Abstract
The increase in huge number of genomic sequences in recent years has contributed to various genetic events such as horizontal gene transfer (HGT), gene duplication and hybridization of species. Among them HGT has played an important role in the genome evolution and was believed to occur only in Bacterial and Archaeal genomes. As a result, genomes were found to be chimeric and the evolution of life was represented in different forms such as forests, networks and species evolution was described more like a rhizome, rather than a tree. However, in the last few years, HGT has also been evidenced in other group such as metazoa (for example in root-knot nematodes, bdelloid rotifers and mammals). In addition to HGT, other genetic events such as transfer by retrotransposons and hybridization between more closely related lineages are also well established. Therefore, in the light of such genetic events, whether the evolution of metazoa exists in the form of a tree, network or rhizome is highly questionable and needs to be determined. In the current review, we will focus on the role of HGT, retrotransposons and hybridization in the metazoan evolution.
Keywords: tree of life, horizontal gene transfer, retrotransposons, hybridization, metazoa
Proposed concepts for tree of life and position of metazoa
Understanding the relationships among all living organisms by phylogenetic tree reconstruction is one of the fundamental challenges in biology. For almost 200 years, Tree of Life (TOL) has been the most powerful metaphors for biologists in depicting the evolutionary history of organisms. One of the first and most explicit form of TOL was presented by German zoologist Ernst Haeckel (1866), but its exact shape has remained elusive. Indeed, several studies to deduce TOL using various methods were carried out (Fox et al., 1980; Doolittle, 1981; Fitz-Gibbon and House, 1999; Snel et al., 1999; Tekaia et al., 1999; Lin and Gerstein, 2000; Brown et al., 2001; Clarke et al., 2002; Korbel et al., 2002; Rokas et al., 2003; Kunin et al., 2005), but its principal existence is heavily debated.
The rapid increase in molecular and genomic data in recent years have contributed to genetic events such as horizontal gene transfer (HGT), that is often considered as a major constraint in the reconstruction of phylogenetic trees. HGT “the non-genealogical transmission of genetic material from one organism to another” (Goldenfeld and Woese, 2007) is an important driving force in genomic evolution. HGT has contributed to early evolution of life to a larger extent than is presently occurring in modern biota (Zillig et al., 1992; Kandler, 1994, 1998; Woese, 2002). Indeed, number of studies have been reported about the genes acquired by HGT in three domains of life, such as Bacteria (Saunders et al., 1999; Ochman et al., 2000), Archaea (Doolittle and Logsdon, 1998; Faguy and Doolittle, 1999) and Eukaryotes (Andersson, 2005) and also between domains, i.e., from Bacteria to Archaea (Gophna et al., 2004), from Archaea to Eukarya (Andersson et al., 2003), from Bacteria to Eukarya (Watkins and Gray, 2006), from Eukarya to Bacteria (Guljamow et al., 2007) and even within Eukarya (Nedelcu et al., 2008).
Conversely, it has been assumed that the role of HGT is not prevalent in other multicellular eukaryotic organisms like in kingdom Animalia or metazoa. However, the possibility of gene transfers among them have increased in recent years and were reported in various groups such as Porifera (Rot et al., 2006), Cnidaria (Chapman et al., 2010), Nematoda (Danchin et al., 2010), Arthropoda (Fenn et al., 2006; Hotopp et al., 2007), Rotifera (Gladyshev et al., 2008), and Craniata (Graham et al., 2008; Pace et al., 2008). In addition to HGT, other genetic events such as transfer by retrotransposons and hybridization (Seehausen, 2004) were also reported, which are proposed to play an important role in the evolution of metazoa.
The studies discussed above support that HGT has played a significant role in modulating the metazoan evolution including prokaryotes and eukaryotes. Consequently, the increase in the prevalence of HGT events in Bacteria, Archaea, and Eukaryotes have resulted in the chimeric nature of genomes, where different parts of the genome can have different evolutionary histories and its difficult to identify a single common ancestor for the gene repertoire of any organism. All these results have conferred to undermine the TOL concept, thereby giving rise to a new paradigm. As a result, many proposals have emerged for the tree-like pattern replacing it with more complex models such as the “reticulate evolution” (Sneath, 1975), “synthesis of life” (Bapteste et al., 2004), “web of life” (Doolittle, 1999), “ring of life” (Rivera and Lake, 2004), “network of life” (Ragan et al., 2009), the “forest” of evolutionary trees (Puigbo et al., 2009; Schliep et al., 2010), the genetic network (Puigbo et al., 2010; Popa et al., 2011). Moreover, the evolution of species was described more like a rhizome (Deleuze and Guattari, 1976; Raoult, 2010), reflecting various origins of genomic sequences in each species (Raoult, 2010).
The phylogenetic relationships among the major groups of animals was represented in the form of a tree (Bergstrom, 1985; Field et al., 1988; Valentine, 1997; Halanych, 2004; Giribet et al., 2007; Dunn et al., 2008). However, the increase in the extent of HGT, retrotransposons, and hybridization events in the metazoan lineage precludes the reconstruction of animal TOL. In addition to these, recent studies on whole genomic sequences from various metazoan phyla suggest a chimeric origin for its major groups due to the presence of HGT and hybridization (Syvanen and Ducore, 2010).
The results from these studies pose new questions about the evolution of metazoa, as whether it exists in the form of network or rhizome of life? Therefore, there is a need for a new pattern to be determined. In the current review, we will discuss the role of HGT, transfer by retrotransposons, hybridization and the challenges that it proposes for the current metazoan evolutionary paradigm.
Metazoa and its classification
Metazoa (animals) represent a group of multicellular eukaryotes (Haeckel, 1874) and constitute a monophyletic clade with heterotrophic organisms. They are believed to have diversified around the beginning of the Cambrian period (∼543 million years ago). According to the recent reports, the metazoan group includes approximately 1.3 million described living species distributed in 35–40 phyla (subjected to the classification) (Edgecombe et al., 2011). This exponential growth in molecular sequence data in the metazoan group has increased the amount of phylogenetic information to study the animal relationships.
According to the Linnaeus (1758) system of classification, kingdom Animalia was classified into six classes: Amphibia, Aves, Mammalia, Pisces, Reptilia, Insecta, and Vermes, the latter divided into Intestina, Mollusca, Testacea, Lithophyta, and Zoophyta. Later on this classification has been revised constantly by biologists to study their evolutionary relationships. The early metazoan tree on animal phylogeny was based on 18S rRNA (Field et al., 1988) and Cnidaria, Ctenophora, Placozoa, Porifera, and Bilateria constituted the basal metazoan groups (Medina et al., 2001; Collins et al., 2005). However, the increase in molecular data has given rise to so-called “new animal phylogeny” (Adoutte et al., 2000; Halanych, 2004; Giribet et al., 2007) supporting the monophyly of Bilateria, which is again divided into two major lineages, Protostomia, and Deuterostomia. These two clades are well resolved by broad taxon sampling (Dunn et al., 2008; Hejnol et al., 2009; Philippe et al., 2009). The diversity within Protostomia led to division of two clades, Ecdysozoa (Aguinaldo et al., 1997; Schmidt-Rhaesa et al., 1998; Giribet, 2003; Telford et al., 2008), and Spiralia (Spiralia or Lophotrochozoa; Halanych et al., 1995; Giribet et al., 2000, 2009; Halanych, 2004; Giribet, 2008). Deuterostomia encompass two clades, Ambulacraria (Winchell et al., 2002; Brown et al., 2008), and Chordata (Delsuc et al., 2006; Mallatt and Winchell, 2007). The present consensus on metazoan phylogeny based on various hypotheses is represented in Figure 1. (Figure 1, Edgecombe et al., 2011) The relationships on deep metazoan groups have been extensively reviewed elsewhere (Edgecombe et al., 2011).
Figure 1.
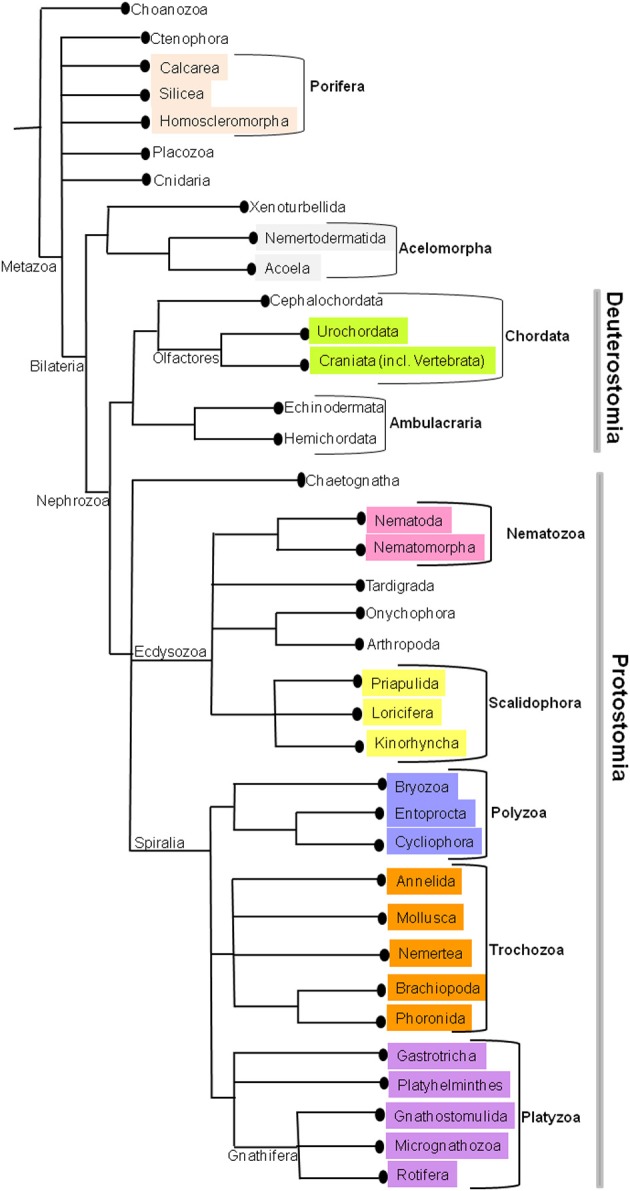
The current phylogenetic relationships in Metazoa. This tree was generated by consensus based on various phylogenetic studies.
Horizontal gene transfer in metazoa
HGT in animals has long been neglected and considered to be rare. However, the increase in molecular data in recent years has contributed to the possibility of gene transfers in various metazoan phyla such as Porifera, Cnidaria, Nematoda, Arthropoda, Rotifera, and Craniata, thereby creating the need to analyze more transfer events in other unidentified metazoan groups. Therefore, we will discuss the transfer events in each of them in the following sections respectively.
Transfer in phylum porifera (sponge)
Poriferans represent the earliest diverging metazoans due to the presence of distinct cell types called choanocytes, which are similar to choanoflagellates, the closest unicellular relatives of metazoans (Medina et al., 2001; Nielsen, 2001; Muller, 2003). Other unique features include the lack of intestinal epithelium and digestive parenchyma (Ereskovsky and Dondua, 2006). The molecular analyses resolved sponges at the base of metazoa (Peterson and Butterfield, 2005).
The mitochondrial genome of Metazoa lacks introns, except in case of corals and sea anemones (Cnidaria), in which group I introns have been discovered in the cox1 and nad5 genes (Beagley et al., 1996; van Oppen et al., 2002). Infact, a recent cross-kingdom HGT of group I intron of cox 1 gene in sponge Tetilla sp. (Spirophorida) mitochondrial genome from fungal origin was reported, indicating the unexpected plasticity of the mitochondrial genomes of basal Metazoa (Rot et al., 2006).
A unique and first known case of HGT event of octocoral mtMuTS gene into animal mitochondrial genome has been reported (Bilewitch and Degnan, 2011), suggesting the need to reconsider the evolution of mitochondrial genome in metazoa.
Transfer in phylum cnidaria
The phylum Cnidaria constitutes a diverse monophyletic group (Collins, 2002). Cnidarians have many different cell types, including gametes and nematocytes, which originate in the adult form from an interstitial cell lineage. Many of them are characterized by a complex, metagenetic life cycle including a sexually produced planula larva that metamorphoses into a sessile polyp stage, which may in turn asexually produce morphologically distinct, free-swimming, sexual medusae (Hyman, 1940).
Few instances of HGT were also seen in Cnidarians. For example, a subunit of bacterial poly-γ-glutamate (PGA) synthase was transferred to metazoan ancestor, suggesting its significant role on the evolution of stinging cells (nematocytes) in cnidarians (sea anemones, jellyfish, corals, etc.) (Denker et al., 2008). Hydra is simple freshwater animal which reproduce asexually by budding. The genome of Hydra magnipapillata contains 71 candidates for HGT, that show closer relationship to bacterial genes than metazoan genes and 70% of these are supported by ESTs (Chapman et al., 2010).
Protostomia
Protostomes are defined as a group of animals in which blastopore typically becomes the future mouth in most of the groups (Nielsen, 2001). It consists of two clades, Ecdysozoa and Spiralia (Spiralia or Lophotrochozoa). Ecdysozoa includes the following phyla: Nematoda, Nematomorpha, Tardigrada, Onychophora, Arthropoda, Priapulida, Loricifera, and Kinorhyncha (Figure 1) and Spiralia include two clades Platyzoa (Cavalier Smith, 1998) and Trochozoa (Roule, 1891).
Ecdysozoa are called as moulting protostomes and two phyla that are included under Ecdysozoa with reported cases of HGT include Nematoda and Arthropoda (Aguinaldo et al., 1997; Schmidt-Rhaesa et al., 1998; Edgecombe et al., 2000; Garey, 2001; Peterson and Eernisse, 2001; Zrzavý, 2003, Figure 1).
Transfer in phylum nematoda
Nematodes represent the largest animal phylum, with an estimated number in the range of one to ten million species (Lambshead, 1993) and are found in virtually all habitats on earth. Many of them are parasites of plants and animals, including humans. The recent increase in nematode genomes has made attributions in comparative genomics to study the impact of HGT on their adaptation to new ecological niches. Although inter-kingdom HGT was initially controversial, it has been established with evidence of such recent events (Richards et al., 2011). Danchin et al. (2010) have studied the whole-genome sequences of root-knot nematodes and cyst-nematodes for the genes encoding proteins involved in the plant cell wall degradation and showed the incorporation of at least six distinct types of bacterial genes encoding proteins that can modify the plant cell wall into their genomes. These have subsequently undergone extensive gene duplication in the nematode lineages.
Furthermore, the cases of HGT from a diverse set of microorganisms into various nematode genomes have also been identified. The genome of Bursaphelenchus xylophilus (the pine wilt nematode) has incorporated six glycoside hydrolase family 16 (GH16) proteins from gammaproteobacteria, two hydrolases from Firmicutes and four aspartic-type endopeptidases and 11 GH45 cellulases from the Ascomycota of fungal origin (Kikuchi et al., 2011). The genome of Pristionchus pacificus (a necronemic nematode) suggests that it contains substantial amount of genes of insect origin (Rödelsperger and Sommer, 2011). The Meloidogyne incognita (the root-knot nematode) contains genes similar to those of actinobacteria, proteobacteria, and fungi (Abad et al., 2008). The plant-parasitic nematode Heterodera glycines contains a biosynthetic pathway for vitamin B6 of bacterial origin (Craig et al., 2008).
Transfer in phylum arthropoda
The members of the phylum Arthropoda are characterized by exoskeleton, segmented bodies and jointed appendages (Valentine, 2004). The appendages form part of an exoskeleton, which is mainly made of α-chitin, a derivative of glucose (Cutler, 1980). They are important members of marine, freshwater, land, and air ecosystems and are one of only two major animal groups that have adapted to life in dry environments (Ruppert et al., 2004). They include insects, arachnids, crustaceans, others and account for over 80% of all known living animal species (Anna, 2008).
The reported cases of HGT in insects include the acquisition of P elements by Drosophila melanogaster from Drosophila willistoni (Daniels et al., 1990; Engels, 1997), transfer of entire genes for carotenoid biosynthetic pathway from fungi (Moran and Jarvik, 2010) and 12 genes from bacteria (IAGC, 2010; Nikoh et al., 2010) to Acyrthosphion pisum. The largest HGT transfer such as the transfer of entire Wolbachia bacterial genome (∼1.4 Mb) into Drosophila ananassae Hawaii 2L chromosome (Hotopp et al., 2007) and also of Wolbachia to a wider range of insects such as pea aphid (Hemiptera), mosquitoes (Diptera), beetle (Coleoptera), fruit flies (Diptera), and parasitoid wasps (Hymenoptera) (Fenn et al., 2006; Hotopp et al., 2007) has been described. Recently, transfers have also been reported in Bombyx mori that has acquired 10 genes from plant and bacteria (Zhu et al., 2011) and 14 types of 22 transferred genes from entomopathogenic bacteria, of which 13 types shared homology with sequences of Lepidopteran insects (Li et al., 2011), thus providing novel insights on the biological significance of HGT in the evolution of metazoan recipients.
Transfer in phylum rotifera
The Rotifera (also called Rotatoria) is a group of aquatic micrometazoans that usually occur in freshwater and marine environments (Wallace, 1998; Wallace et al., 2006; Segers, 2007) and are classified under Gnathifera of Platyzoa (Figure 1). The word “rotifer” is derived from a Latin word meaning “wheel-bearer” (Pechenik, 2005) and commonly called wheel animals due to the presence of corona in the cephalic region that resembles a wheel, which is used for locomotion and food gathering. They form the largest metazoan asexual group where no sexual reproduction has ever been reported and represent an ancient origin of asexuality with great evolutionary success in the diversification of the species (Mark Welch and Meselson, 2000; Fontaneto et al., 2007).
The first evidence of HGT has been reported in bdelloid rotifers to contain many foreign genes from diverse bacterial and eukaryotic origin, which are strongly concentrated in the telomeric regions along with diverse transposable elements (TEs) (Gladyshev et al., 2008, Figure 2). The evidence of extensive HGT in these asexual animals might be due to repeated cycles of desiccation-induced membrane disruption and DNA breakage and repair that occur as part of their life style.
Figure 2.
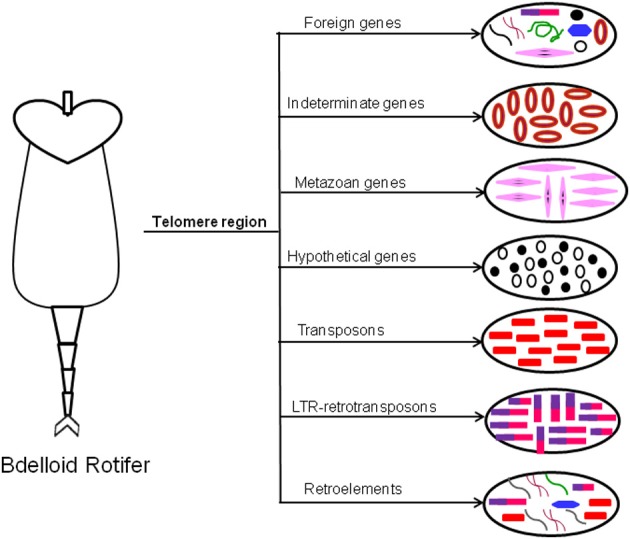
Horizontal gene transfer in Bdelloid Rotifer. Bdelloid rotifer contain diverse array of foreign genes acquired by HGT from various origin (like bacteria, metazoa, retrovirus, etc.). They are strongly concentrated in the telomeric region. The names of the transferred genes are indicated above the arrows.
The HGT in the asexual organisms (i.e., Rotifera and Hydra), suggests that they might have accumulated mutations in an irreversible manner in the absence of recombination through sexual reproduction (Muller, 1932, 1964; Felsenstein, 1974).
Deuterostomia
Deuterostomes are a group of animals in which the blastopore becomes the anus in the adult, while the mouth develops as a new opening from the end of the archenteron (Nielsen, 2001). It includes two main clades, Ambulacraria and Chordata. Phylum Chordata is again classified into Cephalochordata, Urochordata, and Craniata (incl. Vertebrata, see Figure 1).
Craniata
The term “Craniata” was coupled with “Vertebrata” by Linnaeus (1758) to include lampreys, jawed fishes, and terrestrial vertebrates (tetrapods). The Craniata or craniates, are characterized by a skull (or cranium, hence their name). They comprise of all fishes including jawless fishes as hagfishes and lampreys, amphibians, reptiles, birds, and mammals, including Man. Now, the majority of the craniate species are represented by one group of fish, the actinopterygians, and the tetrapods (four-legged vertebrates) (Philippe, 1997).
Transfer in class teleostei (fishes)
Teleostei (the ray-finned fishes) represent one of the three classes of actinopterygii and includes most of the living fishes (Miller and Harley, 2007).
Cases of HGT in fishes are very sparse which include the transfer of lectin-like antifreeze proteins between them (Graham et al., 2008). Although the transfer of retroposons from Schistosoma japonicum (blood fluke) to salmonoid fishes was identified (Matveev and Okada, 2009), it was refuted with no evidence of such transfer between the two clades based on cross-species and vector contamination and was declared as erroneous report of HGT (Grunau and Boissier, 2010).
Transfer in class mammalia
With the burgeoning database of eukaryotic genomic sequences, it is not surprising to see the increasing cases of HGT in mammals. For example, transfer of DNA SPIN trasposons in mammals and other tetrapods (Pace et al., 2008) and transfer in human germ cells (Hecht et al., 2010) were reported. Recent studies have shown that the human body contains more of bacterial cells than human cells (Gill et al., 2006; Lester et al., 2006; Hehemann et al., 2010; Robinson et al., 2010), and many of them are dominated by members of Bacteroidetes and Firmicutes (Eckburg et al., 2005; Xu et al., 2007). Most of these recent transfers were driven by ecology rather than geography or phylogeny (Smillie et al., 2011).
In summary, the currently reported cases of HGT in animal kingdom are relatively low. However, with the availability of many new whole genome sequences, we can expect more incidences of HGT that will enable to gain further knowledge in the metazoan evolution.
Fate of transferred genes in metazoa
The identification of several HGT cases in animals has given rise to many questions such as the function of transferred genes, their evolutionary pathways and the forces governing the transferred genes. Indeed, efforts have been made recently to answer these questions. For example, in nematodes, recent work using 454-sequencing on cellulase functioning genes has shown that they have integrated into receptor genome by providing special functions, which indicates that genes continue to evolve with several gene duplications or deletions and DNA substitution rates after the HGT event (Mayer et al., 2011). Most of them encode enzymes for the cell wall degradation in plants and fungi and play vital role in the biology of the nematodes (Abad et al., 2008; Kikuchi et al., 2011; Mayer et al., 2011). The most recent progress has been made about the functional role of cell wall degrading enzymes of plant-parasitic nematode (Haegeman et al., 2012).
The acquisition of two enzymes for vitamin B6 biosynthesis in plant pathogenic nematode Heterodera glycines suggests host-parasite interactions (Craig et al., 2008). Incorporation of bacterial genes by specialized animal rumen parasites, i.e., Giardia lamblia (Morrison et al., 2007), Trichomonas vaginalis (Carlton et al., 2007) and Entamoeba histolytica (Loftus et al., 2005), that exist in anaerobic environments suggests that adaptation to parasitism might also favor the acquisition of new genes by HGT. Transfer of lectin-like antifreeze proteins in arctic fish might have favored them to survive in cold-conditions (Graham et al., 2008). The presence of PGA synthase genes in cnidarians might have contributed to the evolution of nematocytes, that help in prey capture (Denker et al., 2008). Recent studies on transfer in bdelloid rotifers, which has acquired genes from bacterial and eukaryotic origin, some of them are expressed suggests that they may possibly provide novel metabolic functions to these asexual animals (Gladyshev et al., 2008). The transfer of bacteria to human (Robinson et al., 2010) is interesting because it may be important to human health, have the potential to provide novel functions, there by affecting the evolution.
In summary, these results support that the transferred genes having a functional role are retained, while useless genes are eliminated. The studies outlined on the function of transferred genes in animals are still rudimentary. Hence, there is need to understand the function of transferred genes in new genomes by involving robust phylogenetic investigations and biological disciplines.
Besides HGT, other genetic events such as transfer by retrotransposons and hybridization (Seehausen, 2004) were also reported in metazoa, which may form additional limitations in the reconstruction of animal TOL.
Retrotransposons and their classification
Retrotransposons (retroelements) belong to group of TEs. TEs (also known as “jumping genes”) include a diverse array of DNA sequences and possess the inherent capacity to self-reproduce and move within and between genomes. Ever since their discovery in maize DNA (McClintock, 1956), TEs have been found in genomes of almost all organisms. They constitute more than 50% of maize (Zea mays) genome (Kidwell and Lisch, 1997; Wessler, 1998), 22% of Drosophila genome (Kapitonov and Jurka, 2003) and half of our human genome with just 1.5% coding for protein region (Lander et al., 2001).
TEs are divided into two groups based on their transposition mechanism and sequence organization (Finnegan, 1989; Capy, 1998): 1. DNA transposons (move predominantly via a DNA-mediated mechanism of excision and insertion and constitute appx. 3% of human genome (Craig et al., 2002) and 2. Retroelements (move by reverse transcription of an RNA intermediate (Rogers, 1985) and include the retrotransposons, eukaryotic TEs, group II mitochondrial introns, bacterial retrointrons and retroviruses). The reverse transcriptase of the retroelements is usually encoded by the element itself. They are subdivided into two major groups based on the presence or the absence of long terminal repeats, which flank the body of the element: long terminal repeat (LTR)-containing elements (LTRs) and non-LTR retrotransposons (non-LTR). Again the non-LTRs are subdivided into two classes: LINEs or L1-element (long interspersed elements) and SINEs (short interspersed elements) (Weiner et al., 1986). LINEs, the autonomous elements are widely distributed in eukaryotes. For example, they occur in >500, 000 copies (∼17%) in human genome (Lander et al., 2001), out of which only ∼80–100 were found to be active (Brouha et al., 2003). They are also found in mouse genome (∼3000, Goodier et al., 2001) and Drosophila genome (Priimagi et al., 1988; Levis et al., 1993; Udomkit et al., 1995). Unlike LINEs, SINEs are non-autonomous and occupy about 12% of the human genome, out of which majority of them belong to Alu elements (Lander et al., 2001). The classification of TEs is shown in Figure 3.
Figure 3.
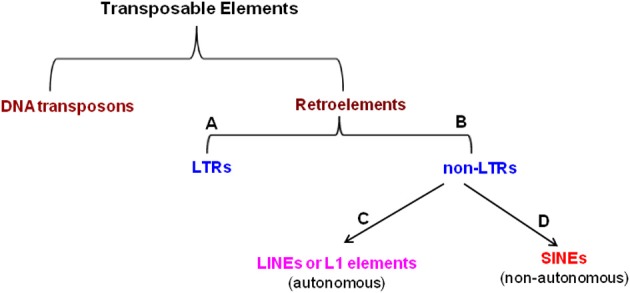
Classification of transposable elements. (A) Long terminal repeats, (B) Non-long terminal repeats, (C) Long interspersed elements, (D) Short interspersed elements.
Transfer by retrotransposons
With the widespread distribution of intergenomic TEs in eukaryotic genomes, it is not surprising to envisage the intriguing feature of HGT among them, a process by which they cross-species boundaries to enter new genomes. In the past decade, substantial evidence of HGT has been reported for all types of TEs in invertebrates as well as vertebrates. Some of the reported cases of HGT involving TEs are summarized in Table 1 as shown below.
Table 1.
Table showing some of the cases of horizontal gene transfer involving Transposable Elements (TEs) among Metazoa.
| Transposable elements | ||
|---|---|---|
| Type | Metazoan group/genome | Reference |
| LINE/jockey | Arthropoda/Drosophila | Mizrokhi and Mazo, 1990 |
| LINE/Bov-B | Mammals | Kordis and Gubensek, 1995 |
| SINE/Smal-cor | Teleostei (coregonid fish) | Hamada et al., 1997 |
| LTR/copia | Arthropoda/Drosophila | Jordan et al., 1999 |
| SURL elements | Echinodermata | Gonzalez and Lessios, 1999. |
| LTR/gypsy | Arthropoda/Drosophila | Vazquez-Manrique et al., 2000 |
| LTR/gypsy | Arthropoda/Drosophila | Terzian et al., 2000 |
| LINE/Rex1 | Teleostei (fish genomes) | Volff et al., 2000 |
| LINE/Bov-B | Reptiles and Mammals | Zupunski et al., 2001 |
| P elements | Arthropoda/Drosophila | Daniels et al., 1990 |
| P elements | Arthropoda/Drosophila | Hagemann et al., 1992 |
| mariner elements | Arthropoda/Drosophila mauritiana and Zaprionus tuberculatus | Maruyama and Hartl, 1991 |
| mariner elements | Arthropoda | Robertson and MacLeod, 1993 |
| mariner elements | Arthropoda/Drosophila | Brunet et al., 1994 |
| mariner elements | Arthropoda/Drosophila | Lohe et al., 1995 |
| mariner elements | Arthropoda | Robertson and Lampe, 1995 |
| mariner elements | Mammals/Homo sapiens | Smit and Riggs, 1996 |
| mariner elements | Amphibians | Lam et al., 1996 |
| mariner elements | Platyzoa and Cnidaria | Robertson, 1997 |
| mariner elements | Mammals | Robertson et al., 2002 |
| DNA SPIN transposons | Mammals and other tetrapods | Pace et al., 2008 |
| Helitrons (rolling circle DNA transposons) | Arthropoda, Reptiles, Teleostei (fish) and Mammals | Thomas et al., 2010 |
In addition to these, the vertebrate genomes also contain numerous copies of retroviral sequences that were acquired over the course of evolution. The majority of them belong to endogenous viral elements, which integrate into the nuclear genome of the host germ line (Tristem, 2000; Lander et al., 2001). As it is exhaustive to provide all the reported cases of retroviral elements, we present few of them. They include the human endogenous retrovirus element HERV-L, that is related distantly by homology to foamy viruses (Cordonnier et al., 1995), and recently reported cases such as the presence of endogenous viral elements in animal genomes (Katzourakis and Gifford, 2010), integration of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes (Belyi et al., 2010), endogenous lentivirus in basal primates (Gifford et al., 2008; Gilbert et al., 2009) and endogenous foamy viruses in the sloth genome (Katzourakis et al., 2009).
All these results suggest that retrotransposons continue to play an active role in shaping the dynamics of metazoan evolution by forming new genes and thus contributing to the chimeric evolution of genomes. The representation of metazoan chimeric evolution of genomes is shown in Figure 4.
Figure 4.
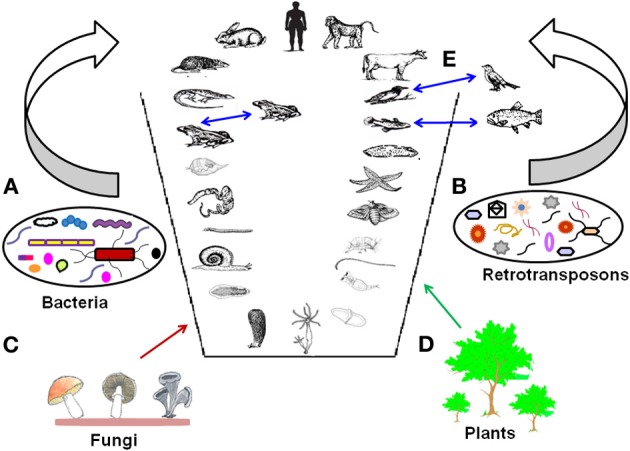
Metazoan chimeric evolution. The figure represents chimeric nature of metazoan genomes with horizontal gene transfer, retrotransposons, and hybridization events. Horizontal gene transfer from (A) Bacteria, (B) Retrotransposons, (C) Fungi, (D) Plants and (E) Hybridization between closely related species (indicated by blue arrows).
Hybridization
Hybridization, the exchange of genes between closely related species by sexual reproduction is a natural evolutionary process. The frequency of hybridization among species is common although it is rare between species on a per-individual basis. About 10–30% of multicellular animal and plant species hybridize regularly (Mallet, 2005). Closely related species tend to hybridize more. Therefore, species-rich groups in rapidly diversifying adaptive radiations tend to be those that hybridize most (Price and Bouvier, 2002; Seehausen, 2004; Gourbière and Mallet, 2010). However, the success of large number of hybridizations that occur among closely related species depends on genetic and environmental factors (Arnold et al., 2012).
Although recent studies have highlighted the events of HGT, information about hybridization is limited and highly unexplored, especially in the case of animals. However, studies have reported the cases of hybridization between more closely related lineages such as fungi, plants, and even vertebrate lineages such as amphibians, fish, and birds (Arnold, 2006). Recently, studies on primates have detected hybridization in not only between species and subspecies but also between genera, including human lineage (Zinner et al., 2011).
Hybridization among species can thus act as a catalyst for the formation of new lineages.
Concluding remarks
The relative amount of HGT events detected in metazoa merely represent just tip of an iceberg. Currently, the reported rate of HGT in animal kingdom is relatively low and still in the state of infancy. There are still many outstanding questions that need to be addressed like: What is the rate of gene transfer within the group? How important is gene transfer in other animal genomes, especially vertebrates? There are many lineages that need to be considered for HGT, such as Rotifers, Aves (birds), Reptiles and Mammals. Why are eukaryotic genes rare in prokaryotic genomes? What is the function of transferred genes in the new genome?
Although, there is substantial evidence on transfer of TEs (especially group I introns) in the mitochondrial genomes of metazoa (ex.sponges), we are unaware on the extent of these transfers in other groups (especially fishes, rotifers, and other vertebrates), which might serve as useful indicators, raising intriguing biological questions related to HGT. The recent evidence of octocoral mtMuTS gene into animal mitochondrial genome also support these findings, thus challenging the evolution of mitochondrial genome in metazoa.
Therefore, there is an emergent need for future HGT studies in Metazoa. Hopefully, with the recent improvements in new sequencing technologies and increased number of diverse vertebrate genomes, we anticipate to gain novel insights into the role played by HGT that will shed light in understanding the metazoan evolution.
Given the extent of gene transfer from different origins (bacteria, plants, fungi, and eukaryotes), transfer by retrotransposons and the fusion of lineages to from new lines of descent (interspecies hybridization) in animals, we believe that majority of the animal genomes exhibit mosaic structure and chimerism, thus replacing the concept of animal TOL with a new paradigm. Therefore, the question still remains unanswered about the evolution of metazoa, as whether it can be represented in the form of a network or rhizome and needs to be determined.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Infectiopole Sud for funding.
References
- Abad P., Gouzy J., Aury J. M., Castagnone-Sereno P., Danchin E. G., Deleury E., Perfus-Barbeoch L., Anthouard V., Artiguenave F., Blok V. C., Caillaud M. C., Coutinho P. M., Dasilva C., De Luca F., Deau F., Esquibet M., Flutre T., Goldstone J. V., Hamamouch N., Hewezi T., Jaillon O., Jubin C., Leonetti P., Magliano M., Maier T. R., Markov G. V., McVeigh P., Pesole G., Poulain J., Robinson-Rechavi M., Sallet E., Ségurens B., Steinbach D., Tytgat T., Ugarte E., van Ghelder C., Veronico P., Baum T. J., Blaxter M., Bleve-Zacheo T., Davis E. L., Ewbank J. J., Favery B., Grenier E., Henrissat B., Jones J. T., Laudet V., Maule A. G., Quesneville H., Rosso M. N., Schiex T., Smant G., Weissenbach J., Wincker P. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915 10.1038/nbt.1482 [DOI] [PubMed] [Google Scholar]
- Adoutte A., Balavoine G., Lartillot N., Lespinet O., Prud'homme B., de Rosa R. (2000). The new animal phylogeny: reliability and implications. Proc. Natl. Acad. Sci. U.S.A. 97, 4453–4456 10.1073/pnas.97.9.4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguinaldo A. M. A., Turbeville J. M., Lindford L. S., Rivera M. C., Garey J. R., Raff R. A., Lake J. A. (1997). Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 387, 489–493 10.1038/387489a0 [DOI] [PubMed] [Google Scholar]
- Andersson J. O. (2005). Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 62, 1182–1197 10.1007/s00018-005-4539-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J. O., Sjogren A. M., Davis L. A. M., Embley T. M., Roger A. J. (2003). Phylogenetic analysis of Diplomonad genes reveals frequent lateral gene transfer affecting eukaryotes. Curr. Biol. 13, 94–104 10.1016/S0960-9822(03)00003-4 [DOI] [PubMed] [Google Scholar]
- Anna T. (2008). The Arthopod Story. Berkeley:University of California; http://evolution.berkeley.edu/evolibrary/article/arthropodstory [Google Scholar]
- Arnold M. (2006). Evolution Through Genetic Exchange. Oxford: Oxford University Press [Google Scholar]
- Arnold M. L., Ballerini E. S., Brothers A. N. (2012). Hybrid fitness, adaptation and evolutionary diversification: lessons learned from Louisiana Irises. Heredity (Edinb.) 108, 159–166 10.1038/hdy.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste E., Boucher Y., Leigh J., Doolittle W. F. (2004). Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 12, 406–411 10.1016/j.tim.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Beagley C. T., Okada N. A., Wolstenholme D. R. (1996). Two mitochondrial group I introns in a metazoan, the sea anemone Metridium senile: one intron contains genes for subunits 1 and 3 of NADH dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 93, 5619–5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi V. A., Levine A. J., Skalka A. M. (2010). Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 6:e1001030 10.1371/journal.ppat.1001030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom J. (1985). Metazoan evolution—a new model. Zool. Scr. 15, 189–200 [Google Scholar]
- Bilewitch J. P., Degnan S. M. (2011). A unique horizontal gene transfer event has provided the octocoral mitochondrial genome with an active mismatch repair gene that has potential for an unusual self-contained function. BMC Evol. Biol. 11, 228 10.1186/1471-2148-11-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B., Schustak J., Badge R. M., Lutz-Prigge S., Farley A. H., Moran J. V., Kazazian H. H. (2003). Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 100, 5280–5285 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet F., Godin F., David J. R., Capy P. (1994). The mariner transposable element in the Drosophilidae family. Heredity (Edinb.) 73, 377–385 [DOI] [PubMed] [Google Scholar]
- Brown F. D., Prendergast A., Swalla B. J. (2008). Man is but a worm: chordate origins. Genesis 46, 605–613 10.1002/dvg.20471 [DOI] [PubMed] [Google Scholar]
- Brown J. R., Douady C. J., Italia M. J., Marshall W. E., Stanhope M. J. (2001). Universal trees based on large combined protein sequence datasets. Nat. Genet. 28, 281–285 10.1038/90129 [DOI] [PubMed] [Google Scholar]
- Capy P. (1998). “Classification of transposable elements,” in Dynamics And Evolution of Transposable Elements, eds Capy P., Bazin C., Higuet D., Langin T. (Austin: Landes Bioscience; ), 37–52 [Google Scholar]
- Carlton J. M., Hirt R. P., Silva J. C., Delcher A. L., Schatz M., Zhao Q., Wortman J. R., Bidwell S. L., Alsmark U. C., Besteiro S., Sicheritz-Ponten T., Noel C. J., Dacks J. B., Foster P. G., Simillion C., Van de Peer Y., Miranda-Saavedra D., Barton G. J., Westrop G. D., Muller S., Dessi D., Fiori P. L., Ren Q., Paulsen I., Zhang H., Bastida-Corcuera F. D., Simoes-Barbosa A., Brown M. T., Hayes R. D., Mukherjee M., Okumura C. Y., Schneider R., Smith A. J., Vanacova S., Villalvazo M., Haas B. J., Pertea M., Feldblyum T. V., Utterback T. R., Shu C. L., Osoegawa K., de Jong P. J., Hrdy I., Horvathova L., Zubacova Z., Dolezal P., Malik S. B., Logsdon J. M. Jr., Henze K., Gupta A., Wang C. C., Dunne R. L., Upcroft J. A., Upcroft P., White O., Salzberg S. L., Tang P., Chiu C. H., Lee Y. S., Embley T. M., Coombs G. H., Mottram J. C., Tachezy J., Fraser-Liggett C. M., Johnson P. J. (2007). Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315, 207–212 10.1126/science.1132894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier Smith T. (1998). A revised six-kingdom system of life. Biol. Rev. 73, 203–266 [DOI] [PubMed] [Google Scholar]
- Chapman J. A., Kirkness E. F., Simakov O., Hampson S. E., Mitros T., Weinmaier T., Rattei T., Balasubramanian P. G., Borman J., Busam D., Disbennett K., Pfannkoch C., Sumin N., Sutton G. G., Viswanathan L. D., Walenz B., Goodstein D. M., Hellsten U., Kawashima T., Prochnik S. E., Putnam N. H., Shu S., Blumberg B., Dana C. E., Gee L., Kibler D. F., Law L., Lindgens D., Martinez D. E., Peng J., Wigge P. A., Bertulat B., Guder C., Nakamura Y., Ozbek S., Watanabe H., Khalturin K., Hemmrich G., Franke A., Augustin R., Fraune S., Hayakawa E., Hayakawa S., Hirose M., Hwang J. S., Ikeo K., Nishimiya-Fujisawa C., Ogura A., Takahashi T., Steinmetz P. R., Zhang X., Aufschnaiter R., Eder M. K., Gorny A. K., Salvenmoser W., Heimberg A. M., Wheeler B. M., Peterson K. J., Böttger A., Tischler P., Wolf A., Gojobori T., Remington K. A., Strausberg R. L., Venter J. C., Technau U., Hobmayer B., Bosch T. C., Holstein T. W., Fujisawa T., Bode H. R., David C. N., Rokhsar D. S., Steele R. E. (2010). The dynamic genome of Hydra. Nature 464, 592–596 10.1038/nature08830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G. D., Beiko R. G., Ragan M. A., Charlebois R. L. (2002). Inferring genome trees by using a filter to eliminate phylogenetically discordant sequences and a distance matrix based on mean normalized BLASTP scores. J. Bacteriol. 184, 2072–2080 10.1128/JB.184.8.2072-2080.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A. G. (2002). Phylogeny of Medusozoa and the evolution of cnidarian life cycles. J. Evol. Biol. 15, 418–432 [Google Scholar]
- Collins A. G., Cartwright P., McFadden C. S., Schierwater B. (2005). Phylogenetic context and Basal metazoan model systems. Integr. Comp. Biol. 45, 585–594 10.1093/icb/45.4.585 [DOI] [PubMed] [Google Scholar]
- Cordonnier A., Casella J. F., Heidmann T. (1995). Isolation of novel human endogenous retrovirus-like elements with foamy virus-related pol sequence. J. Virol. 69, 5890–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J. P., Bekal S., Hudson M., Domier L., Niblack T., Lambert K. N. (2008). Analysis of a horizontally transferred pathway involved in vitamin B-6 biosynthesis from the soybean cyst nematode Heterodera glycines. Mol. Biol. Evol. 25, 2085–2098 10.1093/molbev/msn141 [DOI] [PubMed] [Google Scholar]
- Craig N. L., Craigie R., Gellert M., Lambowitz A. M. (2002). Mobile DNA II. Washington: American Society for Microbiology [Google Scholar]
- Cutler B. (1980). Arthropod cuticle features and arthropod monophyly. Cell. Mol. Life Sci. 36, 953 [Google Scholar]
- Danchin E. G., Rosso M. N., Vieira P., de Almeida-Engler J., Coutinho P. M., Henrissat B., Abad P. (2010). Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc. Natl. Acad. Sci. U.S.A. 107, 17651–17656 10.1073/pnas.1008486107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels S. B., Peterson K. R., Strausbaugh L. D., Kidwell M. G., Chovnick A. (1990). Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 124, 339–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleuze G., Guattari F. (1976). Rhizome. Paris: Les éditions de minuit [Google Scholar]
- Delsuc F., Brinkmann H., Chourrout D., Philippe H. (2006). Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–668 [DOI] [PubMed] [Google Scholar]
- Denker E., Bapteste E., Le Guyader H., Manuel M., Rabet N. (2008). Horizontal gene transfer and the evolution of cnidarian stinging cells. Curr. Biol. 18, R858–R859 10.1016/j.cub.2008.07.031 [DOI] [PubMed] [Google Scholar]
- Doolittle R. F. (1981). Similar amino acid sequences: chance or common ancestry? Science 214, 149–159 10.1126/science.7280687 [DOI] [PubMed] [Google Scholar]
- Doolittle W. F. (1999). Phylogenetic classification and the universal tree. Science 284, 2124–2128 [DOI] [PubMed] [Google Scholar]
- Doolittle W. F., Logsdon J. M. Jr. (1998). Do Archaea have a mixed heritage? Curr. Biol. 8, R209–R211 10.1016/S0960-9822(98)70127-7 [DOI] [PubMed] [Google Scholar]
- Dunn C. W., Hejnol A., Matus D. Q., Pang K., Browne W. E., Smith S. A., Seaver E., Rouse G. W., Obst M., Edgecombe G. D., Sørensen M. V., Haddock S. H., Schmidt-Rhaesa A., Okusu A., Kristensen R. M., Wheeler W. C., Martindale M. Q., Giribet G. (2008). Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749 10.1038/nature06614 [DOI] [PubMed] [Google Scholar]
- Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., Gill S. R., Nelson K. E., Relman D. A. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgecombe G. D., Wilson G. D. F., Colgan D. J., Gray M. R., Cassis G. (2000). Arthropod cladistics: combined analysis of histone H3 and U2 snRNA sequences and morphology. Cladistics 16, 155–203 [DOI] [PubMed] [Google Scholar]
- Edgecombe G. D., Giribet G., Dunn C. W., Hejnol A., Kristensen R. M., Neves R. C., Rouse G. W., Worsaae K., Sørensen M. V. (2011). Higher-level metazoan relationships: recent progress and remaining questions. Org. Divers. Evol. 11, 151–172 [Google Scholar]
- Engels W. R. (1997). Invasions of P elements. Genetics 145, 11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ereskovsky A. V., Dondua A. K. (2006). The problem of germ layers in sponges (Porifera) and some issues concerning early metazoan evolution. Zool. Anz. 245, 65–76 [Google Scholar]
- Faguy D. M., Doolittle W. F. (1999). Lessons from the Aeropyrum pernix genome. Curr. Biol. 9, R883–R886 10.1016/S0960-9822(00)80074-3 [DOI] [PubMed] [Google Scholar]
- Fenn K., Conlon C., Jones M., Quail M. A., Holroyd N. E., Parkhill J., Blaxter M. (2006). Phylogenetic relationships of the Wolbachia of nematodes and arthropods. PLoS Pathog. 2:e94 10.1371/journal.ppat.0020094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1974). The evolutionary advantage of recombination. Genetics 78, 737–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field K. G., Olsen G. J., Lane D. J., Giovannoni S. J., Ghiselin M. T., Raff E. C., Pace N. R., Raff R. A. (1988). Molecular phylogeny of the animal kingdom. Science 239, 748–753 10.1126/science.3277277 [DOI] [PubMed] [Google Scholar]
- Finnegan D. J. (1989). Eukaryotic transposable elements and genome evolution. Trends Genet. 5, 103–107 [DOI] [PubMed] [Google Scholar]
- Fitz-Gibbon S. T., House C. H. (1999). Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 27, 4218–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaneto D., Herniou E. A., Boschetti C., Caprioli M., Melone G., Ricci C., Barraclough T. G. (2007). Independently evolving species in asexual bdelloid rotifers. PLoS Biol. 5:e87. 10.1371/journal.pbio.0050087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J., Zablen L. B., Blakemore R., Gupta R., Bonen L., Lewis B. J., Stahl D. A., Luehrsen K. R., Chen K. N., Woese C. R. (1980). The phylogeny of prokaryotes. Science 209, 457–463 10.1126/science.6771870 [DOI] [PubMed] [Google Scholar]
- Garey J. R. (2001). Ecdysozoa: the relationship between Cycloneuralia and Panarthropoda. Zool. Anz. 240, 321–330 [Google Scholar]
- Gifford R. J., Katzourakis A., Tristem M., Pybus O. G., Winter M., Shafer R. W. (2008). A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 20362–20367 10.1073/pnas.0807873105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C., Maxfield D. G., Goodman S. M., Feschotte C. (2009). Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 5:e1000425 10.1371/journal.pgen.1000425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. R., Pop M., Deboy R. T., Eckburg P. B., Turnbaugh P. J., Samuel B. S., Gordon J. I., Relman D. A., Fraser-Liggett C. M., Nelson K. E. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359 10.1126/science.1124234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giribet G. (2003). Molecules, development and fossils in the study of metazoan evolution; Articulata versus Ecdysozoa revisited. Zoology 106, 303–326 10.1078/0944-2006-00131 [DOI] [PubMed] [Google Scholar]
- Giribet G. (2008). Assembling the lophotrochozoan (=spiralian) tree of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 1513–1522 10.1098/rstb.2007.2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giribet G., Distel D. L., Polz M., Sterrer W., Wheeler W. C. (2000). Triploblastic relationships with emphasis on the acoelomates and the position of Gnathostomulida, Cycliophora, Plathelminthes, and Chaetognatha: a combined approach using 18S rDNA sequences and morphology. Syst. Biol. 49, 539–562 [DOI] [PubMed] [Google Scholar]
- Giribet G., Dunn C. W., Edgecombe G. D., Hejnol A., Martindale M. Q., Rouse G. W. (2009). “Assembling the spiralian tree of life,” in Animal Evolution: Genes, Genomes, Fossils and Trees, eds Telford M. J., Littlewood D. T. J. (Oxford: Oxford University Press; ), 53–64 10.1098/rstb.2007.2241 [DOI] [Google Scholar]
- Giribet G., Dunn C. W., Edgecombe G. D., Rouse G. W. (2007). A modern look at the Animal Tree of Life. Zootaxa 1668, 61–79 [Google Scholar]
- Gladyshev E. A., Meselson M., Arkhipova I. R. (2008). Massive horizontal gene transfer in bdelloid rotifers. Science 320, 1210–1213 10.1126/science.1156407 [DOI] [PubMed] [Google Scholar]
- Goldenfeld N., Woese C. (2007). Biology's next revolution. Nature 445, 369 10.1038/445369a [DOI] [PubMed] [Google Scholar]
- Gonzalez P., Lessios H. A. (1999). Evolution of sea urchin retroviral-like (SURL) elements: evidence from 40 echinoid species. Mol. Biol. Evol. 16, 938–952 [DOI] [PubMed] [Google Scholar]
- Goodier J. L., Ostertag E. M., Du K., Kazazian H. H. Jr. (2001). A novel active L1 etrotransposon subfamily in the mouse. Genome Res. 11, 1677–1685 10.1101/gr.198301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U., Charlebois R. L., Doolittle W. F. (2004). Have archaeal genes contributed to bacterial virulence? Trends Microbiol. 12, 213–219 10.1016/j.tim.2004.03.002 [DOI] [PubMed] [Google Scholar]
- Gourbière S., Mallet J. (2010). Are species real? The shape of the species boundary with exponential failure, reinforcement, and the “missing snowball”. Evolution. 64, 1–24 10.1111/j.1558-5646.2009.00844.x [DOI] [PubMed] [Google Scholar]
- Graham L. A., Lougheed S. C., Ewart K. V., Davies P. L. (2008). Lateral transfer of a lectin-like antifreeze protein gene in fishes. PLoS One 3:e2616. 10.1371/journal.pone.0002616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunau C., Boissier J. (2010). No evidence for lateral gene transfer between salmonids and schistosomes. Nat. Genet. 42, 918–919 10.1038/ng1110-918 [DOI] [PubMed] [Google Scholar]
- Guljamow A., Jenke-Kodama H., Saumweber H., Quillardet P., Frangeul L., Castets A. M., Bouchier A. M., Tandeau de Marsac N., Dittmann E. (2007). Horizontal gene transfer of two cytoskeletal elements from a Eukaryote to a Cyanobacterium. Curr. Biol. 17, R757–R759 10.1016/j.cub.2007.06.063 [DOI] [PubMed] [Google Scholar]
- Haeckel E. (1866). Generelle Morphologie der Organismen. Allgemeine Grundzüge der Organischen Formen-Wissen-Schaft, Mechanisch Begründet Durch die von Charles Darwin Reformirte Descendenztheorie, vol. 2 Berlin: Georg Reimer, 574–462 [Google Scholar]
- Haeckel E. (1874). Die Gastraea-Theorie, die phylogenetische Classification des Tierreichs und die Homologie der Keimblätter. Z. Naturwiss. Jena 8, 1–55 [Google Scholar]
- Haegeman A., Mantelin S., Jones J. T., Gheysen G. (2012). Functional roles of effectors of plant-parasitic nematodes. Gene 492, 19–31 10.1016/j.gene.2011.10.040 [DOI] [PubMed] [Google Scholar]
- Hagemann S., Miller W. J., Pinsker W. (1992). Identification of a complete P-elements in the genome of Drosophila bifasciata. Nucleic Acids Res. 20, 409–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halanych K. M. (2004). The new view of animal phylogeny. Ann. Rev. Ecol. Evol. Syst. 35, 229–256 [Google Scholar]
- Halanych K. M., Bacheller J. M., Aguinaldo A. M. A., Liva S. M., Hillis D. M., Lake J. A. (1995). Evidence from 18S ribosomal DNA that lophophorates are protostome animals. Science 267, 1641–1643 10.1126/science.7886451 [DOI] [PubMed] [Google Scholar]
- Hamada M., Kido Y., Himberg M., Reist J. D., Ying C., Hasegawa M., Okada N. (1997). A newly isolated family of short interspersed repetitive elements (SINEs) in coregonid fishes (whitefish) with sequences that are almost identical to those of the SmaI family of repeats: possible evidence for the horizontal transfer of SINEs. Genetics 146, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M. M., Nitz N., Araujo P. F., Sousa A. O., Rosa Ade C., Gomes D. A., Leonardecz E., Teixeira A. R. (2010). Inheritance of DNA transferred from American trypanosomes to human hosts. PLoS One 5:e9181 10.1371/journal.pone.0009181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann J. H., Correc G., Barbeyron T., Helbert W., Czjzek M., Michel G. (2010). Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912 10.1038/nature08937 [DOI] [PubMed] [Google Scholar]
- Hejnol A., Obst M., Stamatakis A., Ott M., Rouse G. W., Edgecombe G. D., Martinez P., Baguñà J., Bailly X., Jondelius U., Wiens M., Müller W. E., Seaver E., Wheeler W. C., Martindale M. Q., Giribet G., Dunn C. W. (2009). Assessing the root of bilaterian animals with scalable phylogenomic methods. Philos. Trans. R. Soc. Lond. B Biol. Sci. 276, 4261–4270 10.1098/rspb.2009.0896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotopp J. C. D., Clark M. E., Oliveira D. C., Foster J. M., Fischer P., Muñoz Torres M. C., Giebel J. D., Kumar N., Ishmael N., Wang S., Ingram J., Nene R. V., Shepard J., Tomkins J., Richards S., Spiro D. J., Ghedin E., Slatko B. E., Tettelin H., Werren J. H. (2007). Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756 10.1126/science.1142490 [DOI] [PubMed] [Google Scholar]
- Hyman L. H. (1940). The Invertebrates: Protozoa Through Ctenophora. New York, NY: McGraw-Hill Book Company, Inc [Google Scholar]
- International Aphid Genomics Consortium (IAGC). (2010). Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313 10.1371/journal.pbio.1000313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I. K., Matyunina L. V., McDonald J. F. (1999). Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proc. Natl. Acad. Sci. U.S.A. 96, 12621–12625 10.1073/pnas.96.22.12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler O. (1994). “The early diversification of life,” in Early Life on Earth, Nobel Symposium, ed Bengston S. (New York, NY: Columbia University Press; ), 152–509 [Google Scholar]
- Kandler O. (1998). “The early diversification of life: a proposal,” in Thermophiles: The Key to Molecular Evolution and the Origin of Life? eds Adams M., Weigel J. (London: Taylor and Francis, 19–31 [Google Scholar]
- Kapitonov V. V., Jurka J. (2003). Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc. Natl. Acad. Sci. U.S.A. 100, 6569–6574 10.1073/pnas.0732024100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A., Gifford R. J., Tristem M., Gilbert M. T., Pybus O. G. (2009). Macroevolution of complex retroviruses. Science 325, 1512 10.1126/science.1174149 [DOI] [PubMed] [Google Scholar]
- Katzourakis K., Gifford R. J. (2010). Endogenous viral elements in animal genomes. PLoS Genet. 6:e1001191 10.1371/journal.pgen.1001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell M. G., Lisch D. (1997). Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. U.S.A. 94, 7704–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T., Cotton J. A., Dalzell J. J., Hasegawa K., Kanzaki N., McVeigh P., Takanashi T., Tsai I. J., Assefa S. A., Cock P. J., Otto T. D., Hunt M., Reid A. J., Sanchez-Flores A., Tsuchihara K., Yokoi T., Larsson M. C., Miwa J., Maule A. G., Sahashi N., Jones J. T., Berriman M. (2011). Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 7:e1002219 10.1371/journal.ppat.1002219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J. O., Snel B., Huynen M. A., Bork P. (2002). SHOT: a web server for the construction of genome phylogenies. Trends Genet. 18, 158–162 10.1016/S0168-9525(01)02597-5 [DOI] [PubMed] [Google Scholar]
- Kordis D., Gubensek F. (1995). Horizontal SINE transfer between vertebrate classes. Nat. Genet. 10, 131–132 10.1038/ng0695-131 [DOI] [PubMed] [Google Scholar]
- Kunin V., Ahren D., Goldovsky L., Janssen P., Ouzounis C. A. (2005). Measuring genome conservation across taxa: divided strains and United Kingdoms. Nucleic Acids Res. 33, 616–621 10.1093/nar/gki181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam W. L., Seo P., Robison K., Virk S., Gilbert W. (1996). Discovery of amphibian Tc1-like transposon families. J. Mol. Biol. 257, 359–366 10.1006/jmbi.1996.0168 [DOI] [PubMed] [Google Scholar]
- Lambshead P. J. (1993). Recent developments in marine benthic biodiversity research. Oceanis 19, 5–24 [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J. P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann N., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J. C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R. H., Wilson R. K., Hillier L. W., McPherson J. D., Marra M. A., Mardis E. R., Fulton L. A., Chinwalla A. T., Pepin K. H., Gish W. R., Chissoe S. L., Wendl M. C., Delehaunty K. D., Miner T. L., Delehaunty A., Kramer J. B., Cook L. L., Fulton R. S., Johnson D. L., Minx P. J., Clifton S. W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J. F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R. A., Muzny D. M., Scherer S. E., Bouck J. B., Sodergren E. J., Worley K. C., Rives C. M., Gorrell J. H., Metzker M. L., Naylor S. L., Kucherlapati R. S., Nelson D. L., Weinstock G. M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D. R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H. M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R. W., Federspiel N. A., Abola A. P., Proctor M. J., Myers R. M., Schmutz J., Dickson M., Grimwood J., Cox D. R., Olson M. V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G. A., Athanasiou M., Schultz R., Roe B. A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W. R., de la Bastide M., Dedhia N., Blöcker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J. A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D. G., Burge C. B., Cerutti L., Chen H. C., Church D., Clamp M., Copley R. R., Doerks T., Eddy S. R., Eichler E. E., Furey T. S., Galagan J., Gilbert J. G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L. S., Jones T. A., Kasif S., Kaspryzk A., Kennedy S., Kent W. J., Kitts P., Koonin E. V., Korf I., Kulp D., Lancet D., Lowe T. M., McLysaght A., Mikkelsen T., Moran J. V., Mulder N., Pollara V. J., Ponting C. P., Schuler G., Schultz J., Slater G., Smit A. F., Stupka E., Szustakowski J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y. I., Wolfe K. H., Yang S. P., Yeh R. F., Collins F., Guyer M. S., Peterson J., Felsenfeld A., Wetterstrand K. A., Patrinos A., Morgan M. J., de Jong P., Catanese J. J., Osoegawa K., Shizuya H., Choi S., Chen Y. J. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lester C. H., Frimodt-Moller N., Sorensen T. L., Monnet D. L., Hammerum A. M. (2006). In vivo transfer of the vanA resistance gene froman Enterococcus faecium isolate of animal origin to an E. faecium isolate of human origin in the intestines of human volunteers. Antimicrob. Agents Chemother. 50, 596–599 10.1128/AAC.50.2.596-599.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R. W., Ganesan R., Houtchens K., Tolar L. A., Sheen F. M. (1993). Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75, 1083–1093 10.1016/0092-8674(93)90318-K [DOI] [PubMed] [Google Scholar]
- Li Z. W., Shen Y. H., Xiang Z. H., Zhang Z. (2011). Pathogen-origin horizontally transferred genes contribute to the evolution of Lepidopteran insects. BMC Evol. Biol. 11, 356 10.1186/1471-2148-11-356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Gerstein M. (2000). Whole-genome trees based on the occurrence of folds and orthologs: implications for comparing genomes on different levels. Genome Res. 10, 808–818 10.1101/gr.10.6.808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnaeus C. (1758). Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis, vol. 1 Holmiae: Laurentii Salvii, 824 [Google Scholar]
- Loftus B., Anderson I., Davies R., Alsmark U. C., Samuelson J., Amedeo P., Roncaglia P., Berriman M., Hirt R. P., Mann B. J., Nozaki T., Suh B., Pop M., Duchene M., Ackers J., Tannich E., Leippe M., Hofer M., Bruchhaus I., Willhoeft U., Bhattacharya A., Chillingworth T., Churcher C., Hance Z., Harris B., Harris D., Jagels K., Moule S., Mungall K., Ormond D., Squares R., Whitehead S., Quail M. A., Rabbinowitsch E., Norbertczak H., Price C., Wang Z., Guillén N., Gilchrist C., Stroup S. E., Bhattacharya S., Lohia A., Foster P. G., Sicheritz-Ponten T., Weber C., Singh U., Mukherjee C., El-Sayed N. M., Petri W. A. Jr., Clark C. G., Embley T. M., Barrell B., Fraser C. M., Hall N. (2005). The genome of the protist parasite Entamoeba histolytica. Nature 433, 865–868 10.1038/nature03291 [DOI] [PubMed] [Google Scholar]
- Lohe A. R., Moriyama E. N., Lidholm D. A., Hartl D. L. (1995). Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol. Biol. Evol. 12, 62–72 [DOI] [PubMed] [Google Scholar]
- Mallatt J., Winchell C. J. (2007). Ribosomal RNA genes and deuterostome phylogeny revisited: more cyclostomes, elasmobranchs, reptiles, and a brittle star. Mol. Phylogenet. Evol. 43, 1005–1022 10.1016/j.ympev.2006.11.023 [DOI] [PubMed] [Google Scholar]
- Mallet J. (2005). Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237 10.1016/j.tree.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Mark Welch D., Meselson M. (2000). Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science 288, 1211–1215 [DOI] [PubMed] [Google Scholar]
- Maruyama K., Hartl D. L. (1991). Evidence for interspecific transfer of the transposable element mariner between Drosophila and Zaprionus. J. Mol. Evol. 33, 514–524 [DOI] [PubMed] [Google Scholar]
- Matveev V., Okada N. (2009). Retroposons of salmonoid fishes (Actinopterygii: Salmonoidei) and their evolution. Gene 434, 16–28 10.1016/j.gene.2008.04.022 [DOI] [PubMed] [Google Scholar]
- Mayer W. E., Schuster L. N., Bartelmes G., Dieterich C., Sommer R. J. (2011). Horizontal gene transfer of microbial cellulases into nematode genomes is associated with functional assimilation and gene turnover. BMC Evol. Biol. 11, 13 10.1186/1471-2148-11-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. (1956). Controlling elements and the gene. Cold Spring Harb. Symp. Quant. Biol. 21, 197–216 [DOI] [PubMed] [Google Scholar]
- Medina M., Collins A. G., Silberman J. D., Sogin M. L. (2001). Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc. Natl. Acad. Sci. U.S.A. 98, 9707–9712 10.1073/pnas.171316998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. A., Harley J. P. (2007). Zoology, 7th Edn. New York, NY: McGraw-Hill Higher Education, 297 [Google Scholar]
- Mizrokhi L. J., Mazo A. M. (1990). Evidence for horizontal transmission of the mobile element jockey between distant Drosophila species. Proc. Natl. Acad. Sci. U.S.A. 87, 9216–9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J. (1932). Some genetic aspects of sex. Am. Nat. 66, 118–138 [Google Scholar]
- Muller H. J. (1964). The relation of recombination to mutational advance. Mutat. Res. 106, 2–9 [DOI] [PubMed] [Google Scholar]
- Muller W. E. G. (2003). The origin of metazoan complexity: Porifera as integrated animals. Integr. Comp. Biol. 43, 3–10 10.1093/icb/43.1.3 [DOI] [PubMed] [Google Scholar]
- Moran N. A., Jarvik T. (2010). Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 328, 624–627 10.1126/science.1187113 [DOI] [PubMed] [Google Scholar]
- Morrison H. G., McArthur A. G., Gillin F. D., Aley S. B., Adam R. D., Olsen G. J., Best A. A., Cande W. Z., Chen F., Cipriano M. J., Davids B. J., Dawson S. C., Elmendorf H. G., Hehl A. B., Holder M. E., Huse S. M., Kim U. U., Lasek-Nesselquist E., Manning G., Nigam A., Nixon J. E., Palm D., Passamaneck N. E., Prabhu A., Reich C. I., Reiner D. S., Samuelson J., Svard S. G., Sogin M. L. (2007). Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317, 1921–1926 10.1126/science.1143837 [DOI] [PubMed] [Google Scholar]
- Nedelcu A. M., Miles I. H., Fagir A. M., Karol K. (2008). Adaptive eukaryote-to-eukaryote lateral gene transfer: stress-related genes of algal origin in the closest unicellular relatives of animals. J. Evol. Biol. 21, 1852–1860 10.1111/j.1420-9101.2008.01605.x [DOI] [PubMed] [Google Scholar]
- Nielsen C. (2001). Animal Evolution, Interrelationships of the Living Phyla, 2nd edn. Oxford: Oxford University Press, 563 [Google Scholar]
- Nikoh N., McCutcheon J. P., Kudo T., Miyagishima S. Y., Moran N. A., Nakabachi A. (2010). Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 6:e1000827 10.1371/journal.pgen.1000827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Lawrence J. G., Grolsman E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- Pace J. K. I. I., Gilbert C., Clark M. S., Feschotte C. (2008). Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc. Natl. Acad. Sci. U.S.A. 105, 17023–17028 10.1073/pnas.0806548105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechenik J. A. (2005). Biology of the Invertebrates. Boston: McGraw-Hill, Higher Education, 178 [Google Scholar]
- Philippe J. (1997). “Craniata. Animals with skulls,” in The Tree of Life Web Project, http://tolweb.org [Google Scholar]
- Philippe H., Derelle R., Lopez P., Pick K., Borchiellini C., Boury-Esnault N., Vacelet J., Renard E., Houliston E., Quéinnec E., Da Silva C., Wincker P., Le Guyader H., Leys S., Jackson D. J., Schreiber F., Erpenbeck D., Morgenstern B., Wörheide G., Manuel M. (2009). Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 19, 706–712 10.1016/j.cub.2009.02.052 [DOI] [PubMed] [Google Scholar]
- Peterson K. J., Butterfield N. J. (2005). Origin of the Eumetazoa: testing ecological predictions of molecular clocks against the Proterozoic fossil record. Proc. Natl. Acad. Sci. U.S.A. 102, 9547–9552 10.1073/pnas.0503660102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K. J., Eernisse D. J. (2001). Animal phylogeny and the ancestry of bilaterians: inferences from morphology and 18S rDNA gene sequences. Evol. Dev. 3, 170–205 [DOI] [PubMed] [Google Scholar]
- Price T. D., Bouvier M. M. (2002). The evolution of F1 postzygotic incompatibilities in birds. Evolution 56, 2083–2089 [PubMed] [Google Scholar]
- Priimagi A. F., Mizrokhi L. J., Ilyin Y. V. (1988). The Drosophila mobile element jockey belongs to LINEs and contains coding sequences homologous to some retroviral proteins. Gene 70, 253–262 10.1016/0378-1119(88)90197-7 [DOI] [PubMed] [Google Scholar]
- Puigbo P., Wolf Y. I., Koonin E. V. (2009). Search for a ‘Tree of Life’ in the thicket of the phylogenetic forest. J. Biol. 8, 59 10.1186/jbiol159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P., Wolf Y. I., Koonin E. V. (2010). The tree and net components of prokaryote evolution. Genome Biol. Evol. 2, 745–756 10.1093/gbe/evq062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa O., Hazkani-Covo E., Landan G., Martin W., Dagan T. (2011). Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 21, 599–609 10.1101/gr.115592.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan M. A., McInerney J. O., Lake J. A. (2009). The network of life: genome beginnings and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2169–2175 10.1098/rstb.2009.0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D. (2010). The post-Darwinist rhizome of life. Lancet 375, 104–105 10.1016/S0140-6736(09)61958-9 [DOI] [PubMed] [Google Scholar]
- Richards T. A., Soanes D. M., Jones M. D., Vasieva O., Leonard G., Paszkiewicz K., Foster P. G., Hall N., Talbot N. J. (2011). Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl. Acad. Sci. U.S.A. 108, 15258–15263 10.1073/pnas.1105100108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera M. C., Lake J. A. (2004). The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431, 152–155 10.1038/nature02848 [DOI] [PubMed] [Google Scholar]
- Robertson H. M. (1997). Multiple mariner transposons in flatworms and hydras are related to those of insects. J. Hered. 88, 195–201 [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Lampe D. J. (1995). Recent horizontal transfer of a mariner transposable element among and between Diptera and Neuroptera. Mol. Biol. Evol. 12, 850–862 [DOI] [PubMed] [Google Scholar]
- Robertson H. M., MacLeod E. G. (1993). Five major subfamilies of mariner transposable elements in insects, including the Mediterranean fruit fly, and related arthropods. Insect Mol. Biol. 2, 125–139 [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Soto-Adames F. N., Walden K. K., Avancini R. M., Lampe D. J. (2002). “The mariner transposons of animals: horizontally jumping genes,” in Horizontal Gene Transfer, eds Syvanen M., Kado C. I. (San Diego, CA: Academic Press; ), 173–183 [Google Scholar]
- Robinson C. J., Bohannan B. J., Young V. B. (2010). From structure to function: the ecology of host-associated microbial communities. Microbiol. Mol. Biol. Rev. 74, 453–476 10.1128/MMBR.00014-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C., Sommer R. J. (2011). Computational archaeology of the Pristionchus pacificus genome reveals evidence of horizontal gene transfers from insects. BMC Evol. Biol. 11, 239 10.1186/1471-2148-11-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. (1985). The origin and evolution of retroposons. Int. Rev. Cytol. 93, 187–279 10.1038/372669a0 [DOI] [PubMed] [Google Scholar]
- Rokas A., Williams B. L., King N., Carroll S. B. (2003). Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425, 798–804 10.1038/nature02053 [DOI] [PubMed] [Google Scholar]
- Rot C., Goldfarb I., Ilan M., Huchon D. (2006). Putative cross-kingdom horizontal gene transfer in sponge (Porifera) mitochondria. BMC Evol. Biol. 6, 71 10.1186/1471-2148-6-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roule L. (1891). Considerations sur l'embranchement des Trochozoaires. Ann. Sci. Nat. (Zool.) 7me Série 11, 121–178 [Google Scholar]
- Ruppert E. E., Fox R. S., Barnes R. D. (2004). Invertebrate Zoology, 7th edn. Toronto: Thomson Brooks/Cole [Google Scholar]
- Saunders N. J., Hood D. W., Moxon E. R. (1999). Bacterial evolution: bacteria play pass the gene. Curr. Biol. 9, R180–R183 10.1016/S0960-9822(99)80108-0 [DOI] [PubMed] [Google Scholar]
- Schliep K., Lopez P., Lapointe F. J., Bapteste E. (2010). Harvesting evolutionary signals in a forest of prokaryotic gene trees. Mol. Biol. Evol. 28, 1393–1405 10.1093/molbev/msq323 [DOI] [PubMed] [Google Scholar]
- Schmidt-Rhaesa A., Bartolomaeus T., Lemburg C., Ehlers U., Garey J. R. (1998). The position of the Arthropoda in the phylogenetic system. J. Morphol. 238, 263–285 [DOI] [PubMed] [Google Scholar]
- Seehausen O. (2004). Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207 10.1016/j.tree.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Segers H. (2007). Annotated checklist of the rotifers (Phylum Rotifera), with notes on nomenclature, taxonomy and distribution. Zootaxa 1564, 1–104 10.3897/zookeys.66.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C. S., Smith M. B., Friedman J., Cordero O. X., David L. A., Alm E. J. (2011). Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244 10.1038/nature10571 [DOI] [PubMed] [Google Scholar]
- Smit A. F., Riggs A. D. (1996). Tiggers and DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. U.S.A. 93, 1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath P. H. A. (1975). “Cladistic representation of reticulate evolution,” in Systematic Zoology, (London: Taylor and Francis, Ltd.), 360–368 [Google Scholar]
- Snel B., Bork P., Huynen M. A. (1999). Genome phylogeny based on gene content. Nat. Genet. 21, 108–110 10.1038/5052 [DOI] [PubMed] [Google Scholar]
- Syvanen M., Ducore J. (2010). Whole genome comparisons reveals a possible chimeric origin for a major Metazoan assemblage. J. Biol. Syst. 18, 261–275 [Google Scholar]
- Tekaia F., Lazcano A., Dujon B. (1999). The genomic tree as revealed from whole proteome comparisons. Genome Res. 9, 550–557 10.1101/gr.9.6.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford M. J., Bourlat S. J., Economou A., Papillon D., Rota Stabelli O. (2008). The evolution of the Ecdysozoa. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 1529–1537 10.1098/rstb.2007.2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzian C., Ferraz C., Demaille J., Bucheton A. (2000). Evolution of the Gypsy endogenous retrovirus in the Drosophila melanogaster subgroup. Mol. Biol. Evol. 17, 908–914 [DOI] [PubMed] [Google Scholar]
- Thomas J., Schaack S., Pritham E. J. (2010). Pervasive horizontal transfer of rolling-circle transposons among animals. Genome Biol. Evol. 2, 656–664 10.1093/gbe/evq050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristem M. (2000). Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J. Virol. 74, 3715–3730 10.1128/JVI.74.8.3715-3730.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udomkit A., Forbes S., Dalgleish G., Finnegan D. J. (1995). BS a novel LINE-like element in Drosophila melanogaster. Nucleic Acids Res. 23, 1354–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine J. W. (1997). Cleavage patterns and the topology of the metazoan tree of life. Proc. Natl. Acad. Sci. U.S.A. 94, 8001–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine J. W. (2004). On the Origin of Phyla. Chicago: University of Chicago Press, 33 [Google Scholar]
- van Oppen M. J. H., Catmull J., McDonald B. J., Hislop N. R., Hagerman P. J., Miller D. J. (2002). The mitochondrial genome of Acropora tenuis (Cnidaria: Scleractinia) contains a large group I intron and a candidate control region. J. Mol. Evol. 55, 1–13 10.1007/s00239-001-0075-0 [DOI] [PubMed] [Google Scholar]
- Vazquez-Manrique R. P., Hernandez M., Martínez-Sebastián M. J., de Frutos R. (2000). Evolution of gypsy endogenous retrovirus in the Drosophila obscura species group. Mol. Biol. Evol. 17, 1185–1193 [DOI] [PubMed] [Google Scholar]
- Volff J. N., Korting C., Schartl M. (2000). Multiple lineages of the non-LTR retrotransposon Rex1 with varying success in invading fish genomes. Mol. Biol. Evol. 17, 1673–1684 [DOI] [PubMed] [Google Scholar]
- Wallace R. L. (1998). “Rotifera,” in Encyclopedia of Reproduction, eds Knobil E., Neil J. D. (San Diego: Academic Press; ), 118–129 [Google Scholar]
- Wallace R. L., Snell T. W., Ricci C. (2006). “Rotifera, biology, ecology and systematics,” in Guides to the Identification of the Microinvertebrates of the Continental Waters of the World, ed Segers H. Ghent. (Kenobi Productions, Leiden: Backhuys Publishers; ), 23 [Google Scholar]
- Watkins R. F., Gray M. W. (2006). The frequency of eubacterium-to-eukaryote lateral gene transfer shows significant cross-taxa variation within Amoebozoa. J. Mol. Evol. 63, 801–814 10.1007/s00239-006-0031-0 [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. (1986). Nonviral retroposons: genes, pseudogenes and transposable elements generated by the reverse flow of genetic information. Annu. Rev. Biochem. 55, 631–661 10.1146/annurev.bi.55.070186.003215 [DOI] [PubMed] [Google Scholar]
- Wessler S. R. (1998). Transposable elements and the evolution of gene expression. Symp. Soc. Exp. Biol. 51, 115–122 [PubMed] [Google Scholar]
- Winchell C. J., Sullivan J., Cameron C. B., Swalla B. J., Mallatt J. (2002). Evaluating hypotheses of deuterostome phylogeny and chordate evolution with new LSU and SSU ribosomal DNA data. Mol. Biol. Evol. 19, 762–776 [DOI] [PubMed] [Google Scholar]
- Woese C. R. (2002). On the evolution of cells. Proc. Natl. Acad. Sci. U.S.A. 99, 8742–8874 10.1073/pnas.132266999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Mahowald M. A., Ley R. E., Lozupone C. A., Hamady M., Martens E. C., Henrissat B., Coutinho P. M., Minx P., Latreille P., Cordum H., Van Brunt A., Kim K., Fulton R. S., Fulton L. A., Clifton S. W., Wilson R. K., Knight R. D., Gordon J. I. (2007). Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 5:e156 10.1371/journal.pbio.0050156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Lou M. M., Xie G. L., Zhang G. Q., Zhou X. P., Li B., Jin G. L. (2011). Horizontal gene transfer in silkworm, Bombyx mori. BMC Genomics 12, 248 10.1186/1471-2164-12-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W., Palm P., Klenk H. P. (1992). “A model of the early evolution of organisms: the arisal of the three domains of life from the common ancestor,” in The Origin and Evolution of the Cell, eds Hartman H., Matsuno K. (Singapore: World Scientific Publishing; ), 163–182 [Google Scholar]
- Zinner D., Arnold M. L., Roos C. (2011). The strange blood: natural hybridization in primates. Evol. Anthropol. 20, 96–103 10.1002/evan.20301 [DOI] [PubMed] [Google Scholar]
- Zrzavý J. (2003). Gastrotricha and metazoan phylogeny. Zool. Scr. 32, 61–81 [Google Scholar]
- Zupunski V., Gubensek F., Kordis D. (2001). Evolutionary dynamics and evolutionary history in the RTE clade of non-LTR retrotransposons. Mol. Biol. Evol. 18, 1849–1863 [DOI] [PubMed] [Google Scholar]


