Abstract
The 1,3-dipolar cycloaddition reaction between unactivated azides and acetylenes proceeds exceedingly slowly at room temperature. However, considerable rate acceleration is observed when this reaction occurs inside the active center gorge of acetylcholinesterase (AChE) between certain azide and acetylene reactants, attached via methylene chains to specific inhibitor moieties selective for the active center and peripheral site of the enzyme. AChE catalyzes the formation of its own inhibitor in a highly selective fashion: only a single syn1-triazole regioisomer with defined substitution positions and linker distances is generated from a series of reagent combinations. Inhibition measurements revealed this syn1-triazole isomer to be the highest affinity reversible organic inhibitor of AChE with association rate constants near the diffusion limit. The corresponding anti1 isomer, not formed by the enzyme, proved to be a respectable but weaker inhibitor. The crystal structures of the syn1- and anti1-mouse AChE complexes at 2.45- to 2.65-Å resolution reveal not only substantial binding contributions from the triazole moieties, but also that binding of the syn1 isomer induces large and unprecedented enzyme conformational changes not observed in the anti1 complex nor predicted from structures of the apoenzyme and complexes with the precursor reactants. Hence, the freeze-frame reaction offers both a strategically original approach for drug discovery and a means for kinetically controlled capture, as a high-affinity complex between the enzyme and its self-created inhibitor, of a highly reactive minor abundance conformer of a fluctuating protein template.
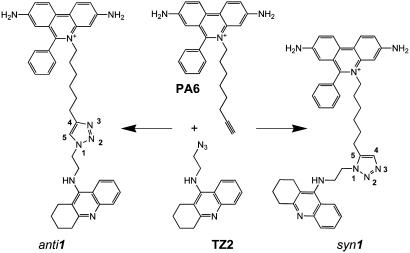
Acetylcholinesterase (AChE) rapidly terminates cholinergic neurotransmission by catalyzing the hydrolysis of the neurotransmitter, acetylcholine, and inhibitors of AChE have been used for over a century in various therapeutic regimens (1, 2). The structure of the target enzyme reveals a narrow gorge ≈20 Å in depth with the catalytic triad of the active center at its base (3). Distinctive inhibitors bind to the active center or to a peripheral anionic site (PAS) located at the rim of the gorge near the enzyme surface (4-6). Previously, we generated a library of active site and PAS inhibitors with respective tacrine and phenanthridinium nuclei, each equipped with an azide or acetylene group at the end of a flexible methylene chain, to enable the reporting 1,3-dipolar cycloaddition to occur (Scheme 1) (7). AChE itself served as the reaction vessel, synthesizing its own inhibitor from these building blocks, in effect, by equilibrium-controlled sampling of various possible pairs of reactants in its active center gorge until irreversible cycloaddition between azide and acetylene ensued at an intersecting point within the gorge, between the two anchoring positions. From 49 building block combinations, the enzyme selected the TZ2/PA6 pair to form, with an enhanced reaction rate, a highly regioselective syn1 triazole as the sole product (Scheme 1). In contrast, chemical synthesis by thermal reaction in the absence of enzyme proceeds very slowly and provides an ≈1:1 mixture of syn1 and anti1 regioisomers, which differ in the nitrogen substitution positions on the 1,2,3-triazole. Although both are high-affinity inhibitors, the syn1 isomer, with a 100-fold greater affinity and a subpicomolar dissociation constant for certain AChEs (7), has a potency greater than all known noncovalent organic AChE inhibitors and high selectivity for individual cholinesterases.
Scheme 1.
Structures of the anti1 and syn1 TZ2PA6 regioisomers formed by 1,3-dipolar cycloaddition (7). The phenylphenanthridinium, triazole, and tacrine moieties are shown from top to bottom.
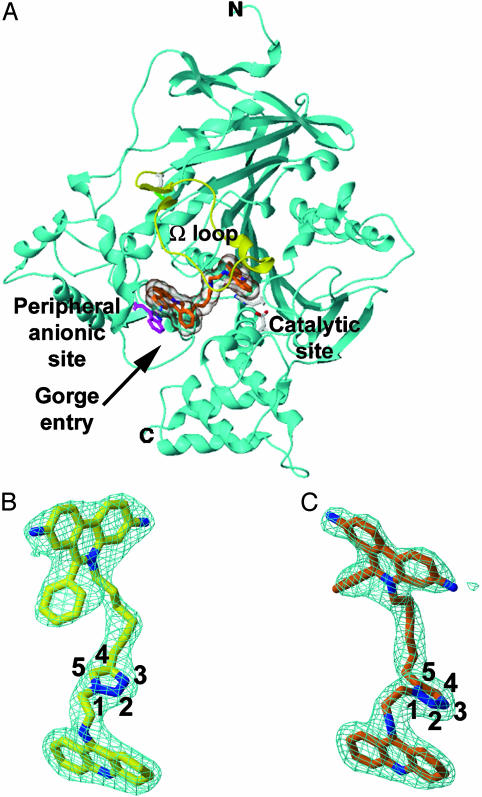
The discovery that enzymes can serve as atomic-scale reaction templates for creating their own inhibitors offers an original approach to drug discovery. In this light, we have solved the crystal structures of complexes of mouse AChE (mAChE) (8, 9) with the TZ2PA6 anti1 and syn1 regioisomers at 2.45- and 2.65-Å resolution (Table 1 and Fig. 1) and have analyzed further their respective binding kinetics and affinities (Table 2). We show that the distinctive binding properties of the two isomers in solution are related to discrete rearrangements in both the ligand and enzyme conformations. Indeed, the active center gorge and PAS conformations for the two crystalline complexes differ greatly, where binding of the higher affinity syn1 isomer unveils a unique enzyme conformation not predicted from the structures of either the apo form or the complexes with the precursor reactants (10, 11). Hence these structures reveal that the syn1 compound specifically formed on the enzyme effectively freezes in frame a highly reactive AChE conformer, and that the two TZ2PA6 regioisomers select distinct conformations from an unliganded enzyme that is presumably fluctuating between multiple conformational states. Because the unique structural features seen for the syn1-mAChE complex likely reflect the unique proximity of reactants causing the higher affinity syn1 isomer to be the sole reaction product in situ, these structures also provide insights on the cycloaddition reaction occurring on a flexible protein template and at a locus remote from the anchoring binding sites of the precursors. Thus, the unique structure of the complex captured by click chemistry leads to an unusual strategy for drug design where the most selective agents induce distinctive conformational states of the target.
Table 1. Data collection and refinement statistics.
| TZ2PA6 isomer complexed to mAChE
|
||
|---|---|---|
| anti1 | syn1 | |
| Data collection* | ||
| Beamline (European Synchrotron Radiation Facility) | ID14-EH1 | ID14-EH2 |
| Wavelength, Å | 0.933 | 0.933 |
| Resolution range, Å | 25-2.45 | 25-2.65 |
| Total observations | 549,476 | 513,851 |
| Unique reflections | 74,834 | 59,650 |
| Multiplicity | 3.9 | 3.6 |
| Completeness, % | 99.8 (99.7) | 99.8 (99.9) |
| I/σ (I) | 9.3 (1.8) | 8.2 (1.8) |
| Rsym† | 5.7 (43.5) | 6.5 (42.0) |
| Refinement‡ | ||
| R factor/Rfree, % | 18.4/21.4 | 18.1/22.0 |
| rms deviation§ | ||
| Bonds, Å/angles, ° | 0.015/1.5 | 0.015/1.5 |
| Chiral volume, Å3 | 0.085 | 0.084 |
| Mean B factors, Å | ||
| Main/side chains | 53.4/55.2 | 49.7/51.6 |
| Solvent/carbohydrate | 50.8/91.5 | 46.7/95.2 |
| Ligand/polyethylene glycol | 63.5/72.2 | 57.2/66.3 |
| rms deviation on B factors, Å2 | ||
| Main/side chains | 0.9/1.6 | 0.9/1.6 |
Values in parentheses are those for the last shell
Rsym = | I - 〈I〉 |/Σ I, where I is an individual reflection measurement and 〈I〉 is the mean intensity for symmetry-related reflections
R factor = Σ | | Fo | - | Fc| |/Σ | Fo |, where Fo and Fc are observed and calculated structure factors, respectively. Rfree is calculated for 2% of randomly selected reflections excluded from refinement
rms deviation from ideal values
Fig. 1.
Overall fold and structural quality of the TZ2PA6-mAChE complexes. (A) Overall view of the mAChE molecule (cyan ribbon) showing the syn1 isomer (orange bonds; transparent molecular surface) bound within the enzyme active-site gorge; the long Ω loop Cys-69-Cys-96 is displayed in yellow. (B and C) Determined structures of the bound anti1 and syn1 isomers (yellow and orange bonds, respectively; blue nitrogens; numbered triazole atoms; same orientation as in Scheme 1), with the respective 2.45- and 2.65-Å resolution final 2Fo-Fc electron density maps contoured at 1 σ (cyan).
Table 2. Kinetic parameters for inhibition of various cholinesterases by the TZ2PA6 isomers.
|
syn1
|
anti1
|
|||||
|---|---|---|---|---|---|---|
| Enzyme | kon, 1010 M-1·min-1 | koff, 10-3 min-1 | Ki, fM | kon, 1010 M-1·min-1 | koff, 10-3 min-1 | Ki, fM |
| mAChE* | 1.7 | 7.1 | 410 | 2.5 | 220 | 8,900 |
| mAChE mutant Trp286Ala | 0.94 | 19 | 2,000 | 0.87 | 1,800 | 210,000 |
| AChE, Electrophorus electricus* | 1.5 | 1.5 | 99 | 1.8 | 250 | 14,000 |
| AChE, Torpedo californica* | 1.4 | 1.1 | 77 | 3.2 | 23 | 720 |
| AChE, Drosophila melanogaster | 2.0 | 72 | 3,600 | 3.4 | 58 | 1,700 |
| Butyrylcholinesterase, mouse | 0.36 | 2.6 | 720 | 0.69 | 3.2 | 460 |
Parameters are means of n ≥ 3 individual values with SD ≤ 20%.
Modified from ref. 7
Experimental Procedures
Preparation and Analysis of the Complexes. The TZ2PA6 anti1 and syn1 isomers were synthesized as described (7). Monomeric mAChE expressed from human embryonic kidney-293 cells (9) was purified by affinity chromatography by using propidium elution (10). The anti1- and syn1-mAChE complexes were prepared by using a 2-fold molar excess of the inhibitors and concentrations well above their Kds (≈135 μM i.e., 15 106 × Ki(anti) and 330 106 × Ki(syn); Table 2) (7, 10). Titration of the isomer stock solutions (ε490nm = 6,000 M-1·cm-1) and analysis of the complex solutions were carried out spectrophotometrically (10).
Crystallization and Data Collection. Crystallization was achieved at 4°C by vapor diffusion by using hanging drops (1-2 μl) and a protein-to-well solution ratio of 1:1 with polyethylene glycol-600 25-32% (vol/vol) in 20-100 mM Hepes, pH 6.0-7.5, as the well solution. The crystals were directly flash-cooled in the nitrogen gas stream (100 K); optimal occupancies of the crystalline PASs were controlled by spectrophotometry before data collection (10). The crystals belonged to the orthorhombic space group P212121 with unit cell dimensions a = 79.7 Å, b = 111.9 Å, and c = 226.5 Å. Oscillation images were integrated with denzo (12), and data were scaled and merged with scala (13).
Structure Determination and Refinement. Coordinates of the anti1 and syn1 molecules were obtained from docked simulations of TZ2PA6-TcAChE complexes (7). The apo-mAChE structure (Protein Data Bank ID code 1J06) (10) without solvent was used as a starting model to refine the anti1- and syn1-mAChE complex structures with cns (14) and refmac (15) (Table 1). Rigid-body refinements on each of the two subunits in the crystalline mAChE dimer (10) gave R factor values of 25.2% and 24.8% (Rfree values of 25.8% and 25.3%) for the anti1 and syn1 complexes, respectively, by using all data in the 25- to 2.45/2.65-Å resolution range. The resulting 2Fo-Fc and Fo-Fc electron density maps were used to position the inhibitors and correct the protein model with the graphics program turbofrodo (16).
The final two TZ2PA6-mAChE structures comprise residues Glu-1-Ala-541 and Glu-4-Thr-540 for the two mAChE molecules in the asymmetric unit (10). High-temperature factors and weak electron densities are associated with residues Cys-257, Pro-258, and Asp-265 in the short Ω loop Cys-257-Cys-272 and with the surface loop region Asp-491-Pro-498. The average rms deviation (rmsd) between the anti1 and syn1 complex structures is 0.24 Å for 535 Cα atoms. Between the anti1 and syn1 complexes and the apo form, the rmsds are 0.19 and 0.23 Å for 534 and 533 Cα atoms, respectively. The stereochemistries of the bound isomers were checked by using the MM2 force field as implemented in macromodel (17). Those of the structures were analyzed with procheck (18); with the exception of the catalytic Ser-203, no residues were found in the disallowed regions of the Ramachandran plot. Figs. 1, 2, 3 were generated with spock (19) and raster3d (20).
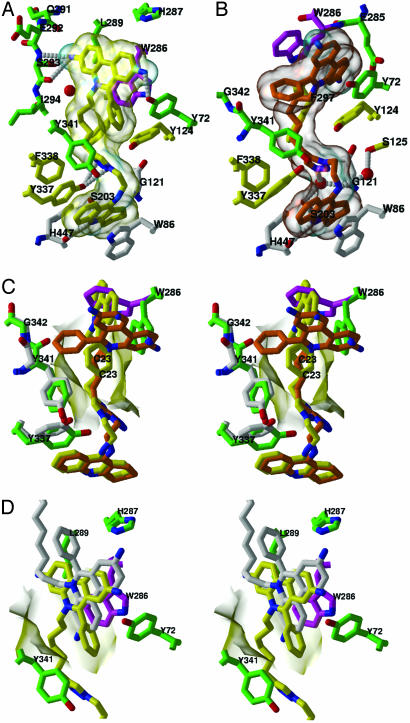
Fig. 2.
Close-up views of the TZ2PA6-mAChE complexes. (A and B) Bound anti1 and syn1 isomers (colored as in Fig. 1) with interacting mAChE side chains colored white, yellow, and green/magenta (blue, nitrogens; red, oxygens) for those that respectively interact with the tacrine, triazole, and phenanthridinium moieties of the isomers. The isomer molecular surfaces are displayed in transparency. The side chains of the catalytic residues Ser-203, Glu-334, and His-447 are shown as white bonds, and hydrogen bonds between mAChE residues and the isomers are shown as white dotted lines. (C) Stereo superimposition of the anti1 and syn1 complexes (colored as in A and B) according to all Cα atoms of mAChE. The side chains of residues Trp-286 and Tyr-337 and of dipeptide Tyr-341-Gly-342, which adopt distinctive positions in the complexes, are shown in magenta and green, respectively. The χ values for the Trp-286 side chain are (anti χ1 =-73°, χ2 = 100°; syn χ1 =-158°, χ2 = 50°). (D) Stereo superimposition of the anti1 complex with the decidium-mAChE complex (Protein Data Bank ID code 1J07; ref. 10) according to all Cα atoms of mAChE in the two complexes. The anti1 and decidium phenylphenanthridinium moieties (yellow and white bonds, respectively; blue, nitrogens) adopt distinct positions and orientations relative to Trp-286 in the PAS, whereas their alkyl chains diverge. The side chains of the PAS residues are highlighted in green (blue, nitrogens; red, oxygens). The mAChE molecular surface buried at the anti1 complex interface is displayed in transparency.
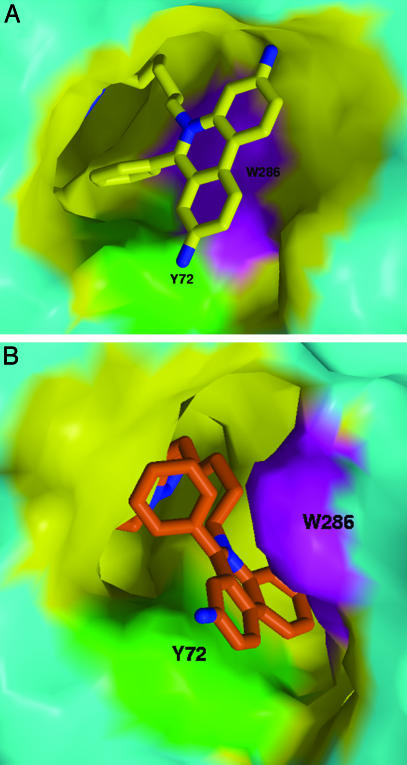
Fig. 3.
Distinctive topographies of the PAS regions in the anti1 and syn1 complexes. Views of the PAS region of mAChE bound to the phenanthridinium moiety present in the anti1 (A) and syn1 (B) isomers (colored as in Figs. 1 and 2). The mAChE molecular surfaces buried at the complex interfaces are shown in yellow, with the Tyr-72 and Trp-286 side chains highlighted in green and magenta, respectively. The mAChE surface areas (Connolly's surfaces) buried to a 1.6-Å radius probe at the anti1 and syn1 complex interfaces by the phenylphenanthridinium and linker first carbon are 256 and 313 Å2 (the double-face burying of the syn1 phenanthridinium being counterbalanced by the deeper burying of the anti1 phenyl). The gorge mouth openings (Richards' surface) for the anti1 and syn1 complexes are 14 and 29 Å2, respectively.
Inhibition Studies. Inhibition constants were measured from the ratio of dissociation and association rates ascertained by conventional mixing and stopped-flow instrumentation (7). The mAChE Trp286Ala mutant was expressed, sequence verified, and concentrated from the expression medium as described (21). Purified AChE from D. melanogaster was a gift from D. Fournier (Institut de Pharmacologie et de Biologie Structurale, Toulouse, France).
Results and Discussion
Overall View of the TZ2PA6-mAChE Complexes. The structures of mAChE in complexes with the anti1 and syn1 regioisomers (Fig. 1) show the canonical catalytic subunit, made up of a 12-stranded central β-sheet surrounded by 14 α-helices (10, 22, 23), and well-ordered bound inhibitor molecules. The isomer-binding site may be deconstructed into three discrete loci: (i) the active center at the base of the gorge that binds the tacrine moiety; (ii) an intervening site in the constricted region within the gorge that associates with the triazole moiety and adjacent methylene groups; and (iii) the PAS at the rim of the gorge that binds the phenylphenanthridinium moiety.
The anti1-mAChE Complex. In the anti1 complex, the tacrine moiety is positioned at the base of the mAChE gorge similar to the T. californica AChE-tacrine complex (11) (Fig. 2 A). However, the density maps clearly reveal a slight bend in the moiety that may enhance π-π stacking of the tetrahydroaminoacridine ring inserted between the Trp-86 and Tyr-337 aromatic side chains. At the region of constriction formed by the side chains of Tyr-124, Phe-297, Tyr-337, and Phe-338, ≈5-8 Å into the gorge, the triazole establishes van der Waals contacts with the Phe-297 and Tyr-341 side chains. The hydroxyl groups of Tyr-337 and Tyr-124, on opposite sides of the gorge, are hydrogenbonded to the triazole N2 and N3 atoms and may interact with the heteroaromatic π-system. The dimethylene linker connecting the tacrine and triazole is well ordered within the gorge and in van der Waals contact with the side chains of the conserved residues Asp-74, Tyr-124, and Tyr-341.
At the PAS, the phenylphenanthridinium moiety is positioned by the hexamethylene chain that links it to the triazole. Major interactions include a near-parallel stacking of the planar phenanthridinium with the Trp-286 indole, an edge-to-face arrangement with the Tyr-72 ring, and stabilizing interactions with the Ser-293 hydoxyl and Gln-291 carbonyl (Figs. 2 A and 3A). The anti1 exocyclic phenyl moiety, nearly buried at the gorge entrance and in a T shaped arrangement with the Tyr-72 and Trp-286 side chains, orients to establish van der Waals contacts with the Asp-74, Tyr-124, and Tyr-341 side chains and hydrogen bonds with the Tyr-72 hydroxyl and Ser-293 N, O, and Oγ atoms. Partial delocalization of electrons between the Trp-286 indole and the phenanthridinium observed in the density maps suggests formation of a charge-transfer complex, similar to that observed in the decidium-mAChE complex (10) (Fig. 2D).
The architecture of the PAS region in the anti1-mAChE complex (Figs. 2 A and 3A) is virtually identical to that seen in the decidium- and propidium-mAChE complexes (10), and the major interactions involved in the phenanthridinium-Trp-286 stacking are retained (Fig. 2D). Yet the phenylphenanthridinium adopts distinctive orientations in the PAS in the three complexes. In the anti1 complex, the phenanthridinium is rotated by ≈180° around its centroid axis, whereas this centroid is translated by 1.5 Å from its position in the other two complexes. This positioning leads to stabilizing interactions of the anti1 phenanthridinium with the Gln-291 and Ser-293 side chains, instead of the His-287 imidazole located across the gorge opening, and to a distinct environment for the exocyclic phenyl group. This versatility in the rotational and translational orientations of the bound phenylphenanthridinium relative to the Trp-286 side chain adds a new dimension to the design of molecules that associate with the PAS region.
The syn1-mAChE Complex. The structure of the syn1-mAChE complex differs considerably from that of the anti1-mAChE complex, due to the respective 1,5- and 1,4-disubstitution of the syn1 and anti1 1,2,3-triazole rings (Scheme 1, Fig. 1). The anti1 isomer adopts an elongated flat shape, whereas the syn1 isomer presents a corkscrew-like topology that provides greater surface complementarity with the gorge walls, resulting in fewer solvent-filled voids around the bound syn1 (Figs. 2 B and C and 3B). Whereas the tacrine moiety in both complexes is positioned similarly within the active site, the triazole in the syn1 complex is shifted 2 Å deeper into the gorge, where it is held in place by the tacrine ring and the Phe-297 and Phe-338 side chains. Consequently, a single π-aromatic interaction may exist between the syn1 triazole and Tyr-124 hydroxyl, and a water-bridged hydrogen bond is created between the triazole N3 and the catalytic Ser-203 Oγ and Gly-121 amide backbone. Additional van der Waals contacts occur between the triazole and the Gly-121-Gly-122 dipeptide backbone and Tyr-124 side chain. The shifted syn1 triazole occludes the Tyr-337 ring, which sweeps out in a 60° arc, resulting in a hydroxyl group displacement of 4.5 Å and in new interactions with the Tyr-341 ring near the gorge entry.
The position and orientation of the syn1 triazole at the gorge constriction influence the phenylphenanthridinium position at the PAS (Figs. 2 B and C and 3B). Compared to the anti1 complex, the overall span of the linker is shortened by 1.5 Å in the more compact syn1 triazole, a value close to the length of a C-C bond (Fig. 1 B and C). As a result of the reduced linking distance, the syn1 phenanthridinium moiety is constrained to a narrow region deep within the gorge where it would sterically clash with the Trp-286 indole. This causes the Trp-286 side chain to be dislodged from the PAS surface and to swing into the solvent, with differences of 85° and 50° in the χ1 and χ2 values, respectively. This distinct side chain conformation enlarges the opening at the gorge rim 2-fold, creating a 10 × 9 Å groove delimited by the Tyr-72 and uniquely positioned Trp-286 side chains on each side and by Glu-285 at its base, wherein the phenanthridinium tightly intercalates (Figs. 2 B and C and 3B). Hence, the aromatic plate of the syn1 phenanthridinium, oriented 90° from its position in the anti1 complex, is wedged neatly between the two aromatic side chains where it is stabilized by π-π interactions with centroids separated by 3.6-3.8 Å. This causes the phenyl group to become solvent exposed and establish discrete van der Waals contacts with residues Leu-76, Tyr-341, and Gly-342 at the gorge rim.
Kinetic Analysis of syn1 and anti1 Isomer Binding. Comparison of the relative rates of association and dissociation shows that the enhanced affinity of the syn1 isomer for mAChE largely results from a slower dissociation rate (Table 2). The small differences seen in the association rates would be anticipated in view of the rates being at or near the diffusion rate limit. Hence the enhanced affinity of the syn1 isomer is reflected in a greater activation barrier for dissociation of the complex. Several conformational changes and positional rearrangements of side chains observed within the PAS and active-site gorge correlate with the syn1 isomer being more tightly sequestered in the active site gorge.
The syn1 isomer displays up to 5-fold greater affinity for E. electricus and T. californica AChEs than for mAChE, and the selectivity of E. electricus AChE for the syn1 isomer is as much as 300-fold higher than that of any other AChE species assayed (Table 2). Only low-resolution structures of E. electricus AChE are available (24), but the limited sequence differences found at the PAS and within the gorge do not reveal particular determinants responsible for the higher selectivity. In contrast, D. melanogaster AChE, with its Glu, Tyr, and Met substitutions for mAChE residues Tyr-72, Asp-74, and Tyr-124, and mouse butyrylcholinesterase, with its Arg and Asn substitutions for Trp-286 and Tyr-72 (see http://bioweb.ensam.inra.fr/ESTHER/general?what=index), show an inverted but small selectivity for the anti1 isomer, consistent with a more open gorge and fewer aromatic side chains at the PAS in these enzymes. Moreover, removal of the indole ring at the rim of the mAChE gorge by a Trp286Ala mutation results in higher dissociation rates and up to 20-fold reduction in affinity for the syn1 and anti1 complexes. These results further emphasize the importance of the peripheral aromatic side chains in trapping the syn1 phenanthridinium and of Trp-286 in stabilizing both complexes despite the distinctive orientations of the indole side chain.
Functional Role of the Central Triazole Moiety. The triazole moieties of the syn1- and anti1-mAChE complexes are both tightly bound within the gorge, due to rearrangements in both ligand position and mAChE conformation (Fig. 2). Hence, the triazoles actively contribute to the binding interactions in the respective complexes instead of acting as passive linkers. The high affinities of the complexes (Table 2) arise not only from proximity reducing entropic contributions for a “divalent” ligand with separate binding groups but also from interactions along the gorge wall. The observation that the alternate synthesis product TZ6PA2 with inverted dimethylene and hexamethylene linkers is a weak AChE inhibitor (7) provides additional evidence for specific position-sensitive triazole contributions to the overall binding energy. The formative adaptability of this five-membered heterocycle is reflected in syn1 cycloaddition occurring with the azide and acetylene extended in a parallel orientation, effectively shortening the intervening linker through the triazole by one bond length, whereas they would lie antiparallel in formation of the anti1 triazole product (7). The triazole's large dipole (>5 Debye), which bisects the ring plane near atoms N3 and C5 (Scheme 1, Fig. 1 B and C), and the capacity of the N2 and N3 electron lone pairs to serve as hydrogen bond acceptors not only enhance binding affinity but also contribute to the efficiency of the 1,3-dipolar-coupling reaction. The conformation of the syn1 complex is congruent with syn1's shorter linker, compared to the anti1 complex. This is evident from the positioning of the phenanthridinium at the PAS, where it resides closer to the tetrahydroaminoacridine in the syn1 than in the anti1 complex (Fig. 1 B and C, Fig. 2C). To achieve this disposition of proximal reactants, a substantial conformational change occurs at the rim of the gorge with a secondary change evident in the vicinity of the triazole formed by the cycloaddition (Figs. 2C and 3). Hence, the reactivity of the azide and acetylene precursors in the gorge and the resulting affinity of the triazole formed could not have been predicted from structures of apo-AChE (10) or of complexes with close congeners to the precursors (10, 11). To date, inhibitors containing substituted 1,2,3-triazoles have been observed only in plant glycolate oxidase inhibitors (25) and β-lactam antimicrobials (26).
Functional Role of the Trp-286 Indole. Most AChE crystal structures show the Trp-286 side chain centered in the PAS with one face of the indole buried at the rim of the gorge and the other occluded by either a symmetry-related AChE molecule or a bound PAS ligand (ref. 3, and subsequent structures derived from the same TcAChE crystal form; refs. 10, 11, 22, 23, 27, and 28). Only the apo-mAChE structure (10), solved from the crystal form used here, shows the Trp-286 external face fully accessible to solvent, providing it with potential mobility. Indeed, binding of the syn1 isomer displaces the Trp-286 side chain from the AChE hydrophobic core toward the solvent, with one face of the indole now stacked with the phenanthridinium and the other face still exposed to the solvent and in van der Waals contact with Leu-289 (Figs. 2C and 3). One of the phenanthridinium rings is buried in a near-parallel stacking interaction with the Tyr-124 ring at the bottom of the groove, whereas the other two rings insert into the Trp-286-Tyr-72 parallel sandwich. The buried phenanthridinium amino group is hydrogen bonded to Glu-285. Whereas the phenanthridinium ring system in the bound anti1 isomer is virtually planar, the syn1 system adopts a slightly curved shape conforming to the induced groove where it intercalates.
Indirect evidence suggests that the PAS region, with its surface-exposed aromatic groups, may be responsible for the adhesion properties of AChE (29). Recognition and adhesion play important roles in synapse maintenance, as seen with the structurally related neuroligins (30). They may also play a role for the AChE-promoted nucleation of amyloid peptides in the Alzheimer's disease pathogenesis (31). The flip in the Trp-286 indole and the newfound π-π and cation-π interactions seen in the syn complex raise the possibility that an open conformation of AChE with distinctive exposure of aromatic groups is involved in these adhesion functions. π-orbital stacking mediated by aromatic side chains is instrumental in other molecular recognition and cell adhesion processes (32). Other intercalations of polycylic aromatic compounds between protein aromatic side chains or nucleotide bases involve the isoalloxazine moiety of the flavin cofactor and flavodoxin (33); ethidium derivatives and the multidrug-resistant-binding protein QacR (34) and adjacent base pairs (see Nucleic Acid Database, code 1JTY); the benzothiazole moiety of thioflavin and DNA (35) and β-sheet amyloid structures (36); and perhaps thioflavin and the PAS of AChE (37).
Role of the AChE Gorge Flexibility in Catalysis and Inhibitor Binding. Controversy surrounds how AChE, with its catalytic triad at the base of a narrow gorge, sustains high catalytic efficiency. Alternate portals for substrate and product access have been proposed (38); however, catalytic and inhibitor-binding parameters are influenced only by mutations in the gorge (21) and not in the vicinity of the putative additional portals (39). Rapid fluctuations giving rise to transient enlargements of the gorge appear critical (40). Ligand binding evidently induces a closed gorge state, whereas the unliganded enzyme seems to fluctuate rapidly between multiple states with varying degrees of gorge openness (41, 42).
Previous structural analysis of PAS ligands associated with mAChE showed that the tips of the long Ω loop Cys-69-Cys-96 and loop Val-340-Gly-342 bordering on the gorge (Fig. 1 A) possessed sufficient mobility to enlarge the gorge entry, thereby facilitating access to and from the active center (10). In fact, only the syn1-mAChE complex exhibits an increase of up to 12 Å in the mean temperature factors for residues at the loop tips where movement of 1.2 Å of Leu-76 and an inversion of the Gly-342 carbonyl carbon occur; the weak electron densities for the Leu-76 and Tyr-341 side chains are also consistent with substantial localized fluctuations. Hence, the flexibility of the AChE long Ω loop differs from the hinge-like motion of a homologous loop that, in the structurally related lipases, forms a rigid flap and opens only in the presence of the lipid substrate (43, 44). Moreover, ligand binding to AChE may cause the gorge to collapse around the ligand, minimizing internal dimensions (41, 42). This notion is supported by the observed repositioning of the Tyr-337 side chain and associated perturbation of Tyr-341, which not only alter the gorge shape but also enlarge its width at the position of constriction to accommodate the syn1 triazole (Fig. 2C). Such large conformational changes involving these residues were not observed for the PAS or active center complexes from which the precursor reactants were designed (10, 11). These computational and experimental results point to concerted fluctuations all along the gorge, which may facilitate access of incoming substrate to the active site at the gorge base and presumably occur in short time frames relative to diffusional translation of substrate (40).
In summary, the use of the enzyme active-site gorge as an atomic-scale template for inhibitor synthesis (7) and of structural analyses of the anti1- and syn1-mAChE complexes has revealed (i)an in situ phemonenon (7) that bears an uncanny resemblance to pioneering studies begun in 1983 by W. L. Mock (45); (ii) inherent flexibility and conformational fluctuations in the AChE molecule; and (iii) a most stable and selective complex that could not have been predicted from the apo-enzyme structure (10). The highly exergonic nature of the 1,3-dipolar-[2 + 3]-cycloaddition (ΔH >50 kcal/mol) has allowed us to immobilize and then identify by structural means an otherwise minor abundance conformation of the enzyme. Because only the higher-affinity syn1-triazole regioisomer is associated with major changes in enzyme conformation, the crystalline syn1-mAChE complex becomes the lead template in the design of selective pharmacologic agents directed toward the catalytic or noncatalytic functions of AChE. If, in fact, AChE through its PAS plays a role in synaptic adhesion processes and in nucleating plaque formation associated with dementia (29), then AChE inhibitors that also influence surface conformation may offer a means of enhancing therapeutic efficacy.
Acknowledgments
We are grateful to D. Fournier for providing the D. melanogaster enzyme, L. Green for synthesis of the regioisomers; F. Grynszpan, M. G. Finn, and W. G. Lewis for discussion; M. Juin for assistance in crystallogenesis; and G. Sulzenbacher, M. Czjzek, and the ID14 staff of the European Synchrotron Radiation Facility for expert assistance in data collection. This work was supported by grants from the Association Française contre les Myopathies (to P.M.); U.S. Public Health Service (R37-GM18360) and Department of Army Medical Defense (17-1-8014) (to P.T.); and National Institutes of Health (GM 28384), National Science Foundation (CHE-9985553), National Institute of General Medical Sciences, and the W. M. Keck Foundation (to K.B.S.).
Abbreviations: AChE, acetylcholinesterase; mAChE, mouse AChE; PAS, peripheral anionic site.
Data deposition: The atomic coordinates and structure factors of the anti1- and syn1-mAChE complex structures have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1Q84 and 1Q83, respectively)
References
- 1.Argyl-Robertson, D. (1863) Edinb. Med. J. 8, 815-820. [Google Scholar]
- 2.Dale, H. H. (1914) J. Pharmacol. Exp. Ther. 6, 147-190. [Google Scholar]
- 3.Sussman, J. L., Harel, M., Frolow, F., Oefner, C., Goldman, A., Toker, L. & Silman, I. (1991) Science 253, 872-879. [DOI] [PubMed] [Google Scholar]
- 4.Changeux, J.-P. (1966) Mol. Pharmacol. 2, 369-392. [PubMed] [Google Scholar]
- 5.Taylor, P. & Lappi, S. (1975) Biochemistry 14, 1989-1997. [DOI] [PubMed] [Google Scholar]
- 6.Taylor, P. (2001) in Goodman and Gilman's Pharmacological Basis of Therapeutics (eds. Hardman, J. G. & Limbird, L. E.), 10th Ed., pp. 175-192.
- 7.Lewis, W. G., Green, L. G., Grynszpan, F., Radić, Z., Carlier, P. R., Taylor, P., Finn, M. G. & Sharpless, K. B. (2002) Angew. Chem. Int. Ed. 41, 1053-1057. [DOI] [PubMed] [Google Scholar]
- 8.Rachinsky, T. L., Camp, S., Li, Y., Ekström, J., Newton, M. & Taylor, P. (1990) Neuron 5, 317-327. [DOI] [PubMed] [Google Scholar]
- 9.Marchot, P., Ravelli, R. B. G., Raves, M. L., Bourne, Y., Vellom, D. C., Kanter, J., Camp, S., Sussman, J. L. & Taylor, P. (1996) Protein Sci. 5, 672-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourne, Y., Taylor, P., Radić, Z. & Marchot, P. (2003) EMBO J. 22, 1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harel, M., Schalk, I., Ehret-Sabatier, L., Bouet, F., Goeldner, M., Hirth, C., Axelsen, P. H., Silman, I. & Sussman, J. L. (1993) Proc. Natl. Acad. Sci. USA 90, 9031-9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 13.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 14.Brünger, A. T., Adams, P. D., Marius Clore, G., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-909. [DOI] [PubMed] [Google Scholar]
- 15.Murshudov, G., Vagin, A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 16.Roussel, A. & Cambillau, C. (1989) in Silicon Graphics Geometry Partners Directory, eds. Silicon Graphics Committee (Silicon Graphics, Mountain View, CA), pp. 77-78.
- 17.Mohamadi, F., Richards, N. G. J., Guida, W. C., Liskamp, R., Lipton, M., Caufield, C., Chang, G., Hendrickson, T. & Still, W. C. (1990) J. Comput. Chem. 11, 440-467. [Google Scholar]
- 18.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 19.Christopher, J. A. (1998) spock (Center for Macromolecular Design, Texas A&M University, College Station, TX).
- 20.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505-524. [DOI] [PubMed] [Google Scholar]
- 21.Radić, Z., Pickering, N. A., Vellom, D. C., Camp, S. & Taylor, P. (1993) Biochemistry 32, 12074-12084. [DOI] [PubMed] [Google Scholar]
- 22.Bourne, Y., Taylor, P. & Marchot, P. (1995) Cell 83, 503-512. [DOI] [PubMed] [Google Scholar]
- 23.Bourne, Y., Taylor, P., Bougis, P. E. & Marchot, P. (1999) J. Biol. Chem. 274, 2963-2970. [DOI] [PubMed] [Google Scholar]
- 24.Bourne, Y., Grassi, J., Bougis, P. E. & Marchot, P. (1999) J. Biol. Chem. 274, 30370-30376. [DOI] [PubMed] [Google Scholar]
- 25.Stenberg, K. & Lindqvist, Y. (1997) Protein Sci. 6, 1009-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuzin, A. P., Nukaga, M., Nukaga, Y., Hujer, A. M., Bonomo, R. A. & Knox, J. R. (2001) Biochemistry 40, 1861-1866. [DOI] [PubMed] [Google Scholar]
- 27.Harel, M., Kleywegt, G. J., Ravelli, R. B., Silman, I. & Sussman, J. L. (1995) Structure (London) 3, 1355-1366. [DOI] [PubMed] [Google Scholar]
- 28.Kryger, G., Silman, I. & Sussman, J. L. (1999) Acta Crystallogr. D 56, 1385-1394. [DOI] [PubMed] [Google Scholar]
- 29.Soreq, H. & Seidman, S. (2001) Nat. Rev. Neurosci. 2, 294-302. [DOI] [PubMed] [Google Scholar]
- 30.Ichtchenko, K., Nguyen, T. & Sudhof, T. C. (1996) J. Biol. Chem. 271, 2676-2682. [DOI] [PubMed] [Google Scholar]
- 31.Inestrosa, N. C., Alvarez, A., Perez, C. A., Moreno, R. D., Vicente, M., Linker, C., Casanueva, O. I., Soto, C. & Garrido, J. (1996) Neuron 16, 881-891. [DOI] [PubMed] [Google Scholar]
- 32.Zacharias, N. & Dougherty, D. A. (2002) Trends Pharmacol. Sci. 23, 281-287. [DOI] [PubMed] [Google Scholar]
- 33.Genzor, C. G., Perales-Alcon, A., Sancho, J. & Romero, A. (1996) Nat. Struct. Biol. 3, 329-332. [DOI] [PubMed] [Google Scholar]
- 34.Schumacher, M. A., Miller, M. C., Grkovic, S., Brown, M. H., Skurray, R. A. & Brennan, R. G. (2001) Science 294, 2158-2163. [DOI] [PubMed] [Google Scholar]
- 35.Canete, M., Villanueva, A., Juarranz, A. & Stockert, J. C. (1987) Cell. Mol. Biol. 33, 191-199. [PubMed] [Google Scholar]
- 36.LeVine, H., III (1999) Methods Enzymol. 309, 274-284. [DOI] [PubMed] [Google Scholar]
- 37.De Ferrari, G. V., Mallender, W. D., Inestrosa, N. C. & Rosenberry, T. L. (2001) J. Biol. Chem. 276, 23282-23287. [DOI] [PubMed] [Google Scholar]
- 38.Gilson, M. K., Straatsma, T. P., McCammon, J. A., Ripoll, D. R., Faerman, C. H., Axelsen, P. H., Silman, I. & Sussman, J. L. (1994) Science 263, 1276-1278. [DOI] [PubMed] [Google Scholar]
- 39.Kronman, C., Ordentlich, A., Barak, D., Velan, B. & Shafferman, A. (1994) J. Biol. Chem. 269, 27819-27822. [PubMed] [Google Scholar]
- 40.Shen, T., Tai, K., Henchman, R. H. & McCammon, J. A. (2002) Acc. Chem. Res. 35, 332-340. [DOI] [PubMed] [Google Scholar]
- 41.Shi, J., Radić, Z. & Taylor, P. (2002) J. Biol. Chem. 277, 43301-43308. [DOI] [PubMed] [Google Scholar]
- 42.Shi, J., Tai, K., McCammon, A. J., Taylor, P. & Johnson, D. A. (2003) J. Biol. Chem. 278, 30905-30911. [DOI] [PubMed] [Google Scholar]
- 43.Cygler, M., Schrag, J., Sussman, J. L., Harel, M., Silman, I., Gentry, M. K. & Doctor, B. P. (1993) Protein Sci. 2, 366-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grochulski, P., Li, Y., Schrag, J. D. & Cygler, M. (1994) Protein Sci. 3, 82-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mock, W.L. (1995) Top. Curr. Chem. 175, 1-24. [Google Scholar]