Abstract
Burkholderia cepacia complex (BCC) bacteria can cause devastating chronic infections in people with cystic fibrosis. Of particular concern is “cepacia syndrome,” a rapidly progressive and usually fatal decline in health, characterized by a necrotizing bacteremic pneumonia. An important component of defense against bloodstream infections is the bactericidal action of serum. Traditional methods to determine the capacity of bacterial isolates to resist the bactericidal effects of serum are relatively low-throughput viability assays. In this study, we developed a novel growth-based assay for serum susceptibility, which allows for high throughput analysis. We applied this assay to a range of clinical isolates of BCC as well as isolates comprising the BCC experimental strain panel. Our data demonstrate that isolates from all species of BCC examined can possess serum resistant or serum sensitive/intermediate phenotypes. Of particular clinical significance, we also found no direct link between the last saved pulmonary isolate from patients who subsequently developed “cepacia syndrome” and their capacity to resist the inhibitory effects of human serum, suggesting serum resistance cannot be used as a marker of an isolate’s capacity to escape from the lung and cause bacteremia.
Keywords: Burkholderia cepacia complex, serum, cystic fibrosis, complement, Bioscreen, cepacia syndrome
Introduction
Bacteria belonging to the Burkholderia cepacia complex (BCC) can cause devastating pulmonary infections in people who have cystic fibrosis (CF). The BCC is composed of at least 17 distinct species, all of which, except B. ubonensis, have been isolated from the lungs of people with CF (Mahenthiralingam et al., 2008). Infection with BCC bacteria can be a frightening prospect for people with CF because these bacteria are known to cause devastating clinical deterioration in a subset of patients, and it is not possible to determine a priori which are at risk. Individuals with BCC respiratory infection can show either no substantive change in pulmonary function, a gradual decline in health or, worst of all, an invasive, and rapidly fatal septicemia known as “cepacia syndrome” (Isles et al., 1984; Glass and Govan, 1986). The reasons for these disparate outcomes are not understood; thus accurate prognoses for CF patients infected with BCC are extremely difficult to define. While a number of different BCC species have been associated with “cepacia syndrome” (Zahariadis et al., 2003; Nash et al., 2011), historically infections caused by Burkholderia cenocepacia have been associated with a higher risk of developing “cepacia syndrome” than the other species (Mahenthiralingam et al., 2001; Jones et al., 2004). A further complication of infection with B. cenocepacia appears to be a higher risk of death following lung transplantation, often as a result of sepsis (Alexander et al., 2008; De Soyza et al., 2010).
Serum-mediated bactericidal activity is an especially important innate immune defense against intravascular invasion of bacterial pathogens and is thought to occur as a result of membrane attack complexes (MAC) that are assembled from terminal complement components in the bacterial cell envelope (Frank et al., 1987; Walport, 2001). Serum resistance of bacteria occurs as a result of the failure of complement-mediated lysis. Most isolates of Pseudomonas aeruginosa from chronically infected CF patients are serum-susceptible; this is thought to explain in part why P. aeruginosa bacteremia/septicemia is essentially never seen in CF (Hoiby and Olling, 1977; Thomassen and Demko, 1981; Speert et al., 1990). In the context of BCC, serum sensitivity/resistance might be an important microbial phenotype, which could conceivably differentiate between invasive and non-invasive strains and isolates. This could be especially important in the context of “cepacia syndrome,” where the invading strain would have to be able to resist the effect of complement either through intrinsic resistance to the formation of the MAC in the bacterial membrane, or by interfering with the complement cascade that leads to the formation of the MAC.
Serum sensitivity and serum resistance phenotypes have both been described previously in BCC isolates from patients with CF; the isolates studied had rough lipopolysaccharide (LPS) rendering them serum sensitive (Butler et al., 1994). The mechanistic basis of serum resistance in BCC is not fully understood; however, consistent with the observations of Butler et al. possession of a complete LPS has been shown to confer resistance to serum (Ortega et al., 2005). Another type of polysaccharide – secreted exopolysaccharides – may also be significant in serum resistance as a recent study on a small number of B. cenocepacia isolates suggested that the “mucus layer” may also contribute to resistance to the bactericidal effects of serum (Savoia et al., 2008). Indeed, BCC are capable of synthesizing at least five different exopolysaccharides (Cescutti et al., 2010). In characterizing a pair of isogenic clonal non-mucoid/mucoid clinical isolates, C8963 (non-mucoid) and C9343 (mucoid), we noticed that the non-mucoid C8963 was serum resistant in spite of its rough LPS, while the mucoid C9343 was serum sensitive (Conway et al., 2004). However, these studies were either performed before BCC had been reclassified into species (Vandamme et al., 1997) or before mucoidy was recognized as a common phenotypic feature of the complex (Cunha et al., 2004; Zlosnik et al., 2008).
The purpose of this study was to develop a higher throughput and automated technique for assessing the capacity of human serum to inhibit the growth of BCC isolates in order to facilitate an improved understanding of mechanisms of virulence in these invasive CF pathogens. Additionally, we applied this technique to determine the serum resistance phenotypes of a range of clinical isolates and representative isolates comprising the BCC experimental strain panel (Mahenthiralingam et al., 2000). These data show that both serum resistant and serum sensitive/intermediate phenotypes can be found in isolates of each of the five species examined. Furthermore, we examined a range of clinical isolates of BCC from patients who died from “cepacia syndrome” and were unable to find a link between the serum resistance phenotype of the last saved pulmonary isolate and subsequent progression to “cepacia syndrome”.
Materials and Methods
Source of bacterial isolates and human serum
Clinical isolates of BCC bacteria were taken from the Canadian BCC research and referral repository (CBCCRRR; Speert et al., 2002). Serum resistant (B. cenocepacia K56-2, and C8963 and P. aeruginosa M2) and serum sensitive (B. cenocepacia J2315, C9343, and P. aeruginosa P1) have been previously described (Dasgupta et al., 1994; Conway et al., 2004; Ortega et al., 2005). Bacteria were grown from cultures stored at −80°C on to Columbia Blood Agar plates for 24–48 h at 37°C and plates were passaged no more than three further times before reverting to the freezer stock. Complement preserved pooled normal human serum (PNHS; Innovative Research, MI, USA) was used in all experiments. For heat inactivated serum assays, PNHS was heated to 65°C for 30 min.
Assessment of bacterial viability after treatment with serum
The viability of bacteria following treatment with serum was determined by a modification of a previously described assay (Dasgupta et al., 1994). Briefly, isolates were grown overnight in LB broth (10 g l−1 tryptone, 5 g l−1 yeast extract, and 10 g l−1 sodium chloride) at 37°C on an orbital rotator, then 1 ml of overnight culture was pelleted at 3,000 rpm (800 × g) for 5 min in a Jouan A14 microcentrifuge, resuspended in Hanks Balanced Salt Solution (GIBCO) with 0.1% gelatin and adjusted to 1 × 108 colony forming units (CFU) ml−1. 106 CFU were then transferred to a 5-ml polypropylene tube and incubated with 10% serum in a final volume of 3 ml for 180 min at 37°C with tumbling. CFU were determined at 0 and 180 min by serial dilution and culture; serum sensitive isolates were defined by a ≥90% reduction in viable count between 0 and 180 min, while serum resistant isolates were classified by <20% loss in viability. Between 20 and 90% loss in viability was classified as indeterminate. For direct comparison with the automated growth assessment, serum resistance was also assessed using the above method, except using an inoculum of 1 × 107 CFU ml−1, and with CFUs determined at 0 and 720 min. All isolates were characterized in experiments on at least three separate days.
Automated growth assessment of serum susceptibility
Overnight cultures were grown in LB broth and then pelleted at 3,000 rpm (800 × g) for 10 min in a Juan A14 microcentrifuge. The cell pellets were resuspended in 1 ml LB broth and adjusted to an OD600 nm of approximately 0.1 (equivalent to 1.0 × 108 CFU/ml) using fresh LB broth. Thirty microliters of the adjusted overnight culture was then inoculated into 270 μl of LB broth with 11% pooled human serum (resulting in a final inoculum of 107 CFU ml−1 in 10% serum) in a Bioscreen 100 well honeycomb plate. An equal number of control wells, without serum, were set up for each isolate in all experiments performed, as were blank wells containing only growth media (with or without serum). Growth at 37°C was then monitored in a Bioscreen C machine (TYPE FP-1100-C, Oy Growth Curves Ab Ltd.) with measurement intervals of 15 min and continuous shaking at medium amplitude, controlled by EZ experiment software.
Data processing
Growth of all isolates was assessed at each time point by subtracting the value for the media blank from the mean OD600 nm of experimental replicates. The serum resistance phenotype for each isolate was then determined by calculating the average percent increase in OD in the wells containing serum compared to wells without serum at 12 h. For the growth-based assay, serum sensitive isolates were considered to be those that had failed to reach 20% growth in PNHS at 12 h compared to no-serum controls. Serum resistant isolates were defined as those that reached 80% or more growth in PNHS at 12 h compared to no-serum controls, while those isolates that grew between 20 and 80% in PNHS compared to no-serum controls were considered intermediate. For each isolate, replicates were performed on at least three different days. All data were processed in MS Excel.
Results
Automated growth analysis to assess serum resistance
The traditional method to determine serum resistance of bacterial isolates is to expose them to human serum, in a medium that does not support growth, for several hours, and then measure viable counts to assess whether or not an isolate is susceptible or resistant (Butler et al., 1994; Ortega et al., 2005). However, although effective and direct, manual counts of viability are laborious and time consuming. Therefore, we sought to evaluate alternative methods. Initially, we assessed both a Live/Dead BacLight™ (Molecular Probes) kit assay, based on staining of live and dead cells, and the BacTiter-Glo™ (Promega) cell viability assay based on the detection of ATP as an indicator of cell viability. Neither technique proved suitable for our needs to assess serum resistance, due to interference by components of human serum or cost of reagents (data not shown). Therefore, we investigated whether or not the inhibitory effects of human serum could be determined using an automated growth curve machine, reasoning that serum sensitive isolates would not grow or display inhibited growth in the presence of serum.
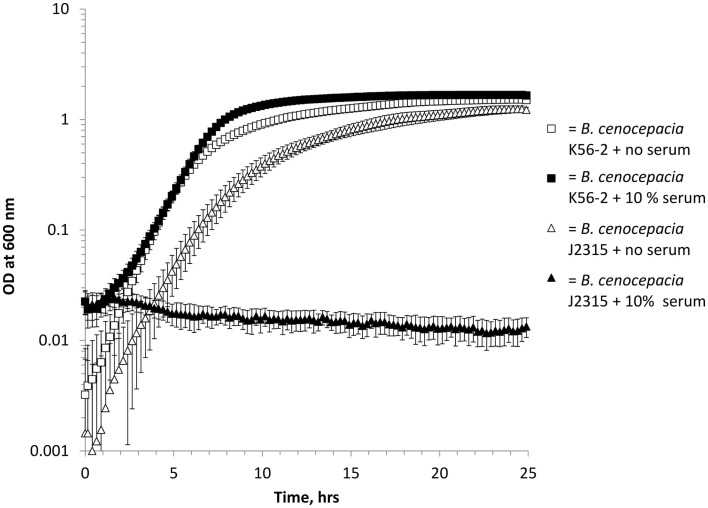
Using a Bioscreen automated growth curve machine we evaluated control isolates, with known serum resistance phenotypes, of both B. cenocepacia (Figure 1) and P. aeruginosa (data not shown). Under standard growth conditions, we found that 10% serum inhibited the growth of serum sensitive isolates (B. cenocepacia J2315 and P. aeruginosa P1) in a highly reproducible manner, while serum resistant (B. cenocepacia K56-2 and P. aeruginosa M2) isolates grew normally (Figure 1, P. aeruginosa isolates data not shown). Other studies report a range of different serum concentrations (between 5 and 40%) to assess serum susceptibility (Hancock et al., 1983; Butler et al., 1994; Ortega et al., 2005; Savoia et al., 2008). Therefore, we investigated the growth of the three control serum resistant isolates in 10, 20, and 40% pooled normal human serum (PNHS). We found that P. aeruginosa M2 isolate grew normally at all concentrations, while serum resistant B. cenocepacia K56-2 grew normally at both 10 and 20% serum but exhibited delayed growth in 40% serum (data not shown). Finally, C8963 grew normally at 10% serum, exhibited delayed growth at 20% serum and did not grow at all in 40% serum (data not shown). We elected to use 10% serum in all further experiments as all previously described serum resistant isolates grew normally at this concentration.
Figure 1.
Growth of B. cenocepacia isolates in LB with (solid shapes) and without (open shapes) 10% pooled normal human serum in a Bioscreen growth curve machine. Squares = B. cenocepacia K56-2 (serum resistant) and triangles = B. cenocepacia J2315 (serum sensitive). All experiments were performed in triplicate with three biological repeats, error bars are standard errors of the mean and where not visible are smaller than the symbols.
In order to create a quantitative measure of the extent of growth inhibition by serum, we normalized all data generated by the Bioscreen C machine, calculating the mean percentage growth of an isolate in serum compared to the mean growth of the isolate in an equal number of control wells containing the same media without serum (Figure 2). An isolate’s serum resistance phenotype was then determined by the percentage growth in PNHS compared to growth in media alone at 12 h. This time point was chosen to allow the control wells to have grown into stationary phase as well as to reduce some of the “noise” generated by the less accurate readings taken at the time of initial inoculum. Serum resistant isolates were characterized as achieving 80% or greater of the OD600 nm of the no-serum control wells at 12 h. Serum sensitive isolates were defined as measuring less than 20% of the OD increase of the no-serum control wells. Isolates that achieved between 20 and 80% of the OD of the no-serum control wells were classified as intermediate. It was notable that the sensitive control isolate P. aeruginosa P1 demonstrates regrowth at 24 h (Figure 2). To assess the prevalence of this phenomenon, we examined a larger range of BCC isolates (Table 1). We found that all isolates classified as intermediate had fully grown at 24 h. Of the strains classified as sensitive, 4/9 (BCC isolates C7376, C8482, and C4365 as well as P. aeruginosa P1) demonstrated growth recovery (to either intermediate or resistant phenotype) at 24 h, while 5/9 (J2315. C9343, C2864, C4380, and C4873) remained classified as sensitive (Table 1).
Figure 2.
Percentage growth of isolates in LB supplemented with 10% pooled normal human serum in a Bioscreen growth curve machine compared to no-serum controls grown in LB. Squares = B. cenocepacia K56-2 (serum resistant), crosses = P. aeruginosa M2 (serum resistant), diamonds = P. aeruginosa P1 (serum sensitive), and triangles = B. cenocepacia J2315 (serum sensitive). All experiments were performed in triplicate with three biological repeats, error bars are standard errors of the mean where not visible are smaller than the symbols.
Table 1.
Serum susceptibilities in B. cenocepacia cases.
| Method |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Conventional assay |
Bioscreen |
||||||||||||||
| CFU |
CFU |
||||||||||||||
| Isolate | Species | RAPD type | Clinical case | EPS | 0 h | 3 h | Serum resistance phenotype | 0 h | 12 h | Serum resistance phenotype | % Growth at 12 h (±SEM) | Serum resistance phenotype | % Growth at 24 h (±SEM) | Comments | |
| CONTROL ISOLATES | |||||||||||||||
| M2 | P. aeruginosa | n/a | n/a | n/a | 1.6 × 106 | 1.2 × 107 | R | 1.32 × 107 | >1 × 109 | R | 97.8 (3.1) | R | 105.0 (3.7) | Dasgupta et al. (1994) | |
| P1 | P. aeruginosa | n/a | n/a | n/a | 2.1 × 106 | 1.0 × 102 | S | 1.45 × 107 | <1 × 104 | S | 8.0 (3.6) | S | 64.0 (20.2) | Dasgupta et al. (1994) | |
| K56-2 | B. cenocepacia | 02 | n/a | − | 2.2 × 106 | 1.7 × 106 | R | 1.33 × 107 | 2.05 × 108 | R | 138.0 (4.8) | R | 110.1 (0.7) | Ortega et al. (2005) | |
| J2315 | B. cenocepacia | 02 | n/a | − | 9.0 × 105 | 1.4 × 102 | S | 1.46 × 107 | <1 × 104 | S | 2.7 (0.4) | S | 1.0 (0.2) | Ortega et al. (2005) | |
| C3921 | B. cenocepacia | 01 | BCC1 | +++ | 1.1 × 106 | 1.2 × 106 | R | 2.08 × 107 | 2.6 × 108 | R | 116.9 (11.2) | R | N.D. | Zlosnik et al. (2011) | |
| C3921-CTZ32G | B. cenocepacia | 01 | BCC1 | − | N.D. | N.D. | N.D. | 1.57 × 107 | 2.32 × 108 | R | 124.0 (18.7) | R | N.D. | Zlosnik et al. (2011) | |
| C8963 | B. cenocepacia | 01 | BCC1 | − | 1.9 × 105 | 1.1 × 105 | R | 1.41 × 107 | 1.13 × 105 | S | 139.3 (26.7) | R | 120.0 (11.1) | Conway et al. (2004) | |
| C9343 | B. cenocepacia | 01 | BCC1 | +++ | 5.9 × 106 | 4.7 × 105 | S | 1.87 × 107 | <1 × 104 | S | 1.7 (1.3) | S | 4.0 (2.2) | Conway et al. (2004) | |
| CEPACIA SYNDROME CASES | |||||||||||||||
| C7376 | B. cenocepacia | 02 | BCC2 | − | 1.8 × 106 | 9.2 × 104 | S | N.D. | N.D. | N.D. | 7.4 (1.0) | S | 79.0 (36.1) | Last isolate prior to death | |
| C8482 | B. cenocepacia | 02 | BCC3 | − | 2.3 × 106 | 1.7 × 105 | S | N.D. | N.D. | N.D. | 6.9 (0.06) | S | 66.7 (39.0) | Last isolate prior to death | |
| C2864 | B. cenocepacia | 02 | BCC4 | − | 2.1 × 106 | 9.0 × 102 | S | N.D. | N.D. | N.D. | 2.2 (1.3) | S | 3.1 (1.2) | First isolate | |
| C4380 | B. cenocepacia | 02 | BCC4 | − | 4.8 × 106 | 1.8 × 104 | S | N.D. | N.D. | N.D. | −0.4 (0.9) | S | 0.3 (0.7) | 1 year before last | |
| C4873 | B. cenocepacia | 02 | BCC4 | − | 1.7 × 106 | 2.5 × 103 | S | N.D. | N.D. | N.D. | 2.0 (2.6) | S | 0.3 (1.0) | Last isolate prior to death | |
| C4871 | B. cenocepacia | 01 | BCC5 | − | 3.1 × 105 | 2.5 × 105 | R | N.D. | N.D. | N.D. | 71.9 (13.2) | I | 118.3 (20.4) | Co-last isolate | |
| C4872 | B. cenocepacia | 01 | BCC5 | − | 1.7 × 106 | 5.3 × 105 | I | N.D. | N.D. | N.D. | 155.0 (19.1) | R | 197.3 (34.8) | Co-last isolate | |
| C4365 | B. cenocepacia | 02 | BCC6 | − | 1.5 × 107 | 6.7 × 104 | S | N.D. | N.D. | N.D. | 8.6 (4.4) | S | 49.0 (25.6) | First isolate | |
| C4825 | B. cenocepacia | 02 | BCC6 | − | 2.5 × 106 | 1.8 × 104 | S | N.D. | N.D. | N.D. | 34.9 (16.8) | I | 135.6 (12.4) | Last isolate prior to death | |
| C7397 | B. cenocepacia | 02 | BCC7 | − | 6.7 × 105 | 7.7 × 104 | I | N.D. | N.D. | N.D. | 31.2 (13.9) | I | 173.8 (30.0) | Last isolate | |
| C3092 | B. cenocepacia | 04 | BCC8 | − | 3.5 × 106 | 5.1 × 105 | I | N.D. | N.D. | N.D. | 157.3 (18.8) | R | 170.2 (14.0) | Last isolate | |
| C4357 | B. cenocepacia | 04 | BCC9 | + | 2.6 × 106 | 6.4 × 104 | S | N.D. | N.D. | N.D. | 54.6 (35.0) | I | 158.1 (6.5) | Last isolate | |
| C3403 | B. cenocepacia | 04 | BCC10 | ++ | 2.9 × 106 | 6.6 × 105 | I | N.D. | N.D. | N.D. | 49.1 (20.5) | I | 145.5 (11.6) | First and only isolate | |
ND, not determined; n/a, not applicable; R, resistant; I, intermediate; S, sensitive. SEM, standard error of the mean. Exopolysaccharide (EPS) production was previously scored as non-mucoid (− or +) or frankly mucoid (++, +++, +++d; Zlosnik et al., 2008). RAPD type refers to strain types that have previously been determined by RAPD typing (Mahenthiralingam et al., 1996). Clinical case numbers denote separate CF patients. Serum resistance phenotypes in the conventional assay were defined as a less than 20% drop in viability, as determined by CFU, while serum sensitivity was defined as a greater than 90% drop in viability and intermediate isolates defined as between 20 and 90% drop in CFU. For the Bioscreen assay, serum resistance was defined as 80% or greater growth in serum, sensitive isolates were defined as <20% growth and intermediate isolates as 20–80% growth in serum compared to the no-serum control. Cepacia syndrome cases are the last stored pulmonary isolates from cases of BCC infections in people with CF who subsequently developed cepacia syndrome.
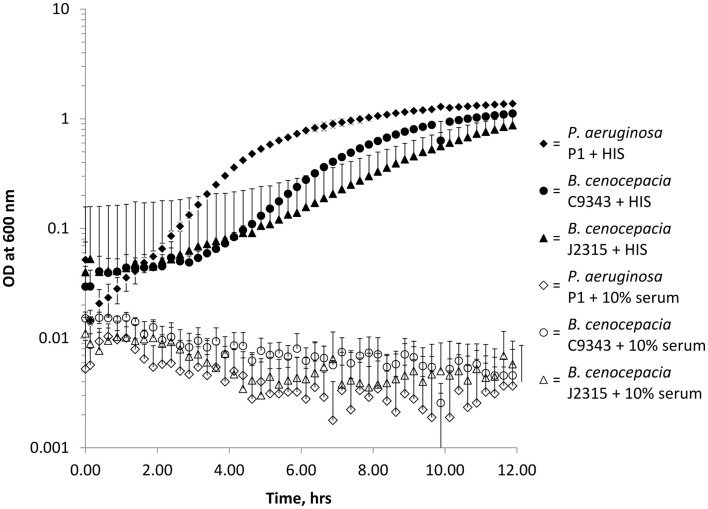
To validate the Bioscreen automated growth curve method for assessing serum resistance we conducted a number of additional experiments. Firstly, to ensure that the growth inhibition of serum sensitive isolates was due to the heat-labile fraction of PNHS, we grew the three control serum sensitive isolates in heat inactivated serum. In all cases, heat inactivation rendered PNHS incapable of inhibiting bacterial growth (Figure 3). Secondly, to demonstrate that the Bioscreen generates equivalent data to the conventional serum assay, we determined the serum killing in the conventional assay at 12 h for the control isolates used in this study. These data (Table 1) showed, with the exception of C8963, agreement with the Bioscreen method. Additionally, it is notable that these data also show that serum resistant isolates of P. aeruginosa (M2) and B. cenocepacia (K56-2, C3921, C3921-CTZ32G) grow in the presence of serum over the course of 12 h with all gaining at least a one log increase in CFU, suggesting they are growing on nutrients present in the serum. Finally, data from the Bioscreen automated growth curve method also showed good concordance with measurements of viability performed using the traditional serum sensitivity assay at 3 h with 75% (±20 at a 95% confidence interval of the mean) isolates giving identical results (Table 1).
Figure 3.
Bioscreen growth curves of serum sensitive isolates in 10% pooled normal human serum (open shapes) and heat inactivated pooled normal human serum (HIS, solid shapes). Diamonds = P. aeruginosa P1, triangles = B. cenocepacia J2315, and circles = B. cenocepacia C9343. All experiments were performed in triplicate with three biological repeats. Error bars are standard errors of the mean, where not visible are smaller than the symbols and only the upper error bars are included for clarity.
Application of the Bioscreen method to a range of BCC bacteria
To demonstrate the utility of the Bioscreen assay, we applied this technique to characterize the isolates comprising the BCC strain panel (Mahenthiralingam et al., 2000). This panel contains representative isolates for the species B. cepacia, B. multivorans, B. cenocepacia, B. stabilis, and B. vietnamiensis. These data identify serum resistant and sensitive isolates in all species except for B. cepacia, where only resistant and intermediate phenotypes were found in the four isolates representing that species (Table 2).
Table 2.
Bioscreen determined serum susceptibility profiles of representative isolates of BCC.
| Species | Isolate | % Growth at 12 h (SEM) | Serum phenotype |
|---|---|---|---|
| B. multivorans | C5393 | 82.5 (0.71) | R |
| LMG13010T | 96.1 (1.02) | R | |
| C1576 | 84.4 (1.58) | R | |
| CF-A1-1 | −0.74 (0.87) | S | |
| JTC | 87.1 (0.91) | R | |
| C1962 | 85.6 (6.82) | R | |
| ATCC17616 | −0.51 (0.47) | S | |
| 249-2 | −1.26 (0.31) | S | |
| B. cenocepacia | J2315 | See Table 1 | S |
| BC7 | 61.7 (11.7) | I | |
| K56-2 | See Table 1 | R | |
| C5424 | 108 (4.15) | R | |
| C6433 | 82.4 (2.5) | R | |
| C1394 | 1.33 (1.33) | S | |
| PC184 | −0.93 (1.24) | S | |
| CEP511 | −1.31 (0.34) | S | |
| J415 | 121 (39.7) | R | |
| ATCC1765 | 69.1 (10.3) | I | |
| B. cepacia | ATCC25416T | 82.8 (2.76) | R |
| ATCC17759 | 81.9 (1.45) | R | |
| CEP509 | 75.0 (1.81) | I | |
| LMG17997 | 61.6 (11.4) | I | |
| B. stabilis | LMG14294 | 1.84 (4.55) | S |
| C7322 | ND | ND | |
| LMG14086 | 109 (11.8) | R | |
| LMG18888 | 27.7 (5.8) | I | |
| B. vietnamiensis | PC259 | −0.64 (0.51) | S |
| LMG16232 | 89.5 (1.71) | R | |
| FC441 | 87.4 (1.41) | R | |
| LMG10929 | 123.7 (35) | R |
Serum resistance phenotypes were determined using the Bioscreen growth method. R, resistant (>80% growth in serum); S, sensitive (<20% growth in serum); and I, intermediate (20–80% growth in serum) at 12 h. ND, not determined. Experiments were performed in triplicate on three separate days.
We have previously shown that a non-mucoid clinical isolate of B. cenocepacia (C8963) is serum resistant, while a mucoid sequential clonal isolate from the same infection (C9343) was serum sensitive. This observation was repeated in both methods used in this study (Table 1). In this study, we also examined a non-mucoid isolate that we have previously derived from a mucoid parent under antibiotic pressure (Zlosnik et al., 2011). This isolate, C3921-CTZ32G, and its parent, C3921, both displayed serum resistance and therefore the absence of exopolysaccharide production capacity is not linked to the serum resistance phenotype in these isolates (Table 1). Additionally, both serum resistant and serum sensitive/intermediate phenotypes were found for mucoid and non-mucoid isolates of B. cenocepacia (Tables 1 and 3). We also examined the serum resistance phenotypes of further B. cenocepacia clinical isolates in order to have assessed a range of examples of the major epidemic clones of B. cenocepacia (Table 3). These data also suggest that there is no direct link between serum resistance and RAPD type; sensitive/intermediate and resistant isolates were found for all RAPD types examined (Tables 1 and 3).
Table 3.
Serum susceptibility phenotypes of additional B. cenocepacia clinical isolates.
| Isolate | RAPD type | % Growth at 12 h (±SEM) | Serum resistance phenotype | EPS |
|---|---|---|---|---|
| C0131 | 04 | 98.7 (2.3) | R | ++ |
| C1257 | 01 | 26.4 (5.7) | I | +++ |
| C2303 | 06 | 1.9 (2.0) | S | ++ |
| C2457 | 06 | 3.53 (2.8) | S | +++ |
| C3883 | 04 | 93.7 (3.3) | R | +++ |
| C4526 | 04 | 70.7 (26.4) | I | +++ |
| C5491 | 01 | 40.2 (16.0) | I | − |
| C5819 | 04 | 177.3 (19.8) | R | ++ |
| C6114 | 04 | 107.9 (42.0) | R | +++ |
| C6144 | 06 | 0.7 (0.5) | S | − |
| C6159 | 073a | 89.4 (6.3) | R | ++ |
| C6345 | 02 | 8.8 (4.9) | S | − |
| C6483 | 02 | 99.4 (10.9) | R | − |
| C6615 | X | 86.7 (5.6) | R | − |
| C7586 | 04 | 149.5 (3.7) | R | +++d |
| C7755 | 067 | 90.5 (1.8) | R | +++d |
| C7788 | 04 | 104.3 (29.7) | R | − |
| D0134 | X | 123.6 (17.3) | R | +++ |
| D0960 | 06 | 99.3 (3.7) | R | +++d |
| D1817 | 04 | 159.0 (3.4) | R | +++ |
| D2282 | 06 | 172.5 (1.5) | R | +++d |
Serum resistance phenotypes were determined using the Bioscreen growth method. R, resistant (>80% growth in serum); S, sensitive (<20% growth in serum); and I, intermediate (20–80% growth in serum) at 12 h. RAPD types are strain types as determined previously by RAPD PCR and exopolysaccharide (EPS) phenotypes were previously determined by growth on yeast extract mannitol media (Mahenthiralingam et al., 1996; Zlosnik et al., 2008). Experiments were performed in triplicate on three separate days. X = RAPD type not assigned but does not match a major existing RAPD type.
“Cepacia syndrome” is a necrotizing pneumonia, often involving a fatal bloodstream invasion from the pulmonary tract by the infecting BCC bacterium, and currently there are no known predictive factors, either microbial or human, of this phenomenon. We reasoned that to result in bloodstream invasion, a BCC isolate would need to be serum resistant. Therefore, we constructed a hypothesis that the last stored pulmonary isolate taken from patients that subsequently develop “cepacia syndrome” would be serum resistant to facilitate intravascular invasion. Should this prove to be the case, it could be used as a risk factor for “cepacia syndrome”. To investigate this hypothesis we retrieved the last stored pulmonary isolate from nine patients that were annotated in our database as having subsequently developed cepacia syndrome or severe pneumonia that led to death (Mahenthiralingam et al., 2001; Zlosnik et al., 2011) and assessed their resistance to serum using both a traditional serum sensitivity assay and our Bioscreen growth-based assay (Table 1). From the nine cases of cepacia syndrome that were analyzed, we found that the last stored isolate was resistant to serum in two cases, susceptible in three cases, and of intermediate resistance in four cases. In one case, BCC5, there were two isolates stored on the same day prior to death and it was notable that these displayed different serum resistance profiles with C4871 being serum intermediate and C4872 being serum resistant.
Discussion
The capacity to resist the bactericidal effects of human serum is thought to be an important adaptation in a number of bacteria capable of causing septicemia, such as Neisseria meningitidis, in otherwise healthy people (Kugelberg et al., 2008; Lambris et al., 2008). Complement activity represents one of the primary lines of defense of the human body against systemic bacterial infection. Indeed, defects in components of the complement cascade are associated with an elevated susceptibility to serious infections caused by a number of bacteria, including Haemophilus influenzae and Streptococcus pneumoniae (Walport, 2001).
Assessing serum resistance in bacterial isolates is a time consuming and low-throughput process involving enumerating CFU before and after exposure to serum in a medium that does not support growth of the bacterium. A number of modifications and alternatives have been described (Provonchee and Zinner, 1974; Hancock et al., 1983; Pelkonen, 1987), but the assays remain dependent upon enumeration of viable bacteria. This study presents a further refinement, demonstrating the applicability of a growth-based method for assessing serum resistance in bacteria that allows for larger numbers of isolates to be assessed at one time. Conceptually, this method differs from others in that it assesses the ability of an isolate to grow in a medium that supports bacterial growth rather than a direct assessment of bacterial killing. This could have a number of advantages; firstly, by using an automated growth machine this method allows for a higher throughput of isolates. Secondly, unlike the traditional serum sensitivity assay, which determines killing of bacteria, a growth-based assay assesses whether or not resistant bacteria have the capacity to continue to thrive in the presence of serum. The Bioscreen automated growth assay proved to be an effective alternative method to assess the capacity of serum to inhibit B. cenocepacia isolates by determining the percentage inhibition of growth at 12 h (Figure 2). In contrast, traditional methods involve serial dilution and plating both before and after exposure to serum, which for even a small number of isolates is a process that is highly time consuming and laborious, requiring a minimum of 24 h (usually 48 h for the slower growing BCC). Furthermore, the Bioscreen automated growth assay provided data consistent with those from the traditional serum sensitivity test (Table 1).
The Bioscreen machine has been successfully used as a higher-throughput method to assess the inhibitory profile of a number of different antimicrobial compounds including antibiotics (Löwdin et al., 1998), disinfectants (Lambert et al., 1998), and biocides (Lambert et al., 1998) as well as to assess the bacteriolytic effect of bacteriophages (Cooper et al., 2011). In addition to throughput of isolate numbers and decreased time to result, one further advantage that the Bioscreen assay offered over the traditional serum resistance was the ability to monitor the dynamics of growth in the presence of serum. Specifically, we were able to show that all isolates classified as intermediate, and a number classified as serum sensitive, eventually do grow in the presence of serum (Table 1). This observation is similar to the recent study that utilized the Bioscreen machine to assess bacteriophage activity against P. aeruginosa, here the authors found the Bioscreen machine to be more stringent and that it identified the regrowth of some P. aeruginosa samples 20 h after treatment with bacteriophage (Cooper et al., 2011). Such observations may have biological significance in vivo. Finally, while we used a Bioscreen machine in this study, because it permits a full characterization of the growth of isolates across a time-span, it should be easy to validate this experimental approach using other plate reader equipment as all that is required is optical density measurements at 0 and 12 h, with shaking at 37°C in between.
The issue of serum resistance in BCC is important due to the capacity of BCC to cause devastating and often quickly fatal necrotizing bacteremic pneumonia (Isles et al., 1984). To date there have been several studies on serum resistance in BCC bacteria, that have shown that clinical isolates from the pulmonary tract may be serum resistant (Butler et al., 1994; Ortega et al., 2005; Savoia et al., 2008). This is the first study to examine serum resistance in the context of strain background in B. cenocepacia, as determined by the PCR based RAPD typing method (Mahenthiralingam et al., 1996). The strain background of clinical isolates also did not seem to be a determining factor in serum resistance as representative isolates from the major epidemic clonal lineages (RAPD types 01, 02, 04, and 06) exhibited both serum sensitivity and serum resistance (Tables 1 and 3). Applying this assay to clinical isolates allowed us to observe that there is apparently no direct link between the capacity of an isolate to elaborate exopolysaccharide, as previously determined on yeast extract mannitol media, and its ability to resist the bactericidal effects of human serum in LB, however this analysis must be caveated with the comment that the quantity of EPS produced by BCC bacteria during growth in LB liquid culture is not clear. Furthermore, this study is limited because it would need to be powered with a very large number of isolates to provide statistically significant proof that there is no link between exopolysaccharide production and serum resistance and given the small numbers of BCC infections, this is difficult to achieve. Further work is required to understand all the mechanisms involved in serum resistance. It has been shown that possession of a full LPS can confer serum resistance on BCC (Ortega et al., 2005), however as isolate C8963 has a rough LPS phenotype and is serum resistant (Table 1 and Conway et al., 2004) there may be alternative mechanisms of resistance available to BCC bacteria, such as other alterations to the outer membrane or the production of factors that could inhibit the assembly of the MAC.
Using both methods, we have observed that the last stored pulmonary isolate from BCC infected CF patients, who subsequently died from cepacia syndrome, most often displayed either a serum sensitive or serum intermediate phenotype (Table 1). However, it is important to note that although this sample set contains over 30 years of BCC cases across two clinics, the small numbers of cepacia syndrome cases makes a statistical link impossible. Nonetheless we consider these observations from clinical isolates to be valuable data because they suggest that in vitro assessment of serum resistance cannot be used as a marker of subsequent fitness for bloodstream invasion. Specifically, the last isolate prior to cepacia syndrome in seven out of the nine cases was assessed as either serum sensitive (three cases) or serum intermediate (four cases; Table 1). To explain the apparent contradiction that the last stored isolate from patients that subsequently develop cepacia syndrome are most often not resistant to serum, there are a number of possibilities that require further investigation. Firstly, BCC bacteria are known for their phenotypic plasticity (Mahenthiralingam et al., 2005); therefore it is conceivable that the bacteria acquire serum resistance between the date of the last pulmonary isolate and subsequent cepacia syndrome. Phenotypic variation is known to be present within the individual sputum samples (Larsen et al., 1993); therefore, another possibility is that a sub-population of the sputum sample, not captured when these isolates were harvested and stored, is ultimately responsible for invasion. Indeed, supportive of these previous observations, phenotypic variation was observed from a sample from the same day in this study; with two separate isolates from the same day for clinical case BCC5 being characterized as serum intermediate and resistant respectively (Table 1). Finally, another limitation of the present study is the use of pooled sera from non-CF donors. Regrettably sera, blood isolates, and DNA from these patients is not available to conduct a study into the competence of the sera from these individuals to kill the isolates of BCC that ultimately resulted in their death.
In conclusion, we have developed a relatively high throughput technique for assessing serum resistance in bacterial isolates. This technique has proven robust in our hands and was effective at screening larger numbers of isolates than the conventional assay for serum resistance. We believe this is the largest survey to date of serum resistance in clinical isolates of BCC bacteria and we have shown that further work is required to understand how, at least in some cases, patients who harbor serum sensitive isolates of BCC can subsequently develop “cepacia syndrome”. The technique described herein should be of considerable utility in assessing the mechanisms by which BCC bacteria resist the inhibitory effects of serum. Furthermore it could be easily adapted and applied for use with other bacterial species, to determine whether they are capable of resisting the complement components of human sera.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded through operating grants from the BC Lung Association (20R46136) and Cystic Fibrosis Canada (20R32976) to David P. Speert. James E. A. Zlosnik was funded by a post-doctoral fellowship from Cystic Fibrosis Canada and L. Cynthia Gunaratnam was funded by a summer studentship from Cystic Fibrosis Canada. We are also grateful for the technical assistance of Trevor Hird and Monica Fraenkel.
References
- Alexander B. D., Petzold E. W., Reller L. B., Palmer S. M., Davis R. D., Woods C. W., Lipuma J. J. (2008). Survival after lung transplantation of cystic fibrosis patients infected with Burkholderia cepacia complex. Am. J. Transplant. 8, 1025–1030 10.1111/j.1600-6143.2008.02186.x [DOI] [PubMed] [Google Scholar]
- Butler S. L., Nelson J. W., Poxton I. R., Govan J. R. (1994). Serum sensitivity of Burkholderia (Pseudomonas) cepacia isolates from patients with cystic fibrosis. FEMS Immunol. Med. Microbiol. 8, 285–292 10.1111/j.1574-695X.1994.tb00454.x [DOI] [PubMed] [Google Scholar]
- Cescutti P., Foschiatti M., Furlanis L., Lagatolla C., Rizzo R. (2010). Isolation and characterisation of the biological repeating unit of cepacian, the exopolysaccharide produced by bacteria of the Burkholderia cepacia complex. Carbohydr. Res. 345, 1455–1460 10.1016/j.carres.2010.03.029 [DOI] [PubMed] [Google Scholar]
- Conway B. A., Chu K. K., Bylund J., Altman E., Speert D. P. (2004). Production of exopolysaccharide by Burkholderia cenocepacia results in altered cell-surface interactions and altered bacterial clearance in mice. J. Infect. Dis. 190, 957–966 10.1086/423141 [DOI] [PubMed] [Google Scholar]
- Cooper C. J., Denyer S. P., Maillard J.-Y. (2011). Rapid and quantitative automated measurement of bacteriophage activity against cystic fibrosis isolates of Pseudomonas aeruginosa. J. Appl. Microbiol. 110, 631–640 10.1111/j.1365-2672.2010.04928.x [DOI] [PubMed] [Google Scholar]
- Cunha M. V., Sousa S. A., Leitão J. H., Moreira L. M., Videira P. A., Sá-Correia I., Leitao J. H., Sa-Correia I. (2004). Studies on the involvement of the exopolysaccharide produced by cystic fibrosis-associated isolates of the Burkholderia cepacia complex in biofilm formation and in persistence of respiratory infections. J. Clin. Microbiol. 42, 3052–3058 10.1128/JCM.42.7.3052-3058.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T., de Kievit T. R., Masoud H., Altman E., Richards J. C., Sadovskaya I., Speert D. P., Lam J. S. (1994). Characterization of lipopolysaccharide-deficient mutants of Pseudomonas aeruginosa derived from serotypes O3, O5, and O6. Infect. Immun. 62, 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Soyza A., Meachery G., Hester K. L. M., Nicholson A., Parry G., Tocewicz K., Pillay T., Clark S., Lordan J. L., Schueler S., Fisher A. J., Dark J. H., Gould F. K., Corris P. A. (2010). Lung transplantation for patients with cystic fibrosis and Burkholderia cepacia complex infection: a single-center experience. J. Heart Lung Transplant. 29, 1395–1404 10.1016/j.healun.2010.06.007 [DOI] [PubMed] [Google Scholar]
- Frank M. M., Joiner K., Hammer C. (1987). The function of antibody and complement in the lysis of bacteria. Rev. Infect. Dis. 9, S37–S45 10.1093/clinids/9.Supplement_5.S537 [DOI] [PubMed] [Google Scholar]
- Glass S., Govan J. R. (1986). Pseudomonas cepacia-fatal pulmonary infection in a patient with cystic fibrosis. J. Infect. 13, 157–158 10.1016/S0163-4453(86)92953-1 [DOI] [PubMed] [Google Scholar]
- Hancock R. E., Mutharia L. M., Chan L., Darveau R. P., Speert D. P., Pier G. B. (1983). Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect. Immun. 42, 170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiby N., Olling S. (1977). Pseudomonas aeruginosa infection in cystic fibrosis: bactericidal effect of serum from normal individuals and patients with cystic fibrosis on P. aeruginosa strains from patients with cystic fibrosis or other diseases. Acta Pathol. Microbiol. Scand. C 85C, 107–114 [PubMed] [Google Scholar]
- Isles A., Maclusky I., Corey M., Gold R., Prober C., Fleming P., Levison H. (1984). Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104, 206–210 [DOI] [PubMed] [Google Scholar]
- Jones A. M., Dodd M. E., Govan J. R. W., Barcus V., Doherty C. J., Morris J., Webb A. K. (2004). Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax 59, 948–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E., Gollan B., Tang C. M. (2008). Mechanisms in Neisseria meningitidis for resistance against complement-mediated killing. Vaccine 26, I34–I39 10.1016/j.vaccine.2008.11.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert R. J., Johnston M. D., Simons E. A. (1998). Disinfectant testing: use of the bioscreen microbiological growth analyser for laboratory biocide screening. Lett. Appl. Microbiol. 26, 288–292 10.1046/j.1472-765X.1998.00334.x [DOI] [PubMed] [Google Scholar]
- Lambris J. D., Ricklin D., Geisbrecht B. V. (2008). Complement evasion by human pathogens. Nat. Rev. Microbiol. 6, 132–142 10.1038/nrmicro1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen G. Y., Stull T. L., Burns J. L. (1993). Marked phenotypic variability in Pseudomonas cepacia isolated from a patient with cystic fibrosis. J. Clin. Microbiol. 31, 788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwdin E., Odenholt I., Cars O. (1998). In vitro studies of pharmacodynamic properties of vancomycin against Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 42, 2739–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E., Baldwin A., Dowson C. G. (2008). Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104, 1539–1551 [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E., Campbell M. E., Henry D. A., Speert D. P. (1996). Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J. Clin. Microbiol. 34, 2914–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E., Coenye T., Chung J. W., Speert D. P., Govan J. R., Taylor P., Vandamme P. (2000). Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38, 910–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E., Urban T. A., Goldberg J. B. (2005). The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3, 144–156 10.1038/nrmicro1085 [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E., Vandamme P., Campbell M. E., Henry D. A., Gravelle A. M., Wong L. T., Davidson A. G., Wilcox P. G., Nakielna B., Speert D. P. (2001). Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin. Infect. Dis. 33, 1469–1475 10.1086/322684 [DOI] [PubMed] [Google Scholar]
- Nash E. F., Thomas A., Whitmill R., Rashid R., Barker B., Rayner R. J., Whitehouse J. L., Honeybourne D. (2011). “Cepacia syndrome” associated with Burkholderia cepacia (genomovar I) infection in an adolescent with cystic fibrosis. Pediatr. Pulmonol. 46, 512–514 [DOI] [PubMed] [Google Scholar]
- Ortega X., Hunt T. A., Loutet S., Vinion-Dubiel A. D., Datta A., Choudhury B., Goldberg J. B., Carlson R., Valvano M. A. (2005). Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol. 187, 1324–1333 10.1128/JB.187.4.1324-1333.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkonen S. (1987). A rapid turbidimetric assay for the study of serum sensitivity of Escherichia coli. FEMS Microbiol. Lett. 42, 53–57 10.1111/j.1574-6968.1987.tb02298.x [DOI] [Google Scholar]
- Provonchee R. B., Zinner S. H. (1974). Rapid method for determining serum bactericidal activity. Appl. Microbiol. 27, 185–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoia D., Deplano C., Zucca M. (2008). Pseudomonas aeruginosa and Burkholderia cenocepacia infections in patients affected by cystic fibrosis: serum resistance and antibody response. Immunol. Invest. 37, 19–27 [DOI] [PubMed] [Google Scholar]
- Speert D. P., Farmer S. W., Campbell M. E., Musser J. M., Selander R. K., Kuo S. (1990). Conversion of Pseudomonas aeruginosa to the phenotype characteristic of strains from patients with cystic fibrosis. J. Clin. Microbiol. 28, 188–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speert D. P., Henry D., Vandamme P., Corey M., Mahenthiralingam E. (2002). Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerging Infect. Dis. 8, 181–187 10.3201/eid0802.010163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomassen M. J., Demko C. A. (1981). Serum bactericidal effect on Pseudomonas aeruginosa isolates from cystic fibrosis patients. Infect. Immun. 33, 512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P., Holmes B., Vancanneyt M., Coenye T., Hoste B., Coopman R., Revets H., Lauwers S., Gillis M., Kersters K., Govan J. R. (1997). Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47, 1188–1200 10.1099/00207713-47-4-1055 [DOI] [PubMed] [Google Scholar]
- Walport M. J. (2001). Complement: first of two parts. N. Engl. J. Med. 344, 1058–1066 10.1056/NEJM200104053441406 [DOI] [PubMed] [Google Scholar]
- Zahariadis G., Levy M. H., Burns J. L. (2003). Cepacia-like syndrome caused by Burkholderia multivorans. Can. J. Infect. Dis. 14, 123–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlosnik J. E. A., Costa P. S., Brant R., Mori P. Y. B., Hird T. J., Fraenkel M. C., Wilcox P. G., Davidson A. G. F., Speert D. P. (2011). Mucoid and nonmucoid Burkholderia cepacia complex bacteria in cystic fibrosis infections. Am. J. Respir. Crit. Care Med. 183, 67–72 10.1164/rccm.201002-0203OC [DOI] [PubMed] [Google Scholar]
- Zlosnik J. E. A., Hird T. J., Fraenkel M. C., Moreira L. M., Henry D. A., Speert D. P. (2008). Differential mucoid exopolysaccharide production by members of the Burkholderia cepacia complex. J. Clin. Microbiol. 46, 1470–1473 10.1128/JCM.02273-07 [DOI] [PMC free article] [PubMed] [Google Scholar]