) supplemented with 10% fetal bovine serum (FBS, Gibco/BRL) for 2 days in 10-cm dishes for culture and then in 12-well plates (5 × 104/well) for gentamicin protection assay and four- or eight-chamber slides (Nunc Lab-Tek II Chamber Slide System)1 for light and TEM.
AGS cells were grown to monolayer in two groups of four plates or chambers and a similar number of H. pylori J99 WT and of the ΔnudA mutant was inoculated into each group of plates/chambers (4 × 103 H. pylori/cell). Two plates and chambers were cultured for 6 h, used as negative control, and infected with H. pylori plus 200 μg/ml gentamicin (Figure 3; controls, left side) for 12 h. One of the two plates or wells was treated with 0.1% saponin for 15 min and then cultured for H. pylori. Saponin was used to permeabilize AGS cell membranes by penetrating the cholesterol monolayer and forming holes or pits (7–9 nm) that is believed to allow H. pylori to exit from AGS cells and be cultured (Bangham et al., 1962; Kwok et al., 2002). The remaining two plates and chambers were used to study H. pylori binding and cell invasion (Figure 3; right side) and were infected with H. pylori for 6 h. One of the two plates or chambers was treated with gentamicin, cultured for 12 h, treated with saponin for 15 min, and then plated on BAP for colony counting or fixed with formaldehyde or glutaraldehyde (see below).
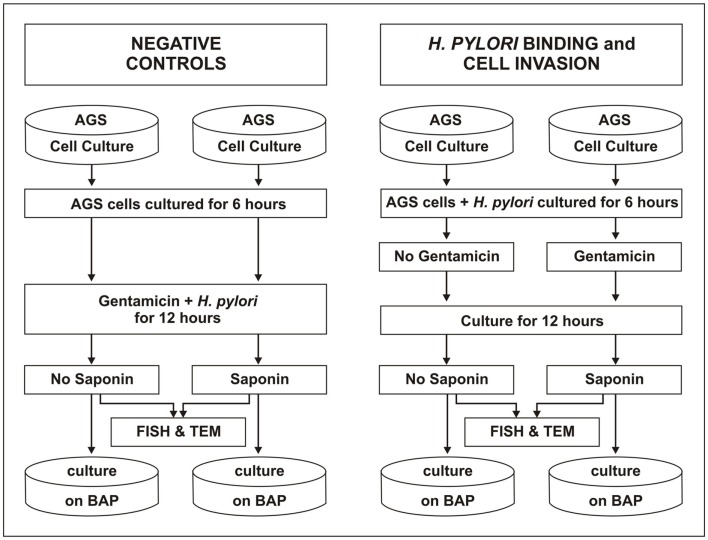
Figure 3.
Experimental design of the gentamicin protection assay. AGS cells were prepared to grow to monolayer in 8 wells in a 12-well plate (four wells for WT strain J99 and four wells for ΔnudA mutant. Each strain was inoculated into the four AGS cell types. Two wells (on the left) were designed as negative control of gentamicin treatment as it was added to H. pylori suspension when inoculated to the cells, and then treated with or without saponin after 12 h inoculation. A third well was used for measurement of H. pylori bound to cell membranes in the conditions without gentamicin and saponin. The last well was designed for measurement of H. pylori invasion into the cell with gentamicin and saponin added. The chamber slides prepared for morphologic observation were exactly the same as above.
Calculation and normalization of H. pylori entering into, or binding to, AGS cells
Counting of H. pylori can be done via OD measurement, but this method is not precise or very accurate. Instead, we counted the number of colonies used for plates and well inoculation by plating and culturing the H. pylori suspension for 3 days. Colony counting after treatment with and without gentamicin and saponin was then determined by culturing H. pylori on plates and then expressing colonization in each compartment as the percentage of H. pylori inoculated onto the plates.
Morphology and morphometry using fluorescence in situ hybridization and transmission electron microscopy
Tissue culture chamber glass slides from each of the four groups were treated using one of the following methods.
For H. pylori dual fluorescence in situ hybridization (FISH), glass chambers were immediately fixed in 4% formaldehyde for 24 h and then processed using probes designed specifically for H. pylori 16S rRNA (digoxigenin–FITC) and nudA (Biotin–Texas red) or 16S rRNA (digoxigenin–FITC) and CAM (biotin–Texas Red) were designed and synthesized as reported (Liu et al., 2008). Control of method was performed as described (Semino-Mora et al., 2003). Chambers were observed using a Nikon Eclipse 80i microscope and pictures were taken using a DS-Qi1MC Nikon camera (Figure 2).
For TEM, plastic chamber slides with four wells were immediately fixed in 2.5% glutaraldehyde for 24 h, the AGS cells were scraped and transferred to Eppendorf tubes, post-fixed in 1% osmium tetroxide at 4°C, and processed as reported until embedding in Spurr Low Viscosity Kit2 to obtain epoxy blocks at 70°C (Semino-Mora et al., 2003). Semi-thin (0.5 μm) sections were stained with toluidine blue and observed by light microscopy using an Eclipse E800 Nikon interfaced with a QCapture digital camera (Micropublisher 5.0, Burnaby, BC, Canada). Grids with thin sections (500 A) were prepared, stained with uranyl acetate and lead citrate, and then observed using a Philips CM100 transmission electron microscope at 80 kV (Biomedical Instrumentation Center, USUHS).
For TEM immunohistochemistry with pre-embedding, plastic chamber slides with eight chambers each were processed inside each chamber slide (in situ method; Tanner et al., 1996). Immunogold was performed using rabbit anti-H. pylori (NeoMarkers, Fremont, CA, USA) and immunogold-labeled secondary antibodies (18 nm colloidal gold, goat anti-rabbit IgG; Jackson ImmunoResearch, West Grove, PA, USA) and after intermediate steps were embedded in Spurr embedding. In other experiments, dual immonogold pre-embedding was performed using both rabbit anti-H. pylori and mouse anti-VacA antigen IgG anti-VacA (Secondary 18 nm colloidal gold, goat anti-rabbit IgG, and 11 nm colloidal gold, respectively, Jackson, InmunoResearch, West Grove, PA, USA). After polymerization, the flat resin blocks were removed by peeling them away from each chamber, mounted in blank mold, and cut in ultramicrotomy. Sections were stained only with uranyl acetate. Control of method was performed in AGS cells infected with WT or ΔnudA, were treated with PBS instead of the anti-H. pylori first antibody, and were processed as described above for pre-embedding immunogold method. Control of H. pylori detection with immunogold with pre-embedding was performed using chamber slides with non-infected AGS cells treated as described above. Importantly, all positive and negative controls were positive and negative, respectively.
A Philips CM100 transmission electron microscope was used at 80 kV for the analysis (Biomedical Instrumentation Center, USUHS) and negatives and photographs were processed. Visibility of colloidal gold particles in the pictures was enhanced using color burn option (Adobe Photoshop 7.0.1) that examines the color information in each channel and darkens the base color to reflect the blend color by increasing the contrast between the two). Nickel grids with five sections were counted at 10,500× in a selection of three mesh areas at a low magnification (2,600×). The number of cells was around 25–30 cells/mesh and two fields were counted in each mesh.
Morphometry
Qualitative fluorescence light microscopy ranged from 0 (negative reaction) to 4 (maximum red, green and yellow). Quantitative bright light microscopy of semi-thin sections stained with toluidine blue was used to count the density of H. pylori located in the extracellular or intracellular compartments, or attached to the cellular membrane of AGS cells using an intraocular grid (Semino-Mora et al., 2003). Three random fields were counted with 1,000× magnification (cell range 77–115/field, mean 96 ± 3/field; total cells counted 300 cells). Elongated cells with hummingbird phenotype were counted as hallmark of infection and compared to normal round cells in uninfected controls (no infection; Schneider et al., 2008).
Quantitative pre-embedding immunogold TEM was performed by counting electron dense gold particles specifically tagging H. pylori located in the extracellular, intracellular, and within cell membrane compartments (Tanner et al., 1996). Three grids of each experiment were analyzed as follows: five sections mounted in the nickel grid were counted at 14,000×. Three meshes were selected at low magnification (2,600×) and two fields were counted in each mesh (number of cells counted ∼25–30 cells/mesh) at higher magnification. A 17.5 cm × 23 cm transparent sheet grid with vertical and horizontal lines 1-cm apart with 374 intersections was placed on each micrograph printed on multi contrast Agfa paper. The point counting method was used to calculate the number of immunogold-tagged H. pylori as coincidences between bacterial electron dense gold particles and point grid intersections and expressed as a fraction of the total points intersections (Olivero et al., 1990).
Statistical analysis
Data are reported as mean ± SEM. Comparisons were performed using one way analysis of variance and subsequent t-tests.
Results
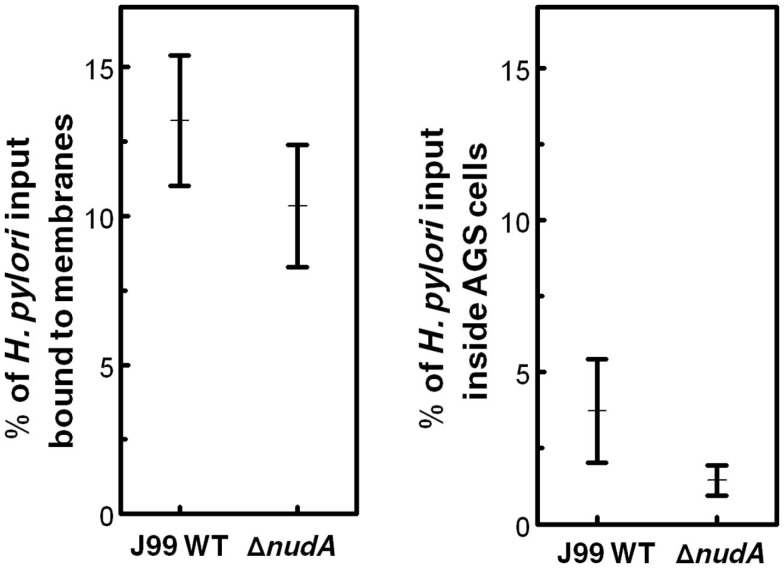
In a “classical” gentamicin protection assay, the density of H. pylori bound to AGS cell membranes and expressed as the percentage of bacteria inoculated into the AGS monolayer was not significantly different in WT J99 and the ΔnudA mutant (Figure 4, left panel). Similarly, the density of intracellular H. pylori expressed as the percentage of bacteria inoculated into the AGS monolayer was not significantly different in WT J99 and the ΔnudA mutant after saponin treatment (Figure 4, right panel).
Figure 4.
Helicobacter pylori NudA invasin and AGS cell entry determined using the “classical” gentamicin protection assay. Values are Means ± SEM. Left panel shows that the density of H. pylori bound to AGS cell membranes and expressed as the percentage of bacteria inoculated into the AGS monolayer was not significantly different in WT J99 and the ΔnudA mutant (p = 0.35). Right panel shows that the density of intracellular H. pylori expressed as the percentage of bacteria inoculated into the AGS monolayer was not significantly different in WT J99 and the ΔnudA mutant after saponin treatment (p = 0.23).
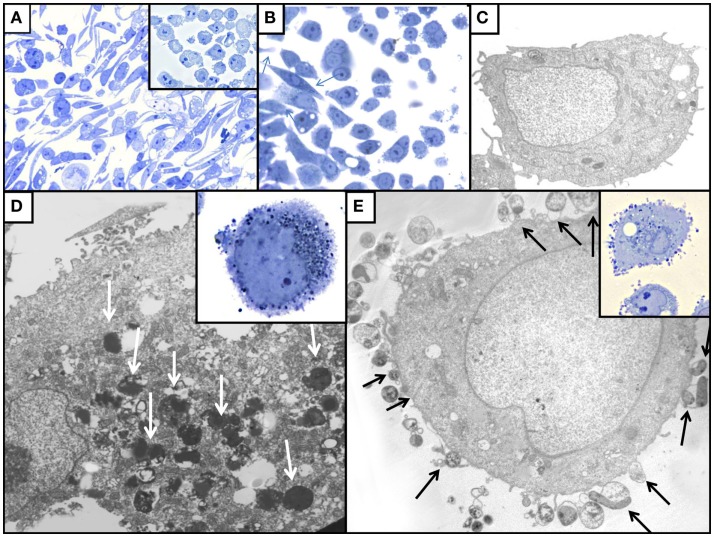
The addition of FISH and ultrastructural techniques to the “classical” gentamicin protection assay used in our study allowed a detailed analysis of the precise location of H. pylori in relation to the AGS cells, as was observed in the case of invasion of erythrocytes by B. bacilliformis (Mitchell and Minnick, 1995). Light microscopy performed on sections stained with toluidine blue demonstrated that elongated cells (“hummingbird” phenotype) were more frequently observed in AGS cells infected with WT J99 strain (Figures 4, left panel and 5) than in control uninfected cells (p < 0.001; Figure 5A insert and Figure 6). In ΔnudA allele infected cells, most H. pylori are associated with the membrane area and cell invasion is minimal but there still are significantly more elongated cells that in the absence of infection (p < 0.01) but less than with J99 WT (Figures 5B and 6; p < 0.01). Remaining cells are round or square. These results suggest that intracellular H. pylori are responsible for the hummingbird transformation of AGS cell in WT-infected cells compared to control uninfected cells, but that membrane associated H. pylori may also play a role in the formation of elongated cells.
Figure 5.
Illustration of the effect of H. pylori colonization on AGS cell monolayers in toluidine blue stained sections [(A,B) inserts of (A,D,E)] and by TEM (C,D,E). (A) WT-infected AGS cells; note the presence of numerous elongated hummingbird cells that are absent among control uninfected AGS cells (insert). (B) AGS cells infected with ΔnudA allele, illustrating that fewer elongated AGS cells (arrows) are present that in WT-infected cells. (C) TEM of uninfected AGS cell with normal ultrastructural aspect. (D) TEM of AGS cell infected with J99 WT H. pylori; note intracellular H. pylori (white arrows). (E) TEM of AGS cell infected with ΔnudA allele; note H. pylori attached to AGS cell (arrows); insert shows the numerous ΔnudA H. pylori attached to AGS cell and relatively few intracellular bacteria. Original magnification of toluidine blue stained pictures (A): 400×; (B) and insert of (A): 1,000×; inserts of (D,E): 1,000×. Original magnification of TEM pictures: (C): 9,800×; (D): 32,500×; (E): 26,000×.
Figure 6.
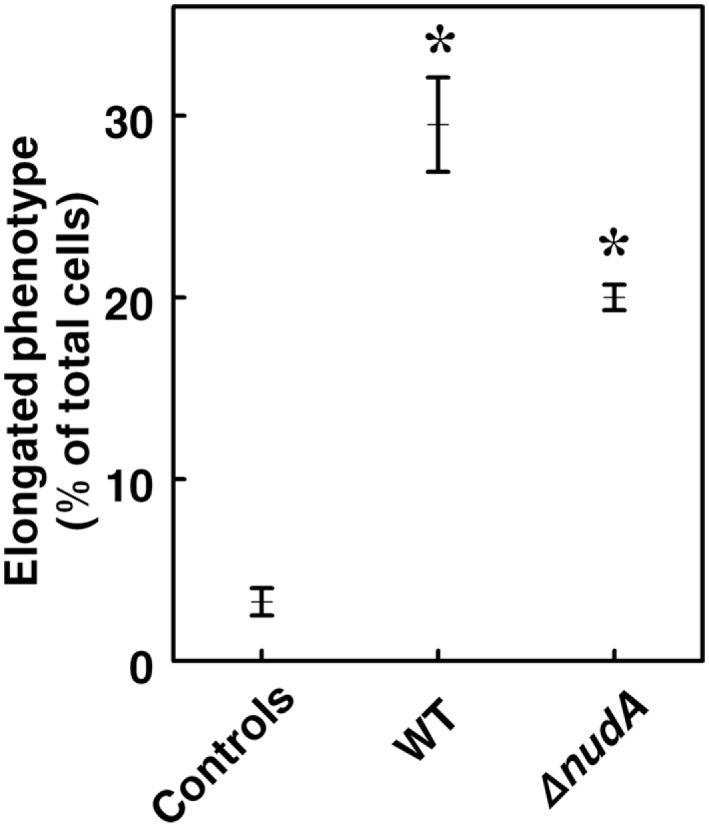
Percentage of AGS cells with elongated (“hummingbird”) phenotype in cells infected with WT J99 and ΔnudAH. pylori compared to control uninfected cells. Values are Means ± SEM. The percentage of cells with abnormal phenotype is higher in both types of infection (*p < 0.001), and also significantly higher in infection with WT H. pylori than with the ΔnudA mutant (p < 0.01).
Transmission electron microscopy indicated that more bacteria were present inside J99 WT-infected AGS cells (Figure 5D) than in ΔnudA allele infected cells and that more bacteria were closely associated with cell membranes in the latter cells (Figure 5E). After infection with J99 WT isolates, more bacteria were observed inside AGS cells (Figure 5D) than with the ΔnudA allele, but more bacteria were observed within the external surface of AGS cell membranes with the ΔnudA allele (Figure 5E). More intracellular H. pylori were observed inside AGS cell vacuoles than in direct contact with the cytosol, free in the cytoplasm (66.7 vs. 16.7%), the remaining H. pylori were partly in a vacuole and partly free in the cytoplasm.
Morphometry of TEM thin sections confirmed these findings, demonstrating that the number of AGS cells infected with J99 WT was significantly greater than with ΔnudA infection (39.8 ± 2.8 vs. 12.3 ± 2.2%, respectively; p < 0.001). In addition, there were significantly more intracellular H. pylori in WT-infected AGS cells than in ΔnudA allele infected cells (15.4 ± 2.1 vs. 6.9 ± 1.9; p < 0.01; Figure 5D), and more H. pylori were attached to AGS cells infected with ΔnudA allele (8.1 ± 2.6 vs. 0.3 ± 0.2, respectively; p < 0.01; Figure 5E). Importantly, the number of extracellular H. pylori not attached to the plasma membrane was similar in those two groups (4.2 ± 0.9 vs. 5.3 ± 2.0 respectively).
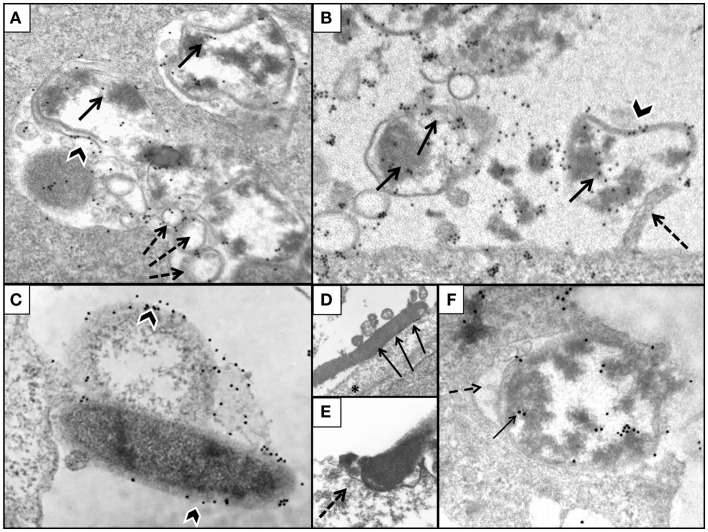
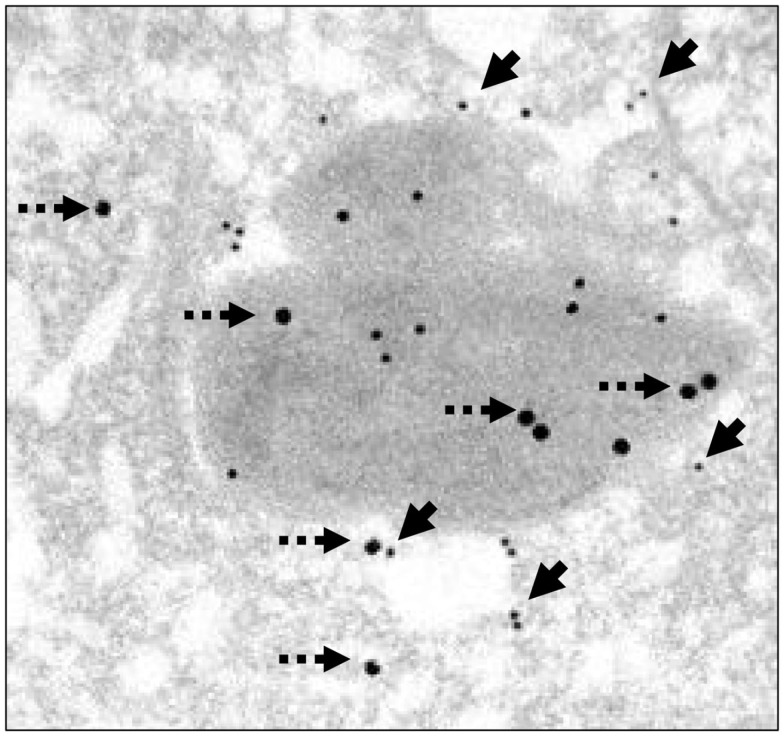
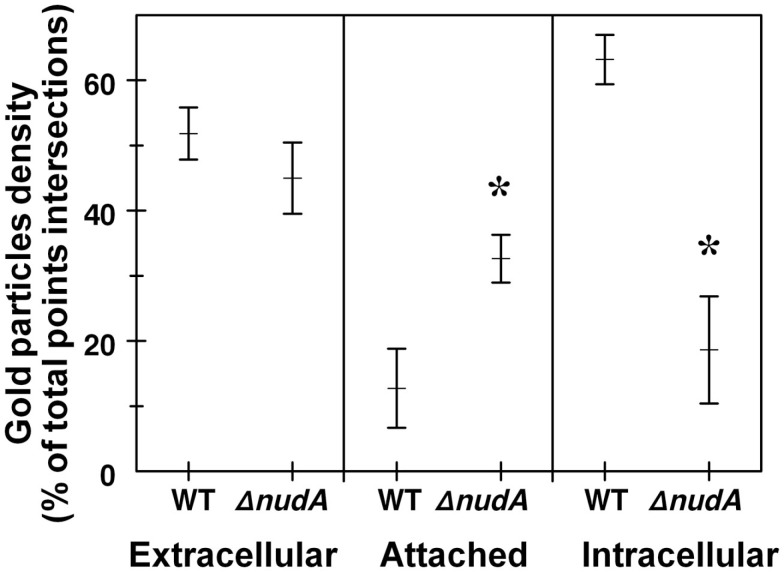
The TEM pre-embedding immunogold method with pre-embedding of H. pylori infected AGS cells was more effective to detect hidden epitopes than post-embedding method as reported (Tanner et al., 1996). The use of this method confirmed observations obtained with light microscopy that more WT H. pylori were intracellular than in ΔnudA allele infection and that more ΔnudA allele were attached to AGS cell membranes than in WT infection. In addition, intracellular H. pylori WT located inside vacuoles had gold-tagged bacterial walls, cytoplasm (Figure 7A), and outer membrane vesicles containing H. pylori VacA (Figure 8; Beveridge, 1999; Fiocca et al., 1999; Viala et al., 2004). Similarly, immunogold-labeled ΔnudA allele H. pylori were closely associated with AGS cell membranes, either attached to cell pedestals (Figure 7B) or to AGS cell membranes (Figure 7C) or undergoing diverse stages of “zipper-like” entry into AGS cell (Figures 7D–F; Noach et al., 1994; Papadogiannakis et al., 2000) Morphometric quantification of these observation demonstrated that most WT H. pylori and very few ΔnudA allele H. pylori were present inside the AGS cells and that the opposite was observed regarding attachment to AGS cell membranes (Figure 9; p < 0.01).
Figure 7.
Illustration of AGS cells with pre-embedded H. pylori immunogold. (A) AGS cell infected with H. pylori WT demonstrating intracellular presence of bacteria located inside vacuoles (gold marked bacterial wall, black arrowhead); note gold particles tagging H. pylori cytoplasm (arrow) and spherical membrane vesicles (dashed arrows). (B) ΔnudA allele with numerous bacteria labeled with gold (arrows) in close association with cell membrane; one of the bacteria is attached to cell pedestal (dashed arrow). (C) Mature and coccoid ΔnudA allele in close association with AGS cell membrane; note bacterial membrane is tagged with gold (arrowheads). (D) ΔnudA allele in early stage of adhesion to AGS cell; note multiple fusions between membranes of bacterium and AGS cell (arrows). (E) Higher magnification of “zipper-like” ΔnudA allele allele entry into AGS cell; note “cup formation” (dashed arrows) identifying H. pylori and AGS cell membranes fusing together during internalization. (F) ΔnudA allele almost completely englobed within invagination of AGS cell membrane; note that this fusion is accompanied by the presence of filamentous strands (fibrils), dense round spheres, vesicles, and amorphous deposits in the space between the membranes of the bacterium and AGS cell (dashed arrow). Immunogold-tagged bacterium (solid arrow) Original magnification (A–C): 41,000×; (D): 24,500×; (E): 32,500×; (F): 41,000×.
Figure 8.
Illustration of AGS cells with intracellular H. pylori WT with H. pylori and VacA immunogold. AGS cell infected using a dual pre-embedding immunogold (H. pylori gold 18 nm and VacA-gold 11 nm) demonstrating the presence of 18 nm H. pylori gold particles (dashed arrow) and 11 nm VacA-gold particles in cytoplasm, in the cell membrane, in membrane vesicles, and in the AGS cell. Original magnification 71,000×.
Figure 9.
Role of H. pylori NudA invasin in AGS cell entry determined using the gentamicin protection assay aided by TEM and anti-H. pylori antibody immunogold. H. pylori density is expressed as the percentage of total points intersections that were immunogold-positive (see Materials and Methods). Values are Means ± SEM. Left panel shows that the extracellular density of H. pylori WT and ΔnudA were not significantly different. In contrast, the middle and right panels show that the density of WT H. pylori attached to AGS cell membranes was significantly lower than that of the ΔnudA mutant (*p < 0.01) and the opposite relation for the density of intracellular H. pylori (*p < 0.01).
Discussion
A major finding of the present study was that H. pylori invasin appears to play a complex role in the entry of the bacterium into AGS cells in vitro if the “classical” gentamicin protection assay is complemented by morphology and morphometry. We first confirmed the previous report that no quantifiable differences were found when using the gentamicin protection assay to compare the invasion frequency of H. pylori wild type (WT) with that of a ΔnudA mutant in AGS cells allele (Lundin et al., 2003). The authors of the report attributed their observation to the fact that the sensitivity of the “classical” assay may not be sufficient to demonstrate differences in invasion capacity, and this lack of sensitivity may be due to the fact that complete killing of extracellular bacteria is rarely obtained (Amieva et al., 2002). In the present study, light and TEM morphology and morphometry of AGS cells demonstrated that a majority of H. pylori were closely associated with cell membranes after infection with the ΔnudA allele whereas most WT J99 bacteria were inside the cells. The TEM immunogold observations confirm this observation (Figures 7 and 9; p < 0.01) and strongly indicate that this difference demonstrates that NudA plays a biologically significant role in H. pylori entry into host cells.
The present study also provides precise information on H. pylori invasion and bacterial cell entry, demonstrating that H. pylori ΔnudA allele appears to be unable to proceed beyond the initial attachment to cell membranes that characterize the zipper-like mechanism of H. pylori cell entry (Griffin Jr. et al., 1975; Kwok et al., 2002). In contrast, J99 WT H. pylori rapidly proceeded from attachment to engulfment and internalization where it was found mostly inside vacuoles formed within AGS cells and, to a lesser extent, free in the cytoplasm. Our observations that gold-tagged outer membrane vesicles containing VacA were closely associated with intracellular H. pylori (Figure 8) demonstrates that H. pylori virulence factors and peptidoglycan may be delivered directly into the AGS cell cytoplasm (Blanke and Ye, 2001; Kaparakis et al., 2010) and suggests an important pathogenic role for cell invasion. This finding is similar to the observation that H. pylori releases small vesicles from its outer membrane by a process similar to the release of membrane vesicles by many other bacterial pathogens (Fiocca et al., 1999). The production of these vesicles may represent an important mechanism for bacterial pathogens to modulate their environment within the host and perhaps cause diseases. Finally, separate experiments showed that the H. pylori nudA (invasin homolog) gene is well expressed only when H. pylori is attached to or inside host cells and that it was closely associated with β1-integrin (Semino-Mora, unpublished).
An additional study of this highly relevant question demonstrated that the extent of adherence and internalization of H. pylori by AGS cells increased continuously for at least 12 h, and that a similar number of H. pylori were internalized by, and adherent to, AGS cells (Kwok et al., 2002) suggesting again that the “classical” gentamicin assay determines the sum of internalized and adherent H. pylori. However, the construction of a ΔnudA mutant performed in this study may have modified the expression of neighboring genes that might affect cell invasion. Since genetic complementation of the mutation and restoration of the WT phenotype was not performed as a control, our observations do not exclude the possibility that the internalization defect we observed is related to factors other than the nudA gene.
Helicobacter pylori invasiveness is important because intracellular bacteria are more resistant to antibiotic treatment and to immune attack by humoral antibodies. In addition, invasive organisms frequently cause common and severe diseases (Isberg et al., 1987) through mechanisms that are presently unknown (Dubois and Boren, 2007). Importantly, it is now recognized that bacteria invading host cells can induce formation of autophagic vacuoles within macrophages and dendritic cells where they can multiply, interfere with MHC class II surface expression, impair antigen presentation, and immunological defenses (Wang et al., 2009). In turn, these mechanisms would explain persistence of the infection and the various diseases caused by H. pylori.
In conclusion, our observations strongly indicate that deletion of the invasin gene limits H. pylori to the AGS cell surface, where it appears to be partially protected against gentamicin (Corthesy-Theulaz et al., 1996). In this site, however, it may be more vulnerable to host defense or therapeutic intervention than if intracellular, and less prone to trigger normal immune, carcinogenic, or other developmental response pathways. These data suggest that H. pylori invasin gene is important for the rare, but we propose potentially biologically significant, uptake of H. pylori by host cells.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Douglas Berg for stimulating discussions and for helpful comments on the results of these experiments and Donald F. Sellitti for reviewing the manuscript. This work was supported in part by NIH grant CA082312.
Footnotes
1www.thermofisher.com
2www.polysciences.com
References
- Alm R. A., Ling L. S., Moir D. T., King B. L., Brown E. D., Doig P. C., Smith D. R., Noonan B., Guild B. C., deJonge B. L., Carmel G., Tummino P. J., Caruso A., Uria-Nickelsen M., Mills D. M., Ives C., Gibson R., Merberg D., Mills S. D., Jiang Q., Taylor D. E., Vovis G. F., Trust T. J. (1999). Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397, 176–180 10.1038/17837 [DOI] [PubMed] [Google Scholar]
- Amieva M. R., Salama N., Tompkins L. S., Falkow S. (2002). Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell. Microbiol. 4, 677–690 10.1046/j.1462-5822.2002.00222.x [DOI] [PubMed] [Google Scholar]
- Bangham A. D., Horned R. W., Glauert A. M., Dingle J. T., Lucy J. A. (1962). Action of saponin on biological cell membranes. Nature 196, 952–955 10.1038/196952a0 [DOI] [PubMed] [Google Scholar]
- Beveridge T. J. (1999). Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181, 4725–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanke S. R., Ye D. (2001). “Alternative mechanisms of protein release,” in Helicobacter pylori: Physiology and Genetics, eds Mobley H. L. T., Mendz G. L., Hazell S. L. (Washington, DC: ASM Press; ), 1–14 [PubMed] [Google Scholar]
- Chalker A. F., Minehart H. W., Hughes N. J., Koretke K. K., Lonetto M. A., Brinkman K. K., Warren P. V., Lupas A., Stanhope M. J., Brown J. R., Hoffman P. S. (2001). Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183, 1259–1268 10.1128/JB.183.4.1259-1268.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthesy-Theulaz I., Porta N., Pringault E., Racine L., Bogdanova A., Kraehenbuhl J. P., Blum A. L., Michetti P. (1996). Adhesion of Helicobacter pylori to polarized T84 human intestinal cell monolayers is pH dependent. Infect. Immun. 64, 3827–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois A., Berg D. E. (1997). The nonhuman primate model for H. pylori infection . Methods Mol. Med. 8, 253–269 [DOI] [PubMed] [Google Scholar]
- Dubois A., Boren T. (2007). Helicobacter pylori is invasive and it may be a facultative intracellular organism. Cell. Microbiol. 9, 1108–1116 10.1111/j.1462-5822.2007.00921.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrand L., Graham D., Scheynius A., Genta R. M., El Zaatari F. (1997). Is the sanctuary where Helicobacter pylori avoids antibacterial treatment intracellular? Am. J. Clin. Pathol. 108, 504–509 [DOI] [PubMed] [Google Scholar]
- Fiocca R., Necchi V., Sommi P., Ricci V., Telford J., Cover T. L., Solcia E. (1999). Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J. Pathol. 188, 220–226 [DOI] [PubMed] [Google Scholar]
- Griffin F. M., Jr., Griffin J. A., Leider J. E., Silverstein S. C. (1975). Studies on the mechanism of phagocytosis. I. Requirements for circumferential attachment of particle-bound ligands to specific receptors on the macrophage plasma membrane. J. Exp. Med. 142, 1263–1282 10.1084/jem.142.5.1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg R. R., Voorhis D. L., Falkow S. (1987). Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50, 769–778 10.1016/0092-8674(87)90335-7 [DOI] [PubMed] [Google Scholar]
- Kaparakis M., Turnbull L., Carneiro L., Firth S., Coleman H. A., Parkington H. C., Le Bourhis L., Karrar A., Viala J., Mak J., Hutton M. L., Davies J. K., Crack P. J., Hertzog P. J., Philpott D. J., Girardin S. E., Whitchurch C. B., Ferrero R. L. (2010). Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell. Microbiol. 12, 372–385 10.1111/j.1462-5822.2009.01404.x [DOI] [PubMed] [Google Scholar]
- Kwok T., Backert S., Schwarz H., Berger J., Meyer T. F. (2002). Specific entry of Helicobacter pylori into cultured gastric epithelial cells via a zipper-like mechanism. Infect. Immun. 70, 2108–2120 10.1128/IAI.70.4.2108-2120.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Rahman A., Semino-Mora C., Doi S. Q., Dubois A. (2008). Specific and sensitive detection of H. pylori in biological specimens by real-time RT-PCR and in situ hybridization. PLoS ONE 3, e2689. 10.1371/journal.pone.0002689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A., Nilsson C., Gerhard M., Andersson D. I., Krabbe M., Engstrand L. (2003). The NudA protein in the gastric pathogen Helicobacter pylori is an ubiquitous and constitutively expressed dinucleoside polyphosphate hydrolase. J. Biol. Chem. 278, 12574–12578 10.1074/jbc.M212542200 [DOI] [PubMed] [Google Scholar]
- Luo Y., Liu Y., Sun D., Ojcius D. M., Zhao J., Lin X., Wu D., Zhang R., Chen M., Li L., Yan J. (2011). InvA protein is a Nudix hydrolase required for infection by pathogenic Leptospira in cell lines and animals. J. Biol. Chem. 286, 36852–36863 10.1074/jbc.M111.251538 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Maki H., Sekiguchi M. (1992). MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355, 273–275 10.1038/355273a0 [DOI] [PubMed] [Google Scholar]
- Mitchell S. J., Minnick M. F. (1995). Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infect. Immun. 63, 1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necchi V., Candusso M. E., Tava F., Lunetti O., Ventura U., Fiocca R., Ricci V., Solcia E. (2007). Intracellular, intercellular and stromal invasion of gastric mucosa, preneoplastic lesions, and cancer by H. pylori. Gastroenterology 132, 1009–1023 10.1053/j.gastro.2007.01.049 [DOI] [PubMed] [Google Scholar]
- Noach L. A., Rolf T. M., Tytgat G. N. (1994). Electron microscopic study of association between Helicobacter pylori and gastric and duodenal mucosa. J. Clin. Pathol. 47, 699–704 10.1136/jcp.47.8.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivero O. A., Semino C., Poirier M. C. (1990). Localization of DNA adducts induced by N-acetoxy-N-2-acetylaminofluorene in Chinese hamster ovary cells using electron microscopy and colloidal gold. Genes Chromosomes Cancer 2, 130–136 10.1002/gcc.2870020209 [DOI] [PubMed] [Google Scholar]
- Papadogiannakis N., Willen R., Carlen B., Sjostedt S., Wadstrom T., Gad A. (2000). Modes of adherence of Helicobacter pylori to gastric surface epithelium in gastroduodenal disease: a possible sequence of events leading to internalisation. APMIS 108, 439–447 10.1034/j.1600-0463.2000.d01-80.x [DOI] [PubMed] [Google Scholar]
- Schneider S., Weydig C., Wessler S. (2008). Targeting focal adhesions: Helicobacter pylori-host communication in cell migration. Cell Commun. Signal. 6, 2. 10.1186/1478-811X-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino-Mora C., Doi S. Q., Marty A., Simko V., Carlstedt I., Dubois A. (2003). Intracellular and interstitial expression of Helicobacter pylori virulence genes in gastric precancerous intestinal metaplasia and adenocarcinoma. J. Infect. Dis. 187, 1165–1177 10.1086/368133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Berg D. E. (2004). Motility of urease-deficient derivatives of Helicobacter pylori. J. Bacteriol. 186, 885–888 10.1128/JB.186.15.5087-5092.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner V. A., Ploug T., Tao-Cheng J. H. (1996). Subcellular localization of SV2 and other secretory vesicle components in PC12 cells by an efficient method of preembedding EM immunocytochemistry for cell cultures. J. Histochem. Cytochem. 44, 1481–1488 10.1177/44.12.8985140 [DOI] [PubMed] [Google Scholar]
- Tomb J. F., White O., Kerlavage A. R., Clayton R. A., Sutton G. G., Fleischmann R. D., Ketchum K. A., Klenk H. P., Gill S., Dougherty B. A., Nelson K., Quackenbush J., Zhou L., Kirkness E. F., Peterson S., Loftus B., Richardson D., Dodson R., Khalak H. G., Glodek A., McKenney K., Fitzegerald L. M., Lee N., Adams M. D., Hickey E. K., Berg D. E., Gocayne J. D., Utterback T. R., Peterson J. D., Kelley J. M., Cotton M. D., Weidman J. M., Fujii C., Bowman C., Watthey L., Wallin E., Hayes W. S., Borodovsky M., Karp P. D., Smith H. O., Fraser C. M., Venter J. C. (1997). The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388, 539–547 10.1038/41483 [DOI] [PubMed] [Google Scholar]
- Viala J., Chaput C., Boneca I. G., Cardona A., Girardin S. E., Moran A. P., Athman R., Mémet S., Huerre M. R., Coyle A. J., DiStefano P. S., Sansonetti P. J., Labigne A., Bertin J., Philpott D. J., Ferrero R. L. (2004). Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 5, 1166–1174 10.1038/ni1131 [DOI] [PubMed] [Google Scholar]
- Wang Y. H., Wu J. J., Lei H. Y. (2009). The autophagic induction in Helicobacter pylori-infected macrophage. Exp. Biol. Med. (Maywood) 234, 171–180 10.3181/0904-MR-134 [DOI] [PubMed] [Google Scholar]