Cellular enzymes that cap, splice, and polyadenylate eukaryotic pre-mRNAs are targeted in vivo to the nascent chains synthesized by RNA polymerase II (pol II). Placing a mammalian pol II transcription unit under the control of a pol III promoter results in a failure to cap, splice, or polyadenylate the transcript (1, 2). How is pre-mRNA “identity” established? Do the various mRNA processing enzymes recognize protein components of the pol II transcription elongation complex or is identity conferred through an initial pol II-specific modification that directs subsequent mRNA fate? We know that acquisition of the m7GpppN cap is the first modification event in mRNA biogenesis and that capping facilitates downstream transactions such as splicing, polyadenylation, transport, and translation. What we don’t understand is how pol II transcripts are specifically singled out for capping. Now, three groups of investigators (the Shatkin and Reinberg laboratories collaboratively, the Bentley and Shuman laboratories collaboratively, and the Buratowski laboratory) have presented findings that offer an elegant solution to the puzzle: the capping enzymes are targeted to pre-mRNA by binding to the phosphorylated C-terminal domain (CTD) of the largest subunit of RNA pol II (3–5). In the paper by Yue et al. (3) in this issue of the Proceedings, the Shatkin and Reinberg group show that mammalian mRNA capping enzyme interacts directly with pol IIO, the hyperphosphorylated form of pol II, but not with pol IIA, the form in which the CTD is either unphosphorylated or hypophosphorylated.
The CTD, which is unique to pol II, consists of a tandem array of a heptapeptide repeat with the consensus sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser. The mammalian pol II large subunit has 52 tandem repeats, whereas the Saccharomyces cerevisiae subunit has 27 copies. The IIA and IIO forms of pol II are interconvertible and functionally distinct. In vivo, the pol IIO enzyme contains as many as 50 phosphorylated amino acids (primarily phosphoserine) within the CTD (6). During transcription initiation, pol IIA is recruited to the DNA template by the general transcription factors TBP (TATA-binding protein), TFIIB (transcription factor IIB), and TFIIF. TBP has been reported to bind to pol IIA, but not pol IIO (7). The pol IIA CTD undergoes extensive phosphorylation and conversion to IIO during the transition from preinitiation complex to stable elongation complex. The CTD kinase activity of transcription factor IIH is implicated in CTD hyperphosphorylation during this step (8). TFIIH contains a cyclin and cyclin-dependent kinase subunit pair (cdk7 and cyclin H) that catalyzes phosphorylation of Ser-5 of the CTD heptapeptide (9). TFIIH is recruited to the preinitiation complex by TFIIE, which binds specifically to pol IIA (10). TFIIE and TFIIH dissociate from the transcription complex shortly after initiation (11). Capping occurs when nascent RNA chains grow to ≈30 nucleotides in length, at which point their 5′ ends are extruded from the RNA binding pocket of the polymerase and are thereby accessible to the capping enzymes (12, 13).
Cap formation entails a series of three reactions catalyzed by RNA triphosphatase, RNA guanylyltransferase, and RNA (guanine-7-) methyltransferase. The triphosphatase hydrolyzes the 5′ triphosphate end of the primary transcript to a diphosphate. Guanylyltransferase adds GMP from GTP to the diphosphate RNA end to form a blocked G(5′)ppp(5′)N structure. The methyltransferase adds a methyl group from S-adenosylmethionine to the cap guanosine to form m7GpppN. This pathway was elucidated more than 20 years ago by the Moss and Shatkin laboratories through studies of the capping enzymes encoded by vaccinia virus and reovirus (reviewed in ref. 14). Only in the past few years have cellular genes encoding the capping enzymes been cloned (15–19). The fact that the cap guanylyltransferase and cap methyltransferase activities are both essential for yeast cell growth underscores the critical role of the cap in mRNA metabolism.
In their paper, Yue et al. (3) identify cDNAs encoding the human and mouse guanylyltransferases—the first examples of cloned capping enzymes from mammals. The mouse and human capping enzymes are 597-aa polypeptides with 95% amino acid sequence identity. Like the 573-aa Caenorhabditis elegans capping enzyme (18, 19), the mammalian proteins are bifunctional and consist of an N-terminal RNA triphosphatase domain linked to a C-terminal guanylyltransferase domain. The metazoan guanylyltransferase domains are structurally similar to the monofunctional guanylyltransferase proteins encoded by yeast and Chlorella virus and to the guanylyltransferase domain of vaccinia capping enzyme. The N-terminal triphosphatase domain contains the signature motif of the protein tyrosine phosphatase superfamily. The authors provide clear biochemical evidence that the mouse cDNA encodes a catalytically active triphosphatase-guanylyltransferase. Moreover, they find that the mouse cDNA rescues growth of a yeast strain lacking the endogenous guanylyltransferase gene (20).
To examine the polymerase-capping connection, Yue et al. (3) incubated mammalian capping enzyme with partially purified RNA pol II consisting of a mixture of the IIO and IIA isoforms, and then immunoprecipitated the sample with antibody to the guanylyltransferase domain. Analysis of the precipitate by Western blotting using an antibody against the largest subunit of RNA pol II revealed that pol IIO, but not pol IIA, was precipitated by the capping enzyme antibody. The guanylyltransferase domain alone was sufficient for selective binding of pol IIO. This simple experiment has broad implications.
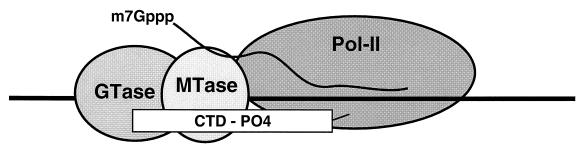
Does the capping enzyme interact directly with the CTD? McCracken et al. (4) have shown by CTD affinity chromatography that mammalian guanylyltransferase binds to a recombinant glutathione-S-transferase–CTD fusion protein containing 15 tandem copies of the heptapeptide, provided that the CTD has been phosphorylated in vitro by HeLa cell extract, recombinant cdk7/cyclin H kinase, or cdc2 kinase. The guanylyltransferase does not bind to nonphosphorylated CTD or to in vitro phosphorylated mutant CTD in which residue Ser-5 of each heptapeptide repeat was replaced by alanine. This engenders a model whereby phosphorylation of Ser-5 suffices for recruitment of mammalian guanylyltransferase to the elongating pol II. The interaction of guanylyltransferase with CTD–PO4 is conserved throughout eukaryotic evolution. S. cerevisiae and Schizosaccharomyces pombe guanylyltransferases bind to GST–CTD–PO4 but not to unphosphorylated glutathione-S-transferase–CTD (4).
Does access of guanylyltransferase to the transcription complex require CTD phosphorylation? Cho et al. (5) detect the presence of the S. cerevisiae guanylyltransferase Ceg1 in pol II transcription complexes assembled in vitro on an immobilized DNA template. Association of Ceg1 with the complexes is promoter-dependent, stimulated by ATP (a substrate for the CTD kinase), and blocked by the kinase inhibitor H8.
Cap methylation is essential for cell viability (21). The S. cerevisiae cap methyltransferase Abd1 is a 436-aa monomeric protein that resembles the methyltransferase domain of vaccinia capping enzyme. Metazoan homologues of Abd1 have been identified (22). Although the cap methyltransferase and guanylyltransferase enzymes are not physically associated during their isolation from yeast or mammalian cell extracts, it is likely that their actions are coordinated in vivo. How is this achieved? McCracken et al. (4) find that recombinant yeast Abd1 protein binds to the phosphorylated CTD, but not to unphosphorylated CTD or to Ser-5 → Ala mutant CTD. Hence the guanylyltransferase and methyltransferase components bind independently to CTD–PO4 (Fig. 1).
Recruitment of the capping apparatus to the phosphorylated CTD neatly accounts for pol II specificity of capping and provides an elegant means of traffic control whereby CTD-interacting factors bind and dissociate from polymerase at the appropriate times in the transcription cycle without getting in each other’s way. During preinitiation complex formation, the unphosphorylated CTD interacts with TBP, the SRB/mediator component of the pol II holoenzyme, and TFIIE. Phosphorylation of the CTD by TFIIH and/or other CTD kinases [e.g., the cdk8/cylinC pair found in the pol II holoenzyme (23, 24) and CTDK-I, a heterotrimeric kinase with cdk-like and cyclin-like subunits (25)] presumably destabilizes these contacts and makes the CTD available for a novel set of interactions with the capping enzymes. A prediction of the model is that alterations of the CTD should impact on cap formation. Indeed, McCracken et al. (4) find that the efficiency of cap formation in mammalian cells is diminished when genes are transcribed by a mutant version of RNA pol II containing only 5 heptapeptide repeats of the CTD instead of the normal complement of 52 repeats. Genetic interactions between the CTD and the guanylyltransferase Ceg1 are evident in S. cerevisiae, i.e., nonlethal rpb1 mutations with a truncated CTD exacerbate the conditional growth phenotype of ceg1 mutations in an allele-specific fashion (ref. 5; B. Schwer and S. Shuman, unpublished work).
The findings contribute to an emerging picture of the CTD as a landing pad for macromolecular assemblies that regulate mRNA synthesis and processing (26). Other recent studies indicate that protein components of the pre-mRNA splicing and 3′ cleavage-polyadenylation assemblies also bind to the CTD (27, 28). The role of CTD phosphorylation in those interactions remains to be clarified. Thus far, only the capping enzymes display a strict requirement for CTD phosphorylation. In light of the new data reviewed here, studies of the impact of CTD mutations or CTD-kinase mutations on mRNA biogenesis must take into account effects on cap formation. Nonetheless, it seems reasonable to invoke overlapping mechanisms of achieving mRNA identity, whereby processing enzymes are targeted to the nascent pre-mRNA by recognition of a cap binding complex bound at the 5′ end (29) and also via contacts with the CTD.
Given the abundance of phosphorylation sites on the CTD, the substantial number of cellular kinases capable of phosphorylating the CTD (some in a site-specific fashion), and the existence of CTD-specific phosphatases, there is ample potential to customize the CTD landing pad through display of different phosphorylation arrays to cellular ligands at different stages of the polymerase elongation cycle. Dynamic remodeling of the CTD docking site by phosphatase and kinase activities would facilitate dissociation of CTD-associated proteins once their task has been executed. It is not at all clear how that process would be coordinated. At present, we do not know if the cellular capping enzymes remain associated with the elongating RNA polymerase after the cap structure has been formed [as is the case for vaccinia capping enzyme and vaccinia RNA polymerase elongation complexes (30)] or if they are jettisoned to make room for other processing assemblies. One way to disengage the capping apparatus from pol IIO in a timely fashion would be to couple its departure to the binding of the nuclear cap binding protein complex (29) to the newly formed 5′ cap of the nascent chain. RNA-bound cap binding protein complex might compete with the capping enzymes for a common binding site on CTD or else recruit factors that remodel the CTD.
Issues of immediate interest concerning the recruitment of the capping enzymes to pol IIO include (i) identification of the moieties on the guanylyltransferase and methyltransferase proteins responsible for CTD binding; (ii) delineation of the CTD side of the interface, i.e., the number of heptamer repeats and phosphates that are required to bind guanylyltransferase and methyltransferase; and (iii) development of quantitative assays of CTD binding affinity and association rate. A high-resolution structure of guanylyltransferase has provided considerable insight into the mechanism of catalysis (31). A structure of capping enzyme complexed with a defined CTD–PO4 ligand would illuminate a key step in the establishment of mRNA identity.
References
- 1.Sisodia S S, Sollner-Webb B, Cleveland D W. Mol Cell Biol. 1987;7:3602–3612. doi: 10.1128/mcb.7.10.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gunnery S, Mathews M B. Mol Cell Biol. 1995;15:3597–3607. doi: 10.1128/mcb.15.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCracken, S., Fong, N., Rosonina, E., Yankulov, K., Brothers, G., Siderovski, D., Hessel, A., Foster, S., Shuman, S. & Bentley, D. L. (1997) Genes Dev., in press. [DOI] [PMC free article] [PubMed]
- 5.Cho, E., Takagi, T., Moore, C. R. & Buratowski, S. (1997) Genes Dev., in press. [DOI] [PMC free article] [PubMed]
- 6.Dahmus M E. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 7.Usheva A, Maldonado E, Goldring A, Lu H, Houbavi C, Reinberg D, Aloni Y. Cell. 1992;69:871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- 8.Lu H, Zawel L, Fisher L, Egly J M, Reinberg D. Nature (London) 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 9.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J P, Schaeffer L, Nigg E A, Hoeijmakers J, Egly J M. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 10.Maxon M E, Goodrich J A, Tjian R. Genes Dev. 1994;8:515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- 11.Zawel L, Kumar K P, Reinberg D. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 12.Hagler J, Shuman S. Science. 1992;255:983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- 13.Rasmussen E B, Lis J T. Proc Natl Acad Sci USA. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shuman S. Prog Nucleic Acid Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 15.Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. J Biol Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- 16.Shuman S, Liu Y, Schwer B. Proc Natl Acad Sci USA. 1994;91:12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao X, Schwer B, Shuman S. Mol Cell Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takagi T, Moore C, Diehn F, Buratowski S. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 19.Wang S P, Deng L, Ho C K, Shuman S. Proc Natl Acad Sci USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwer B, Shuman S. Proc Natl Acad Sci USA. 1994;91:4328–4332. doi: 10.1073/pnas.91.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao X, Schwer B, Shuman S. Mol Cell Biol. 1996;16:475–480. doi: 10.1128/mcb.16.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S P, Shuman S. J Biol Chem. 1997;272:14683–14689. doi: 10.1074/jbc.272.23.14683. [DOI] [PubMed] [Google Scholar]
- 23.Liao S, Zhang J, Jeffrey D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J J, Yound R A. Nature (London) 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 24.Leclerc V, Tassan J P, O’Farrell P H, Nigg E A, Leopold P. Mol Biol Cell. 1997;7:505–513. doi: 10.1091/mbc.7.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sterner D E, Lee J M, Hardin S E, Greenleaf A L. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinmetz E J. Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 27.Yuryev A, Patturajan M, Litingtung Y, Joshi R, Gentile C, Gebara M, Corden J. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G H, Greenblatt J, Patterson S D, Wickens M, Bentley D L. Nature (London) 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 29.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj I W. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 30.Hagler J, Luo Y, Shuman S. J Biol Chem. 1994;269:10050–10060. [PubMed] [Google Scholar]
- 31.Hakansson K, Doherty A J, Shuman S, Wigley D B. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]