Abstract
Of 17 genes annotated in the Arabidopsis genome database as cinnamyl alcohol dehydrogenase (CAD) homologues, an in silico analysis revealed that 8 genes were misannotated. Of the remaining nine, six were catalytically competent for NADPH-dependent reduction of p-coumaryl, caffeyl, coniferyl, 5-hydroxyconiferyl, and sinapyl aldehydes, whereas three displayed very low activity and only at very high substrate concentrations. Of the nine putative CADs, two (AtCAD5 and AtCAD4) had the highest activity and homology (≈83% similarity) relative to bona fide CADs from other species. AtCAD5 used all five substrates effectively, whereas AtCAD4 (of lower overall catalytic capacity) poorly used sinapyl aldehyde; the corresponding 270-fold decrease in kenz resulted from higher Km and lower kcat values, respectively. No CAD homologue displayed a specific requirement for sinapyl aldehyde, which was in direct contrast with unfounded claims for a so-called sinapyl alcohol dehydrogenase in angiosperms. AtCAD2, 3, as well as AtCAD7 and 8 (highest homology to sinapyl alcohol dehydrogenase) were catalytically less active overall by at least an order of magnitude, due to increased Km and lower kcat values. Accordingly, alternative and/or bifunctional metabolic roles of these proteins in plant defense cannot be ruled out. Comprehensive analyses of lignified tissues of various Arabidopsis knockout mutants (for AtCAD5, 6, and 9) at different stages of growth/development indicated the presence of functionally redundant CAD metabolic networks. Moreover, disruption of AtCAD5 expression had only a small effect on either overall lignin amounts deposited, or on syringyl-guaiacyl compositions, despite being the most catalytically active form in vitro.
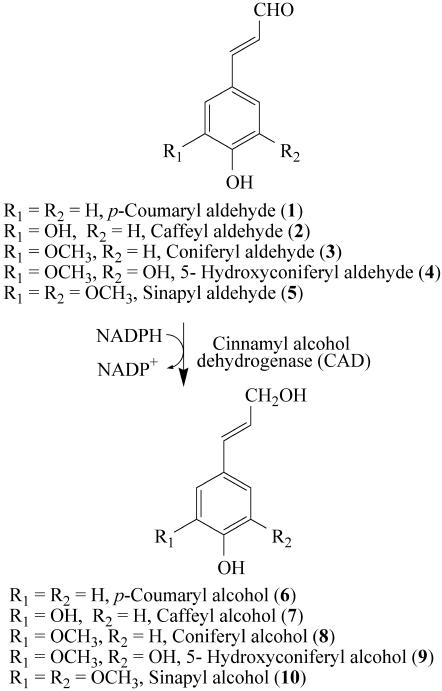
Cinnamyl alcohol dehydrogenase (CAD), discovered by Zenk and coworkers in 1973 (1, 2), depicts a class of NADPH-dependent enzymes catalyzing reduction of various phenylpropenyl aldehyde derivatives 1–5 (Fig. 1). Generically, this conversion affords monolignols 6–10, with the latter mainly being precursors of lignins and lignans (3–5). Since its discovery, various CAD/CAD homologues were reported as multigene families in many plant species. Yet, despite numerous CAD-like genes being described over nearly 15 years, the paucity of detailed protein biochemical characterization is enigmatic. Frequently, they are arbitrarily assigned roles in monolignols 6–10 and lignin formation (6–13), regardless of, for example, low degree of homology to established CADs, and lack of demonstration of biochemical function in vitro and/or physiological role(s).
Fig. 1.
CAD substrates and products.
With the Arabidopsis genome completed (14), 17 genes were annotated as CAD homologues (see Table 1 and Table 3, which is published as supporting information on the PNAS web site). Accordingly, with all putative members potentially identified (15), it was possible to comprehensively examine kinetic properties and substrate versatilities in vitro as a first step to establishing true physiological function(s) (16, 17).
Table 1. Sequence comparisons of an annotated nine-membered CAD multigene family in Arabidopsis to bona fide CADs and a putative SAD.
| Gene annotation
|
Promoter size, kb
|
cDNA, kb
|
Amino acid no.
|
Mm, kDa
|
NtCAD, %
|
PtCAD, %
|
AsCAD, %
|
AsSAD, %
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus no. | Similarity | Identity | Similarity | Identity | Similarity | Identity | Similarity | Identity | |||||
| At4g34230 | AtCAD5 | 1.20 | 1.07 | 357 | 38.7 | 82.9 | 76.5 | 76.5 | 65.8 | 82.9 | 77.9 | 62.6 | 53.1 |
| At3g19450 | AtCAD4 | 1.80 | 1.10 | 365 | 39.1 | 81.5 | 75.1 | 74.0 | 65.8 | 83.5 | 78.7 | 62.5 | 53.3 |
| At4g37990 | AtCAD8 | 1.72 | 1.08 | 359 | 38.9 | 63.4 | 52.7 | 64.6 | 53.4 | 63.8 | 52.0 | 78.0 | 72.1 |
| At2g21730 | AtCAD2 | 2.05 | 1.13 | 376 | 40.9 | 62.4 | 51.7 | 64.7 | 53.1 | 64.0 | 52.7 | 71.1 | 62.6 |
| At2g21890 | AtCAD3 | 2.03 | 1.13 | 375 | 40.9 | 64.0 | 52.1 | 64.4 | 52.8 | 65.6 | 53.1 | 70.2 | 61.5 |
| At4g37980 | AtCAD7 | 0.60 | 1.07 | 357 | 38.2 | 61.4 | 50.1 | 62.9 | 52.0 | 61.4 | 50.4 | 77.6 | 71.4 |
| At4g37970 | AtCAD6 | 0.59 | 1.09 | 363 | 39.0 | 61.3 | 51.0 | 65.5 | 53.5 | 62.5 | 49.3 | 77.3 | 68.5 |
| At4g39330 | AtCAD9 | 1.64 | 1.08 | 360 | 38.9 | 60.2 | 50.7 | 63.6 | 54.1 | 63.9 | 53.8 | 74.4 | 67.5 |
| At1g72680 | AtCAD1 | 0.40 | 1.07 | 355 | 38.7 | 57.1 | 46.0 | 61.0 | 47.5 | 55.1 | 41.2 | 58.6 | 49.3 |
Initially, in silico analysis (18) revealed that only 9 of the 17 genes displayed sufficient homology for consideration as bona fide CADs, six of which are demonstrated herein as being biochemically competent for phenylpropenyl aldehyde 1–5 reduction. These findings, together with analysis of selected CAD knockout lines, strongly suggest that lignin, and presumably also lignan, formation are not dependent on a single CAD isoform, due to the presence of functionally redundant, complex metabolic, networks in different tissues, organs, and cell types in Arabidopsis. Indeed, the data provide more evidence that CAD does not fulfill a key (rate-limiting) regulatory role in lignin deposition, in contrast to various claims (19) that have consistently lacked supporting scientific data (3). Nor did any homologue display a specific substrate preference [i.e., for sinapyl aldehyde (5) or any other aldehyde] as previously claimed essential for syringyl lignin formation in angiosperms (20). Indeed, genes with closest homology to the so-called sinapyl alcohol dehydrogenase (SAD) isoform displayed the poorest ability to use any of the aldehyde substrates examined, and may instead be involved in unrelated defense functions. The present study, thus, also underscores the ongoing difficulties arising from arbitrary annotations [in The Arabidopsis Information Resource (TAIR) database, which can be accessed at www.arabidopsis.org)] of presumed catalytic/physiological functions to various multigene families, such as with CAD, in the Arabidopsis genome (18).
Materials and Methods
Plant Materials. Wild-type and T-DNA mutant Arabidopsis thaliana plants were grown in Washington State University greenhouses.
Materials, Instrumentation, and Chemical Syntheses. For more information, see Supporting Methods, which is published as supporting information on the PNAS web site.
CAD Homologues. The 17 putative CAD homologues were designated AtCAD1–AtCAD9 and AtCAD101–AtCAD108 (see Tables 1 and 3). Each cDNA sequence was obtained from TAIR, and mRNA sequences from this study were deposited in GenBank.
Cloning and Heterologous Expression/Purification of A. thaliana CAD Homologues. For more information, see Supporting Methods.
Enzyme Characterization. Standard assays consisted of 1,3-bis[tris(hydroxymethyl)methylamino]propane (Bistris propane) buffer (100 mM, pH 6.25/100 μl)/130 μl (2–90 μg) of purified CAD in Tris·HCl (20 mM, pH 7.5)/0.4 mM aldehyde 1–5/0.4 mM NADPH, in a total volume of 250 μl. Enzymatic reactions were initiated by enzyme addition and, after 4 min incubation at 30°C, were stopped by adding glacial acetic acid (10 μl); cinnamic acid (6.25 μM, 10 μl) was added as internal standard. An aliquot (80 μl) of each assay mixture was subjected to reversed-phase HPLC analysis with liquid chromatography-MS and UV detection for product identification (data not shown; see Supporting Methods). Assays with individual substrates were performed in triplicate or tetraplicate, with controls in the absence of NADPH.
Optimum pH and temperature were individually determined for each CAD homologue and each aldehyde 1–5 by using standard assay conditions. For pH optima, incubations were carried out at 30°C with Mes buffer (100 mM, pH 5.1–6.8) or Bistris propane buffer (100 mM, pH 6.2–8.0); for temperature optima, incubations were performed at pH 6.25, but with various temperatures (20–50°C). Initial velocity kinetics were determined by individually assaying CAD1–CAD9 under standard conditions at pH 6.25 with 15 different aldehyde 1–5 concentrations (76–400 μM) and at 30°C for 4 min (or 2 min for AtCAD5). Assays were also individually performed by using [4R-3H]NADPH or [4S-3H]NADPH (0.4 mM, 4.5 kBq; ref. 21), respectively, and were analyzed for radiochemical incorporation through stereospecific [3H] transfer into monolignols 6–10 (see Table 4, which is published as supporting information on the PNAS web site).
Knockout Mutants. Insertion mutant information was obtained from the SIGnAL web site, which can be accessed at http://signal.salk.edu (22). After searching the TAIR database, all available T-DNA insertion lines (T3 seeds) of interest were ordered from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University (Columbus, OH): SALK_019355 and SALK_040062 (AtCAD5), and SALK_030496 (AtCAD6) and SALK_037853 (AtCAD9). Homozygous plant lines were obtained as follows: Total genomic DNA was first isolated by using a REDExtract-NAMP plant PCR kit according to the manufacturer's instructions. Left and right gene-specific primers, LP and RP (Table 5, which is published as supporting information on the PNAS web site), were then designed for each CAD gene of interest, as described at the SIGnAL website. Two sets of PCRs were carried out by using the touch-down method (ref. 23; see Supporting Methods): the first with LP and RP primers specific for each gene, as well as with the T-DNA left border primer (LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′) and the second with LP and RP primers only. Homozygous lines were verified by the presence of single bands (≈500 base pairs), which were individually cloned into a pCR4-TOPO vector and sequenced to confirm the T-DNA insertion.
Results and Discussion
Arabidopsis Genome Analysis and Reclassification of Putative CAD Homologues. The first gene purportedly encoding a CAD was reported in 1988 from bean (Phaseolus vulgaris; ref. 6). However, it actually encoded malic enzyme (24, 25), being misidentified in part because of no biochemical confirmation. Other research groups used the malic enzyme cDNA in the mistaken belief that it encoded CAD, such as to study monolignol pathway induction in the rubber tree (Hevea brasiliensis; ref. 26); others still refer to malic enzyme cDNA as encoding CAD (10). The first bona fide CAD gene was reported in 1992 (27) from tobacco (Nicotiana tabacum) stems, with another obtained in 1994 from loblolly pine (Pinus taeda; GenBank accession no. Z37992).
The TAIR database has 17 genes annotated as CAD-like (Tables 1 and 3). In the present study (Figs. 8 and 9, which are published as supporting information on the PNAS web site), the corresponding amino acid sequences for each were aligned against those of both bona fide tobacco (27) and loblolly pine (28) CADs, as well as to a putative CAD and a claimed SAD from aspen (20).
Nine of the 17 putative CADs had relatively high levels of similarity (57.1–82.9% and 61.0–76.5%) and identity (46.0–76.5% and 47.5–65.8%) to both N. tabacum and P. taeda CADs (Table 1), as well as to the putative aspen CAD (≈55–83%) and to the claimed aspen SAD (≈59–78%). Of those nine CADs, AtCAD6–8 had highest similarity (≈78%) and identity (68.5–72.1%) to the claimed aspen SAD. By contrast, the remaining eight (Table 3) shared essentially no homology to either N. tabacum or P. taeda CADs (0.9–1.6% similarity).
Only AtCAD1–AtCAD9 with highest homology to bona fide CADs were chosen for detailed study (see Table 1 and Fig. 8). Relative to tobacco CAD, all had the highly conserved Zn1 catalytic center (C47, H69, and C163), the Zn-binding signature GHEXXGXXXXXGXXV, the Zn2 structural motif (C100, C103, C106, and C114), and the NADPH-binding domain [GLGGV(L)G] motif (so-called Rossmann fold; refs. 4 and 29). Based on homology (18), AtCAD1–9 could be subclassified into two subgroups, with AtCAD5 and AtCAD4 (74% to ≈83% similarity) being most homologous to the bona fide P. taeda/N. tabacum CAD genes; the remaining seven were CAD-like but, of significantly lower homology (57.1–65.5% similarity).
The other eight annotated Arabidopsis CADs [including SAG 26 (11)], had essentially no homology to either tobacco or loblolly pine CADs (see Table 3 and Fig. 9), an observation requiring explanation. In Arabidopsis, SAG 26 annotation with a CAD function was based solely on a 67% similarity/56% identity to an apple (Malus domestica) cDNA claimed to encode a CAD (10), but whose function was actually unknown. Furthermore, whereas gene expression of SAG 26 was considered tightly correlated with senescence onset, as well as being defense-inducible (salicylic acid; ref. 11), there was again no biochemical context established.
Annotation of the eight Arabidopsis genes as being CADs was instead based on a relative degree of homology (82.5–49.8% similarity, see Table 3) to an alcohol dehydrogenase from Eucalyptus gunnii (30), whose precise physiological/biochemical function was actually unknown. The latter was CAD-annotated, even though the corresponding protein had very broad substrate specificity for a variety of aromatic substrates (30). Furthermore, both this gene, and the eight other annotated CADs, encode proteins lacking both the Zn catalytic center and the Zn-binding signature found in bona fide CAD (Fig. 9). It was thus concluded that this CAD annotation was misleading and, accordingly, the eight Arabidopsis genes were not considered further.
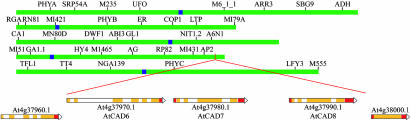
Chromosomal Organization and Cloning. AtCAD1–9 genes are distributed in four of five Arabidopsis chromosomes: chromosome 1 (AtCAD1), chromosome 2 (AtCAD2 and 3), chromosome 3 (AtCAD4), and chromosome 4 (AtCAD5–9), with AtCAD6–8 genes being clustered together (Fig. 2), the significance of which is as yet unknown but potentially most interesting in terms of a defense response (discussed below).
Fig. 2.
Position of AtCAD6–AtCAD8 on chromosome 4 of A. thaliana.
Gene-specific primer sets (Table 6, which is published as supporting information on the PNAS web site) were next designed for each cDNA sequence, these being individually used in PCR amplifications to isolate target genes. To achieve this end, Arabidopsis total RNA was initially isolated from aerial tissues of 41-day-old plants, and by using this as a template, first-strand cDNA was synthesized. PCR amplification by using the touch-down method (23) was then individually performed with first-strand cDNA template, with this method ultimately giving seven of the nine target CAD-like genes (AtCAD1 and AtCAD4–9). AtCAD2 and AtCAD3 genes were also obtained as above, but with total RNA isolated from 26-day-old plants.
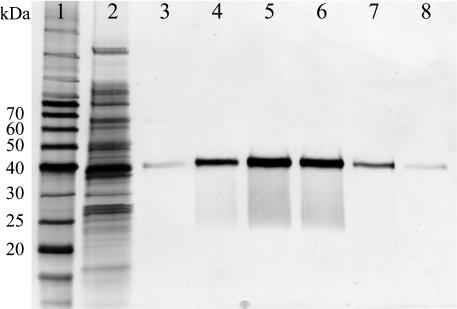
CAD Recombinant Protein Expression and Kinetic Characterization. Crude protein extracts for each CAD homologue (see Supporting Methods) were prepared from cell pellets (derived from 250 ml of culture medium) with, in most cases, ≈150 mg of total protein obtained. Each CAD was individually purified to apparent homogeneity (evaluated by SDS/PAGE with silver staining) after metal chelate affinity column chromatography, with each C-terminal His6-tagged target protein individually eluted between 70 and 230 mM imidazole in Tris·HCl buffer. Fractions were analyzed by SDS/PAGE and those containing the target protein were combined, concentrated, and dialyzed. Typically, ≈2.0 mg of each pure protein was obtained (see Fig. 3, with AtCAD5 as an example). Purified proteins were directly used for enzyme characterization, with those catalytically active being stable up to 5 days at 4°C.
Fig. 3.
Representative purification of CAD, using AtCAD5 for illustrative purposes. Lanes: 1, molecular mass ladder; 2, Escherichia coli crude extract; 3–8, fractions eluted from the metal chelate affinity column between 70 and 230 mM imidazole in Tris·HCl buffer contained pure AtCAD5. Proteins were visualized by silver staining.
Enzyme assays and kinetic measurements were carried out by using all possible substrates 1–5 and products 6–10, in the presence of NADPH (100 μmol), with enzyme activities verified by HPLC analyses with both UV and electron impact MS detection (21, 31–33). [Syntheses of substrates 1–5 and products 6–10 were based on our previous strategies for both monolignols and lignans (see Supporting Methods and Scheme 1, which is published as supporting information on the PNAS web site).] Of the CAD homologues, AtCAD2–5, 7, and 8 were biochemically competent to reduce phenylpropenaldehyde substrates 1–5, with pH and temperature optima of ≈6.25 and 30–40°C, respectively, for each substrate (see Table 4). On the other hand, AtCAD1, 6, and 9 were of very low catalytic activity and only at high protein levels (≈140 μg); i.e., 0.031–0.224, 0.037–0.078, and 0–0.062 pKat per μg of protein, respectively. Furthermore, in an attempt to establish whether low catalytic activity was somehow due to attachment of the C-terminal His tag, N-terminal His-tagged protein (AtCAD1) was also studied. However, this variation had no measurable effect on either substrate versatility or on catalytic efficacy relative to the C-terminal His-tagged analogue. Indeed, because the isoforms (AtCAD1, 6, and 9) were also of the lowest homology relative to AtCAD4 and 5, their low catalytic activity was therefore not unexpected (see Table 1). All of the catalytically active CAD homologues were, however, type A dehydrogenases; i.e., abstracting the pro-R [3H]hydride from [4R-3H]NADPH, but not the corresponding [3H]hydride from [4S-3H]NADPH (ref. 21 and Table 4), which is in agreement with earlier studies by using CAD from Forsythia suspensa (2).
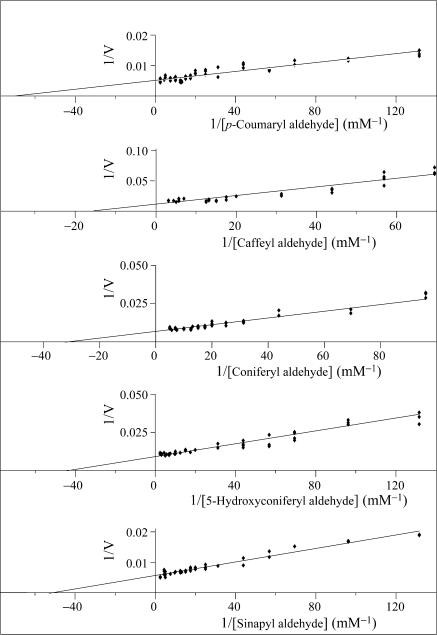
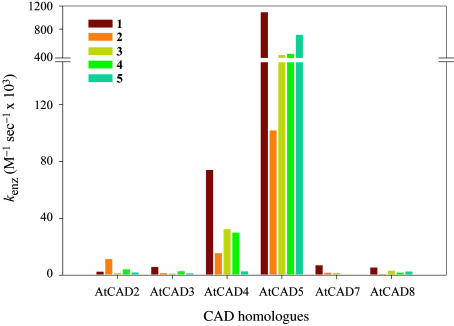
AtCAD5 and AtCAD4 were the most catalytically active overall (see Table 2), with p-coumaryl aldehyde (1) being the preferred substrate for both enzymes. AtCAD5 also effectively used sinapyl (5), coniferyl (3), and 5-hydroxyconiferyl (4) aldehydes, as well to a lesser extent, caffeyl aldehyde (2) (see Figs. 4 and 5), in accordance with the relatively broad substrate versatility of this enzyme class. AtCAD4 also quite readily used substrates 2–4, whereas sinapyl aldehyde (5) was, by comparison, a poor substrate; i.e., indicative of a somewhat more restricted substrate specificity.
Table 2. Kinetic parameters for CAD homologues AtCAD2-5, 7, and 8.
| Substrate | Km, μM | Vmax, pKat/μg | kcat, sec-1 | kenz, M-1·sec-1 | |
|---|---|---|---|---|---|
| AtCAD4 | 1 | 47 | 44.0 | 3.44 | 74,000 |
| 2 | 87 | 17.1 | 1.33 | 15,000 | |
| 3 | 65 | 26.8 | 2.10 | 32,000 | |
| 4 | 85 | 32.3 | 2.52 | 30,000 | |
| 5 | 274 | 9.2 | 0.72 | 2,600 | |
| AtCAD5 | 1 | 13 | 187.3 | 14.52 | 1,091,000 |
| 2 | 68 | 94.1 | 7.30 | 107,000 | |
| 3 | 35 | 157.4 | 12.19 | 348,000 | |
| 4 | 22 | 106.9 | 8.28 | 370,000 | |
| 5 | 20 | 177.0 | 13.72 | 700,000 | |
| AtCAD2 | 1 | 114 | 3.3 | 0.27 | 2,400 |
| 2 | 161 | 22.2 | 1.81 | 11,000 | |
| 3 | 452 | 8.0 | 0.65 | 1,400 | |
| 4 | 336 | 16.4 | 1.34 | 4,000 | |
| 5 | 2,161 | 48.1 | 3.93 | 1,800 | |
| AtCAD3 | 1 | 292 | 20.1 | 1.65 | 5,600 |
| 2 | 581 | 9.7 | 0.80 | 1,400 | |
| 3 | 362 | 4.8 | 0.39 | 1,100 | |
| 4 | 534 | 17.9 | 1.47 | 2,800 | |
| 5 | 629 | 9.1 | 0.74 | 1,200 | |
| AtCAD7 | 1 | 320 | 28.6 | 2.18 | 6,800 |
| 2 | 3,685 | 79.0 | 6.04 | 1,600 | |
| 3 | 675 | 13.0 | 0.99 | 1,500 | |
| 4 | 756 | 2.7 | 0.21 | 280 | |
| 5 | 313 | 0.6 | 0.05 | 150 | |
| AtCAD8 | 1 | 302 | 20.4 | 1.59 | 5,300 |
| 2 | 683 | 7.0 | 0.54 | 800 | |
| 3 | 141 | 5.6 | 0.44 | 3,100 | |
| 4 | 457 | 10.4 | 0.81 | 1,800 | |
| 5 | 898 | 28.9 | 2.25 | 2,500 |
Fig. 4.
Lineweaver–Burk plots of AtCAD5 for substrates 1–5.
Fig. 5.
Relative substrate 1–5 efficacy of Arabidopsis recombinant CAD homologues.
However, AtCAD5 and AtCAD4 differed substantially in their overall catalytic properties, as revealed by kenz values (calculated by dividing Kcat by Km): with substrates 1–5, AtCAD5 was more effective than AtCAD4 by factors of 15, 7, 11, 12, and 270 times, respectively (i.e., differences of ≈1 to 2 orders of magnitude; Table 2). For p-coumaryl aldehyde (1), coniferyl aldehyde (3) and 5-hydroxyconiferyl aldehyde (4), these differences can be explained on the basis of both binding (Km values increasing ≈2- to 4-fold with AtCAD4) and lower turnover numbers (kcat decreases ≈3- to 6-fold with AtCAD4). Differences in kenz for caffeyl aldehyde (2), on the other hand, resulted from a small decrease in Km and a substantial increase in kcat between AtCAD5 and AtCAD4. The most profound difference between AtCAD5 and AtCAD4, however, occurred with sinapyl aldehyde (5), with a >10-fold increase in Km and a ≈20-fold increase in kcat. Based on the substrate versatility preference of AtCAD4 for p-coumaryl (1), coniferyl (3), and sinapyl (5) aldehydes, this isoform thus displays quite similar characteristics to that of a partially purified gymnosperm CAD from spruce (34), in as far as the latter also poorly used sinapyl aldehyde (5).
Arabidopsis AtCAD5 is also catalytically more active for sinapyl aldehyde (5) than the purported aspen SAD, claimed solely responsible for syringyl lignin formation (20). However, subsequent reanalysis (3) of the kinetic data for the claimed sinapyl aldehyde (5) specific SAD revealed that the claims were unfounded, because the so-called SAD could reduce each of the substrates 1–5. Indeed, overall it was actually catalytically more active for substrates 1–5 than a putative CAD isoform, which was also present in aspen (summarized in 3). Clearly, with catalytic properties of both Arabidopsis AtCAD4 and AtCAD5 as determined, there is no need to implicate a distinct SAD for specific generation of sinapyl alcohol 10 in angiosperms, including Arabidopsis.
By contrast, the AtCAD1–3 and 6–9 homologues were catalytically less active, with Km values for potential substrates 1–5 typically higher (relative to AtCAD5) and overall turnover numbers (kcat) lower by at least one order of magnitude, respectively. AtCAD2, however, used caffeyl aldehyde (2) most effectively as a substrate, whereas AtCAD3 had a slight preference for p-coumaryl (1) and 5-hydroxyconiferyl (4) aldehydes. On the other hand, AtCAD7 and 8 both used p-coumaryl aldehyde (1) as the best substrate, albeit only somewhat better than other potential substrates. Whereas AtCAD6–8 have the highest homology (≈ 78% similarity) to the claimed aspen SAD in aspen, neither AtCAD7 nor 8 displayed any substrate preference for sinapyl aldehyde (5). Thus, there is no biochemical data, for any of the CAD homologues, supporting the notion of a specific SAD isoform; i.e., in terms of binding, turnover, substrate preferences, etc.
It is also important to note that whereas AtCAD2, 3, 7, and 8 display more moderate levels of CAD activities (relative to AtCAD5/AtCAD4), none of their physiological functions are unambiguously established to this point. Gene expression profiles suggested previously indicated that AtCAD7 was inducible in response to Pseudomonas syringae infection, perhaps suggesting an alternative metabolic role in plant defense (7); indeed, another study (35) indicated that AtCAD7 (Eli-3) may be an aromatic alcohol dehydrogenase of broad substrate specificity. (However, no characterization of ELI-3 with purified recombinant protein was carried out.)
Molecular Modeling of AtCAD4 and AtCAD5. With the marked differences in substrate preferences of AtCAD5 and AtCAD4 regarding sinapyl aldehyde (5), a molecular modeling study was conducted to provisionally explain such profound differences. A blastp search of the Protein Data Bank (PDB) database revealed that AtCAD4 and AtCAD5 had highest sequence similarities (58.3% and 59.1% respectively) to that of the Sulfolobus solfataricus NAD-dependent alcohol dehydrogenase, whose crystal structure is established (1JVB; ref. 36). Positions of the second structural elements in its crystal structure also matched well with the predicted positions for those elements in AtCAD4 and AtCAD5, based on psipred, which can be accessed at http://bioinf.cs.ucl.ac.uk/psipred and phd, which can be accessed at http://npsa-pbil.ibcp.fr.
Amino acid substitutions, insertions, and deletions were next performed by using the graphics program o (37), starting from the S. solfataricus alcohol dehydrogenase coordinates (PDB ID code 1JVB), followed by quick-energy minimization by using x-plor (38) with potential function parameters of charmm19 as described (39). The initial positions of substrates and NADPH were obtained through the solid docking module on quanta (BioSYM/Micron Separations), which is based on conformational space, followed by a quick energy minimization by x-plor (38).
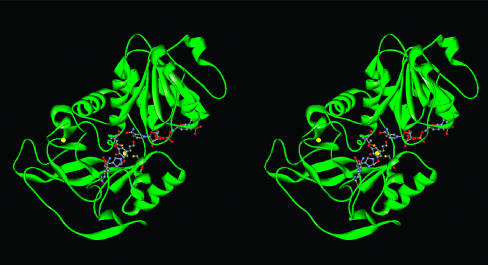
The backbones of AtCAD4- and AtCAD5-modeled structures did not change significantly (Fig. 6) and clearly showed properly sized binding pockets for NADPH and the cinnamyl aldehyde substrate near the catalytic zinc site made of C47, H69, and C163 (see Fig. 8, with AtCAD4 as example). In this modeled structure, the two methoxyl groups of sinapyl aldehyde (5) can fit snugly into the hydrophobic environment of both AtCAD5- and AtCAD4-active sites. Significantly, all residues within contact or potentially interacting distances to the substrate are conserved between AtCAD4 and AtCAD5, except for residue 129/130 (Gln and Lys in AtCAD5 and AtCAD4, respectively). A provisional explanation is that these residues help account for differences in overall catalytic behavior, because the side-chain amide group of Gln is capable of both donating and accepting hydrogen atoms through H-bonding to the phenolic hydroxyl group of 5, whereas the primary amine group in the Lys residue, ionizable at neutral pH, can withdraw electrons. However, a possible indirect contribution of residue 127/128, which are Asn and Asp in AtCAD5 and AtCAD4, respectively, cannot be ruled out, even though it is apparently not within an interacting distance to the substrate. Both residues (128 and 130 in AtCAD4 and 127 and 129 in AtCAD5) are located immediately after the major insertion of 121SYNDVY126, as compared with the S. solfataricus alcohol dehydrogenase. Therefore, due to this uncertainty, the exact distances between the substrate and those two residues cannot be estimated from this modeling study.
Fig. 6.
A stereoview showing the distribution of structural elements of AtCAD4. The two Zn atoms are shown in yellow together with three coordinating amino acids (C47, H69, and C163), Q130, NADP(H), and sinapyl aldehyde (5). This figure was prepared by using web viewer.
Identifying CAD Metabolic Networks Through Mutant Analysis. With four knockout lines for AtCAD5, AtCAD6, and AtCAD9 available from ABRC (see Table 5 for transgene orientations), we investigated whether any would affect either lignin deposition and/or lignin monomer composition, or whether various CAD metabolic networks were present thus conferring functional redundancy.
Comparative analyses of both mutant and wild-type Arabidopsis plants were therefore next carried out from germination to maturation, with sampling at regular points in the life cycle. By contrast, studies of various mutant Arabidopsis lines, including effects on lignin deposition (13, 40), have long been compromised by lack of such detailed analyses, and, surprisingly, typically monitor only one time point (harvesting; ref. 13). Accordingly, such studies often ignore the effects of, for example, phenotypical changes in growth/development rates and differential rates of lignin deposition per cell wall maturation (3). Plants were thus harvested at weekly intervals (from 4 to 10 weeks) for both wild type and knockouts, with bolting stems extracted (see Supporting Methods) and subjected to estimations of total lignin contents and monomer compositions. The acetyl bromide lignin method (see Supporting Methods) was chosen over Klason lignin analysis, because the latter frequently gives overestimations of lignin contents (>26–40% of cell wall residue) with Arabidopsis due to contaminating non-lignin components (data not shown and ref. 3). Monomer compositions were estimated by using both nitrobenzene oxidation (not specific to monolignol moieties in lignin) and thioacidolysis (presumed specific to 8-O-aryl-linked moieties in lignin). However, these methods only give a crude estimate of syringyl:guaiacyl (S:G) ratios, because they typically only account for ≈20–30% of the estimated lignin present (3) and, in some instances, even less (13).
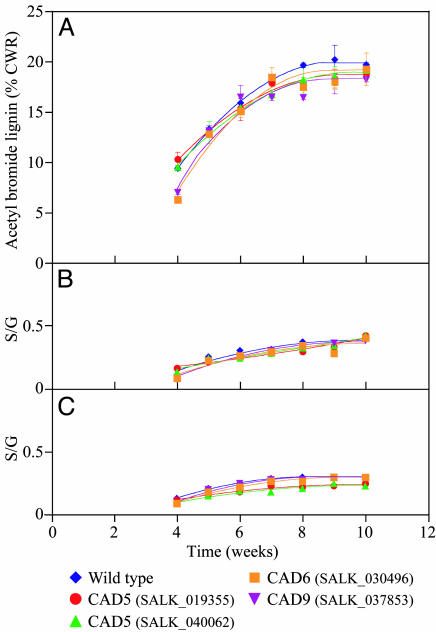
The acetyl bromide lignin analyses of Arabidopsis wild-type and CAD knockout homozygous lines indicated a small level of variability in lignin deposition throughout growth and development; however, at maturity (≈10 weeks), the maximum values were more or less equivalent to that of wild-type plants (Fig. 7A). AtCAD6 and AtCAD9 initially showed very slight decreases in lignin deposition, but the variability was quite minor and not significantly different at plant maturation, nor did the AtCAD5 mutant lines differ significantly from wild-type plants by maturation. Estimations of S:G ratios, by using both nitrobenzene oxidation (Fig. 7B) and thioacidolysis (Fig. 7C) analyses, also revealed some small differences in the S:G ratios during early stages of growth and development, but, which at maturation was largely insignificant, except for a very small decrease in the releasable S component by thioacidolysis; however, it is noteworthy that the S levels increased slowly but steadily up to the maturation point, again being quite similar to wild type at this developmental stage.
Fig. 7.
Time course of Arabidopsis (Columbia) growth and development versus lignin content/monomer composition ratios. (A) Acetyl bromide lignin contents. (B) S:G ratios by nitrobenzene oxidation. (C) S:G ratios by thioacidolysis. CWR, cell wall residue.
Thus, based on these data, AtCAD5 gene disruption apparently results only in slightly delayed syringyl lignin deposition during early phases of growth/development in the various cell types (xylem and interfascicular fibers) and, hence, presumably reflects decreased rates of cell wall maturation. However, syringyl lignin deposition is never “knocked out,” despite this isoform being the most catalytically active for sinapyl aldehyde (5). Indeed, these data reveal that by maturation, the presence of the other CAD homologues eventually permits lignin formation to essentially fully occur (including reaching the wild-type S:G ratios).
Taken together, these data provide support for CAD multigene networks ensuring that monolignols 6, 8, and 10 are formed, even if, for example, expression of one gene (such as AtCAD5) is disrupted and the syringyl lignin deposition rate is slightly delayed. Whether the genes in the proposed networks in the knockouts are expressed similarly as for wild-type plants, or differentially to compensate, remains to be determined.
Surprisingly, CAD is often reported to have a rate-limiting (regulatory) capacity during monolignol biosynthesis (19), which is an enigmatic assertion given that it is the final step in monolignol 6, 8, and 10 formation. Furthermore, Fell's (41) definition of rate-limiting (regulatory) steps as “fractional change in metabolic flux effected by a fractional change in amount (or activity) of the enzyme” does not support this notion. Indeed, we previously established (28), by detailed metabolic flux/transcriptional regulation studies, and comprehensive reevaluation of data obtained from various transgenic (CAD-down-regulated) plant lines (3), that CAD unequivocally does not have a key (rate-limiting) capacity relative to other steps in terms of carbon allocation. This finding is again demonstrated in this study. That is, even under conditions of a single gene disruption, which potentially could have become a rate-limiting step in the S-monolignol forming pathway, other isoforms were capable of compensating, i.e., to ensure that sinapyl alcohol (10) formation could essentially still occur.
In summary, based on the findings of this study, it is tempting to speculate that AtCAD4 is mainly used in p-coumaryl (6) and coniferyl (8) alcohol formation, whereas AtCAD5 can be used to normally generate all three monolignols [i.e., including sinapyl alcohol (10)]. Indeed, the lignin in interfascicular fibers in Arabidopsis is generally considered to have a larger sinapyl alcohol (10)-derived content than xylem cells, perhaps suggesting dominant expression of AtCAD5 in these cells (42). However, β-glucuronidase-promoter expression analysis for each of the CAD homologues revealed a complex pattern of expression, including significant expressional overlap both temporally and spatially during Arabidopsis growth and development (e.g., AtCAD4 and AtCAD5) (S.-J. K., H. W. Kim, L. B. Davin, V. R. Franceschi, and L. N. G., unpublished data). In this way, the organism ensures that lignin deposition occurs, even in the case of disruption of AtCAD5, again reflecting the exquisite control of lignin assembly.
Supplementary Material
Acknowledgments
This work was supported in part by National Science Foundation Arabidopsis 2010 Project Grant MCB-0117260, Department of Energy Grant DE-FG03-97ER20259, National Aeronautics and Space Administration Grant NAG 2-1198, and the G. Thomas and Anita Hargrove Center for Plant Genomic Research.
Abbreviations: CAD, cinnamyl alcohol dehydrogenase; SAD, sinapyl alcohol dehydrogenase; S:G, syringyl:guaiacyl.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY288079 (AtCAD1), AY302077 (AtCAD2), AY302078 (AtCAD3), AY302081 (AtCAD4), AY302082 (AtCAD5), AY302075 (AtCAD6), AY302079 (AtCAD7), AY302080 (AtCAD8), and AY302076 (AtCAD9)].
References
- 1.Gross, G. G., Stöckigt, J., Mansell, R. L. & Zenk, M. H. (1973) FEBS Lett. 31, 283-286. [Google Scholar]
- 2.Mansell, R. L., Gross, G. G., Stöckigt, J., Franke, H. & Zenk, M. H. (1974) Phytochemistry 13, 2427-2435. [Google Scholar]
- 3.Anterola, A. M. & Lewis, N. G. (2002) Phytochemistry 61, 221-294. [DOI] [PubMed] [Google Scholar]
- 4.Lewis, N. G., Davin, L. B. & Sarkanen, S. (1999) in Comprehensive Natural Products Chemistry, eds. Barton, D. H. R., Nakanishi, K. & Meth-Cohn, O. (Elsevier, London), Vol. 3, pp. 617-745. [Google Scholar]
- 5.Lewis, N. G. & Davin, L. B. (1999) in Comprehensive Natural Products Chemistry, eds. Barton, D. H. R., Nakanishi, K. & Meth-Cohn, O. (Elsevier, London), Vol. 1, pp. 639-712. [Google Scholar]
- 6.Walter, M. H., Grima-Pettenati, J., Grand, C., Boudet, A. M. & Lamb, C. J. (1988) Proc. Natl. Acad. Sci., USA 85, 5546-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiedrowski, S., Kawalleck, P., Hahlbrock, K., Somssich, I. E. & Dangl, J. L. (1992) EMBO J. 11, 4677-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somers, D. A., Nourse, J. P., Manners, J. M., Abrahams, S. & Watson, J. M. (1995) Plant Physiol. 108, 1309-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baucher, M., Van Doorsselaere, J., Gielen, J., Van Montagu, M., Inzé, D. & Boerjan, W. (1995) Plant Physiol. 107, 285-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, S.-H., Lee, J.-R., Shin, Y.-U., An, G. & Kim, S.-R. (1999) J. Microbiol. Biotechnol. 9, 475-481. [Google Scholar]
- 11.Quirino, B. F., Normanly, J. & Amasino, R. M. (1999) Plant Mol. Biol. 40, 267-278. [DOI] [PubMed] [Google Scholar]
- 12.Tavares, R., Aubourg, S., Lecharny, A. & Kreis, M. (2000) Plant Mol. Biol. 42, 703-717. [DOI] [PubMed] [Google Scholar]
- 13.Sibout, R., Eudes, A., Pollet, B., Goujon, T., Mila, I., Granier, F., Séguin, A., Lapierre, C. & Jouanin, L. (2003) Plant Physiol. 132, 848-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Arabidopsis Genome Initiative (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 15.Lewis, N. G., Davin, L. B. & Franceschi, V. (2002) in Plant Physiol., ed. Ausubel, F. M. (American Society of Plant Biologists, Rockville, MD), Vol. 129, pp. 394-437. [Google Scholar]
- 16.Kim, S.-J., Bedgar, D. L., Moinuddin, S. G., Kim, M.-R., Cardenas, C., Davin, L. B. & Lewis, N. G. (2003) 14th International Conference on Arabidopsis Research (Univ. of Wisconsin, Madison, WI), p. 179.
- 17.Anterola, A. M., Chung, B.-Y., Kim, S.-J., Bedgar, D. L., Davin, L. B. & Lewis, N. G. (2003) First International Meeting on Phytochemistry and Biology of Lignans, Bornheim-Walberberg, Germany, Phytochemical Society of Europe (Kluwer, Dordrecht, The Netherlands), pp. 2-3.
- 18.Costa, M. A., Collins, R. E., Anterola, A. M., Cochrane, F. C., Davin, L. B. & Lewis, N. G. (2003) Phytochemistry 64, 1097-1112. [DOI] [PubMed] [Google Scholar]
- 19.Baucher, M., Chabbert, B., Pilate, G., Van Doorsselaere, J., Tollier, M.-T., Petit-Conil, M., Cornu, D., Monties, B., Van Montagu, M., Inzé, D., et al. (1996) Plant Physiol. 112, 1479-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, L., Cheng, X.-F., Leshkevich, J., Umezawa, T., Harding, S. A. & Chiang, V. L. (2001) Plant Cell 13, 1567-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinkova-Kostova, A. T., Gang, D. R., Davin, L. B., Bedgar, D. L., Chu, A. & Lewis, N. G. (1996) J. Biol. Chem. 271, 29473-29482. [DOI] [PubMed] [Google Scholar]
- 22.Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- 23.Don, R. H., Cox, P. T., Wainwright, B. J., Baker, K. & Mattick, J. S. (1991) Nucleic Acids Res. 19, 4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter, M. H., Grima-Pettenati, J., Grand, C., Boudet, A. M. & Lamb, C. J. (1990) Plant Mol. Biol. 15, 525-526. [DOI] [PubMed] [Google Scholar]
- 25.Walter, M. H., Grima-Pettenati, J. & Feuillet, C. (1994) Eur. J. Biochem. 224, 999-1009. [DOI] [PubMed] [Google Scholar]
- 26.Kush, A., Goyvaerts, E., Chye, M.-L. & Chua, N.-H. (1990) Proc. Natl. Acad. Sci. USA 87, 1787-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knight, M. E., Halpin, C. & Schuch, W. (1992) Plant Mol. Biol. 19, 793-801. [DOI] [PubMed] [Google Scholar]
- 28.Anterola, A. M., Jeon, J.-H., Davin, L. B. & Lewis, N. G. (2002) J. Biol. Chem. 277, 18272-18280. [DOI] [PubMed] [Google Scholar]
- 29.Rossmann, M. G., Moras, D. & Olsen, K. W. (1974) Nature 250, 194-199. [DOI] [PubMed] [Google Scholar]
- 30.Goffner, D., Van Doorsselaere, J., Yahiaoui, N., Samaj, J., Grima-Pettenati, J. & Boudet, A. M. (1998) Plant Mol. Biol. 36, 755-765. [DOI] [PubMed] [Google Scholar]
- 31.Katayama, T., Davin, L. B., Chu, A. & Lewis, N. G. (1993) Phytochemistry 33, 581-591. [Google Scholar]
- 32.Nose, M., Bernards, M. A., Furlan, M., Zajicek, J., Eberhardt, T. L. & Lewis, N. G. (1995) Phytochemistry 39, 71-79. [DOI] [PubMed] [Google Scholar]
- 33.Anterola, A. M., van Rensburg, H., van Heerden, P. S., Davin, L. B. & Lewis, N. G. (1999) Biochem. Biophys. Res. Commun. 261, 652-657. [DOI] [PubMed] [Google Scholar]
- 34.Lüderitz, T. & Grisebach, H. (1981) Eur. J. Biochem. 119, 115-124. [DOI] [PubMed] [Google Scholar]
- 35.Somssich, I. E., Wernert, P., Kiedrowski, S. & Hahlbrock, K. (1996) Proc. Natl. Acad. Sci. USA 93, 14199-14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esposito, L., Sica, F., Raia, C. A., Giordano, A., Rossi, M., Mazzarella, L. & Zagari, A. (2002) J. Mol. Biol. 318, 463-477. [DOI] [PubMed] [Google Scholar]
- 37.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 38.Brunger, A. T. (1992) x-plor, a System for Crystallography and NMR (Yale Univ. Press., New Haven, CT), Version 3.1.
- 39.Min, T., Kasahara, H., Bedgar, D. L., Youn, B., Lawrence, P. K., Gang, D. R., Halls, S. C., Park, Hilsenbeck, J. L., Davin, L. B., et al. (2003) J. Biol. Chem. 278, 50714-50723. [DOI] [PubMed] [Google Scholar]
- 40.Jones, L., Ennos, A. R. & Turner, S. R. (2001) Plant J. 26, 205-216. [DOI] [PubMed] [Google Scholar]
- 41.Fell, D. (1997) Understanding the Control of Metabolism (Portland Press, London).
- 42.Meyer, K., Shirley, A. M., Cusumano, J. C., Bell-Lelong, D. A. & Chapple, C. (1998) Proc. Natl. Acad. Sci. USA 95, 6619-6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.