Abstract
Most of the small ribozymes, including those that have been investigated as potential therapeutic agents, appear to be rather poor catalysts. These RNAs use an internal phosphoester transfer mechanism to catalyze site-specific RNA cleavage with apparent cleavage rate constants typically <2 min-1. We have identified variants of one of these, the Neurospora Varkud satellite ribozyme, that self-cleaves with experimentally measured apparent rate constants of up to 10 s-1 (600 min-1), ≈2 orders of magnitude faster than any previously characterized self-cleaving RNA. We describe structural features of the cleavage site loop and an adjacent helix that affect the apparent rate constants for cleavage and ligation and the equilibrium between them. These data show that the phosphoester transfer ribozymes can catalyze reactions with rate constants much larger than previously appreciated and in the range of those of protein enzymes that perform similar reactions.
Sequence- or structure-specific cleavage of RNA phosphodiester bonds by many protein enzymes is quite rapid: for example, ribonuclease III cleaves its target RNA structure with an apparent rate constant (kobs) of 6.4 s-1, and RNaseA can cleave its preferred dinucleotide sequence even faster, from 15.2 to 675 s-1, depending on the source of the enzyme (1, 2). Site-specific hydrolytic cleavage of RNA by the RNA subunit of Bacillus RNaseP or the Tetrahymena self-splicing group I intron has been observed (3) or calculated (4) to be fast, in the range of 6 s-1. A rate constant of ≈10 s-1 was measured for a ligase ribozyme obtained by in vitro selection to catalyze the attack of a 3′ hydroxyl on a 5′ triphosphate (5).
In contrast, most ribozymes appear to be rather poor catalysts. The “small ribozymes,” comprising the naturally occurring hammerhead, hairpin, hepatitis delta virus, and Neurospora Varkud satellite (VS) ribozymes, catalyze a transesterification reaction, yielding cleavage products with 2′3′ cyclic phosphate and 5′ hydroxyl termini like those produced by many protein ribonucleases. The vast majority of ribozymes selected in vitro to cleave RNA phosphodiester bonds also use this same phosphoester transfer chemistry and, like their natural counterparts, have cleavage rate constants of <≈2 min-1 (0.033 s-1) (6, 7). A variety of enzymological considerations that affect ribozyme reaction rates have been discussed (8), and it has been recently proposed that chemical principles may limit the rates of certain small ribozymes (9, 10).
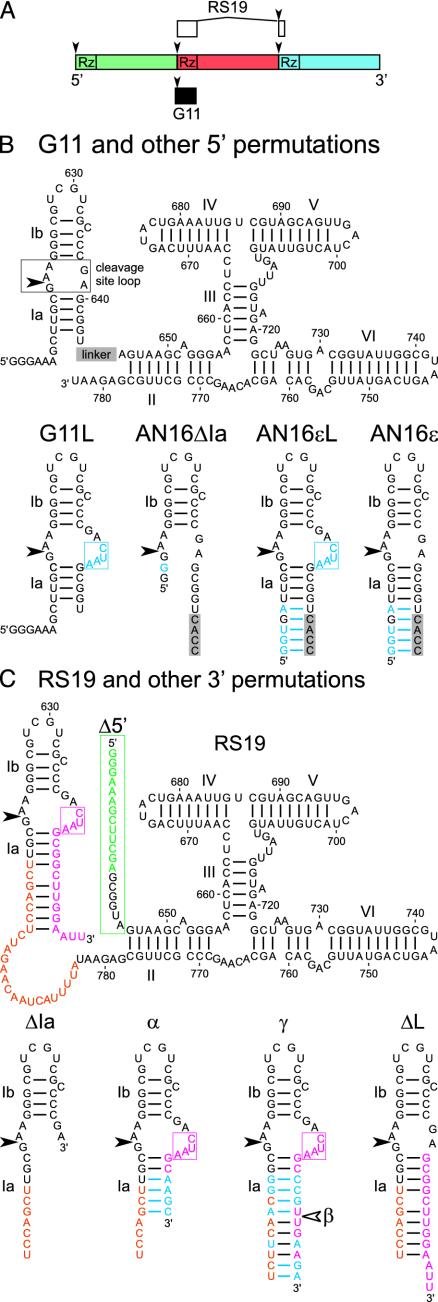
The VS ribozyme is found in RNA transcripts of a plasmid in the mitochondria of certain natural isolates of the fungus Neurospora (11). It catalyzes site-specific cleavage and ligation reactions, similar to those performed by hammerhead, hairpin, and hepatitis delta virus ribozymes that are involved in the replication of the RNAs that contain the ribozyme (reviewed in refs. 12 and 13). Cleavage in VS RNA occurs after nucleotide G620 in an internal loop between helices Ia and Ib (Fig. 1B) (14). Biophysical, crosslinking, mutational, and nucleotide analog interference experiments implicate the 730 loop in helix VI, and especially nucleotide A756 in that loop, as contributing to the active site of the ribozyme (15–18).
Fig. 1.
Sequences and secondary structure diagrams of VS ribozyme variants. (A) Three tandem repeats, each represented by a different color, of the natural 881-nt VS RNA monomer. The cleavage-ligation sites are indicated by the arrowheads. The portions of VS RNA contained in clones G11 and RS19 are indicated by the filled and open boxes, respectively. (B) Secondary structure of G11 and other 5′ permutations. Stem loop I, comprising helices Ia, Ib, and the cleavage site loop, is attached to the 5′ end of helix II. The linker (gray box) at the indicated position is5′CACCUAGCAACUCUAG3′ in the AN16 series of variants and is absent in G11 and G11L. (C) Secondary structure of RS19 and other 3′ permutations. Non-VS nucleotides at the 5′ and 3′ ends are indicated in green and pink, respectively; linker nucleotides are indicated in orange. (B and C) Sequences and secondary structures of stem loop I from mutant RNAs are shown below the complete secondary structure diagrams of their parents. Portions of the linker sequences are included in the secondary structure diagrams of stem loop I. Nucleotides that differ in the mutants from those in their parent are indicated in blue. The four extra nucleotides on the 3′ side of the cleavage site loop are boxed. In 3′ permutations, whose designation includes Δ5′, e.g., RS19Δ5′, the nucleotides in the green box comprising the 5′ tail are replaced with a single G. 3′ permutation mutants that lack the extra CUAA sequence in the cleavage site loop include ΔL in their name. 5′ permutation mutants into which the extra CUAA sequence has been added include L in their name. Different sequences in helix Ia are indicated by suffixes α through ε; β is identical to γ except that it is shortened on the 3′ side to the position indicated by the unfilled arrowhead.
Methods
Plasmid clones G11 and RS19 have been described (19, 20). The mutant versions of each described here (Fig. 1) were constructed by incorporating the desired sequences into primers for PCR amplification from plasmid templates. RNAs were synthesized by in vitro transcription in the presence of [α-32P]GTP from plasmids digested with appropriate restriction enzymes (SspI for 5′ permutations; EcoRI for RS19, ΔL, and Δ5′; AvaI for ΔIa; or downstream type IIS restriction enzymes BsmAI, SapI, or BbsI for α, β, and γ, respectively). To minimize self-cleavage during transcription, reactions were altered from the standard T7 conditions (21) by decreasing the concentration of MgCl2 to 2.5–3 mM in the presence of 1 mM of each unlabeled NTP (22, 23). In some cases an antisense oligodeoxynucleotide (0.5–2 μM) complementary to nucleotides 645–662 was added to interfere with RNA folding. Transcription reactions were loaded on denaturing polyacrylamide gels, and full-length RNAs were identified by UV shadowing and eluted as described (23).
Unless otherwise indicated, cleavage reactions were performed by using a Kintek (Clarence, PA) RQF-3 rapid quench flow instrument at 37°C following the manufacturer's instructions. Briefly, 10 μl of RNA (20 nM) in one times standard cleavage buffer [SCB, 40 mM Tris·HCl, pH 8.0 (measured at room temperature)/50 mM KCl/2 mM spermidine] plus 2 mM EDTA and 10 μl of reaction buffer (1× SCB, 400 mM MgCl2) were preincubated separately for ≥1 min in the sample loading loops, mixed for a specified reaction time, and stopped with 10 vol of quench solution (80% formamide, 200 mM EDTA, 0.01% each xylene cyanol and bromophenol blue). Aliquots from each time point were separated by denaturing gel electrophoresis and exposed to a PhosphorImager screen. Band intensities were quantified by using imagequant software (Molecular Dynamics/Amersham Biosciences).
Cleavage data (fraction cleaved, fclv, at time t) were normalized (see below) and fit to the first-order equation:
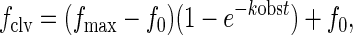 |
[1] |
which corrects for the presence of any cleaved RNA in the starting material, f0, and allows estimation of the apparent maximal extent of cleavage, fmax, and the apparent first-order cleavage rate constant, kobs.
We estimated the individual apparent cleavage and ligation rate constants, k1 and k-1, respectively, by fitting the normalized cleavage data to an equation derived by solving the system of differential equations describing the simplest approach-to-equilibrium model:
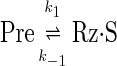 |
[2] |
with the assumption that 100% of the precursor (Pre) is cleavable and in equilibrium with the cleaved ribozyme–substrate complex (Rz·S). This equation is equivalent to Eq. 1, where
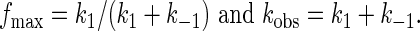 |
[3,4] |
To estimate k1 and k-1 for experiments in which chase (Ch) was added before starting the reaction (Table 1), we fit cleavage data to an equation derived by solving the system of differential equations describing a simplified version of the kinetic scheme seen in Fig. 3A:
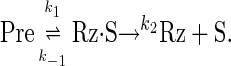 |
[5] |
Table 1. Estimated kinetic parameters (mean ± standard deviation).
| Extra nt in cleavage site loop | Helix la | kobs, s-1 | fmax | k1, s-1 | k-1, s-1 | |
|---|---|---|---|---|---|---|
| 3′ permutations | ||||||
| RS19 | CUAA | + | 3.0 ± 0.19 | 0.89 ± 0.023 | ≥2.7 ± 0.20 | ≤0.32 ± 0.065 |
| RS19Δ5′ | CUAA | + | 4.2 ± 0.25 | 0.91 ± 0.015 | ≥3.8 ± 0.24 | ≤0.37 ± 0.060 |
| RS19Δla | — | - | 0.33 ± 0.036 | 0.91 ± 0.011 | ≥0.31 ± 0.029 | ≤0.029 ± 0.0071 |
| RS19Δ5′α | CUAA | + | 2.0 ± 0.22 | 0.77 ± 0.010 | ≥1.6 ± 0.20 | ≤0.45 ± 0.027 |
| RS19β | CUAA | + | 1.7 ± 0.24 | 0.85 ± 0.029 | ≥1.5 ± 0.16 | ≤0.27 ± 0.082 |
| RS19γ | CUAA | + | 1.8 ± 0.13 | 0.87 ± 0.015 | ≥1.6 ± 0.084 | ≤0.24 ± 0.044 |
| RS19Δ5′γ | CUAA | + | 2.9 ± 0.25 | 0.87 ± 0.017 | ≥2.5 ± 0.27 | ≤0.38 ± 0.016 |
| RS19ΔL | — | + | 1.9 ± 0.38 | 0.11 ± 0.0055 | 0.21 ± 0.031 | 1.7 ± 0.35 |
| RS19ΔL (Ch) | — | + | 1.9 ± 0.42* | 0.12 ± 0.0049* | 0.22 ± 0.049 | 1.7 ± 0.37 |
| RS19L5 | CUAAA | + | 3.7 | 0.88 | ≥3.3 | ≤0.41 |
| RS19L4a | CAAA | + | 2.1 | 0.86 | ≥1.8 | ≤0.28 |
| RS19L4b | AUAA | + | 2.4 | 0.86 | ≥2.1 | ≤0.33 |
| RS19L4c | AAAA | + | 2.8 | 0.90 | ≥2.5 | ≤0.26 |
| RS19L3 | AAA | + | 1.0 | 0.88 | ≥0.93 | ≤0.12 |
| RS19L2 | AA | + | 0.63 | 0.82 | ≥0.51 | ≤0.11 |
| RS19L1 | A | + | 10 ± 1.6 | 0.26 ± 0.012 | 2.6 ± 0.46 | 7.4 ± 1.2 |
| RS19L1 (Ch) | A | + | 6.2 ± 1.4* | 0.32 ± 0.03* | 2.0 ± 0.51 | 4.2 ± 0.95 |
| 5′ permutations | ||||||
| G11 | — | + | 0.0021 ± 0.00031 | 0.91 ± 0.0030 | n/a | n/a |
| G11L | CUAA | + | 0.14 ± 0.0045 | 0.94 ± 0.046 | n/a | n/a |
| AN16 | — | + | 0.11 ± 0.010 | 0.93 ± 0.012 | n/a | n/a |
| AN16Δla | — | - | 0.31 ± 0.016 | 0.91 ± 0.038 | n/a | n/a |
| AN16εL | CUAA | + | 2.2 ± 0.18 | 0.86 ± 0.014 | ≥1.9 ± 0.13 | ≤0.33 ± 0.050 |
| AN16ε | — | + | 1.9 ± 0.21 | 0.15 ± 0.0045 | ≥0.24 ± 0.047 | ≤1.6 ± 0.17 |
n/a, Not available.
Calculated first-order parameters of burst phase; fmax = k1/(k1 + k-1); kobs = k1 + k-1
Fig. 3.
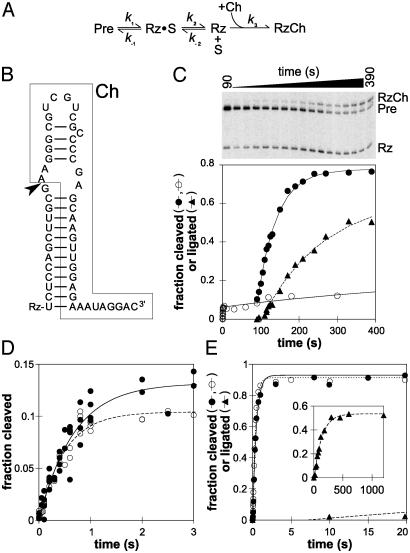
The natural cleavage site loop favors ligation. (A) Minimal kinetic scheme for self-cleavage in the presence of Ch. Cleavage of Pre yields 5′ and 3′ products designated Rz and S, respectively (see text). (B) Sequence and secondary structure of Ch (boxed) bound to Rz; only the nucleotides in Rz that pair with Ch are indicated. The arrowhead marks the site of ligation. (C) Time course of a RS19ΔL Ch experiment. The fraction of Pre cleaved in the absence of Ch (○) or after addition of Ch (to 1 μM) at t = 90 s (•); the fraction of Pre converted to RzCh, i.e., fraction ligated (▴); the smaller cleavage product, S, has run off the gel. (D) The burst phase of RS19ΔL cleavage in the absence of Ch (○) or with Ch added from the start of the experiment (•). Data from multiple experiments are shown. (E) Cleavage of RS19 in the absence (○) or presence (•)ofCh; ▴, fraction ligated. (Inset) Longer time course to show the appearance of RzCh; x axis, time (s); y axis, fraction ligated.
In this model, reassociation of Rz and S (seen as k-2 in Fig. 3A) is negligible because of an excess of Ch. Hence, once S dissociates from Rz·S it cannot religate to form Pre, and cleavage of Pre should proceed to completion. The model yields a double exponential equation, which allows estimation of k1, k-1, and fmax. In this model, all of the active Pre will be converted to Rz + S; therefore, we interpret fmax in this case to represent the fraction of Pre that is active. We calculated fmax and kobs of the burst phase by substituting the estimated values of k1 and k-1 into Eqs. 3 and 4.
To determine k1a and k1b in the on-pathway intermediate model (see Fig. 4 and text), data were fit to an equation derived in the same manner as those above.
Fig. 4.
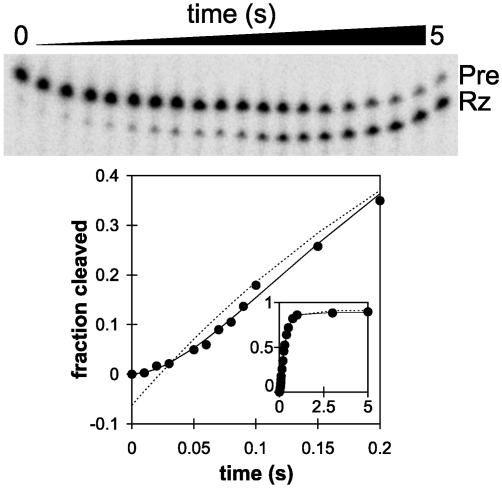
Fast-cleaving RNAs reveal an even faster on-pathway intermediate (I). Time course of self-cleavage of RS19 fittoa first-order equation (dashed line) or the Pre → I → Rz·S on-pathway intermediate model (solid line; see text). (Inset) Complete time course demonstrates that the data fit well to both equations after the lag phase; x axis, time (s); y axis, fraction cleaved.
Cleavage data were normalized by initially fitting to the on-pathway intermediate model to obtain an estimate of f0 and then normalized between f0 and 1 (or the estimated fmax from Ch experiments where applicable) before subsequent fitting.
We used dynafit (24) for simulations and curve fitting that aided us in devising and testing kinetic models. maple 9.01 (Maple-Soft, Waterloo, Canada) was used to help solve differential equations. Curve fitting was performed by nonlinear least-squares regression with custom macros in Microsoft excel xp.
Results and Discussion
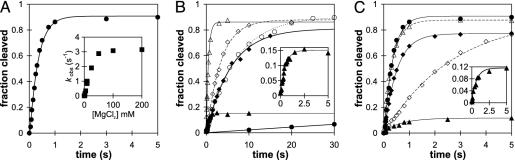
RS19 Self-Cleaves Much More Rapidly Than Previously Characterized VS Ribozymes. Previous deletion experiments identified a contiguous region of the Neurospora VS sequence, designated G11 in Fig. 1 A and B, that is sufficient for self-cleavage. This region comprises the cleavage site and 163 nt immediately downstream (reviewed in ref. 15). However, the natural VS RNA consists of a multimeric series of head-to-tail tandem repeats, so it is possible that any given copy of the ribozyme, although capable of acting on the cleavage site immediately upstream, might prefer to act on the cleavage site many hundreds of nucleotides downstream. Indeed, a version of the VS ribozyme called RS19, in which the cleavage site was tethered via a linker sequence to the 3′ end of the ribozyme (Fig. 1 A and C), showed exceptionally rapid cleavage (Fig. 2A and Table 1). The cleavage data are reasonably well described by a single exponential analysis typical of a first-order self-cleavage reaction (but, see below), and indicate kobs = 3.0 s-1 (180 min-1) at saturating concentrations of MgCl2. This apparent cleavage rate constant is >1,000-fold faster than for the well characterized G11 version of the VS ribozyme (22) and is more than an order of magnitude faster than observed for any other self-cleaving ribozyme (7, 25). Insertion of a linker between helices Ia and II into the G11 version of the ribozyme increased kobs >50-fold: from 0.0021 to 0.11 s-1 (mutant AN16; Figs. 1B and 2B and Table 1). These data suggest that a linker may relieve a steric constraint that limits the cleavage rate of G11. However, the kobs of RS19, at 3 s-1, is still almost 30-fold faster than AN16, indicating that a structural feature in addition to the linker contributes to this exceptionally fast cleavage rate.
Fig. 2.
Fast cleavage of RS19. (A) Time course of self-cleavage of RS19. (Inset) Cleavage reactions were performed over a range of concentrations of MgCl2, and the apparent first-order rate constant was plotted as a function of MgCl2 concentration. (B) Time course of self-cleavage of mutants of G11 (see Fig. 1B): G11, •; G11L, ○; AN16, ♦; AN16ΔIa, ⋄; AN16εL, ▵; and AN16ε, ▴. (Inset) The burst phase of AN16ε cleavage. (C) Time course of self-cleavage of RS19 and 3′ permutations (see Fig. 1C): RS19, •; RS19ΔIa, ⋄; RS19Δ5′α, ♦; RS19Δ5′γ, ▵; and RS19ΔL, ▴.(Inset) The burst phase of RS19ΔL cleavage. (B and C Insets) The x axis is time (s), and the y axis is fraction cleaved. The lines show the nonlinear least-squares fittoa first-order rate equation. Kinetic parameters are summarized in Table 1.
Helix Ia Contributes to Fast Cleavage. Additional features of RS19 that might contribute to the observed rapid cleavage could derive from non-VS sequences of 13 and 17 nt at the 5′ and 3′ ends of the RNA, respectively, that served as primer binding sites in previous experiments (20). Mutants of RS19 in which the 5′ primer binding site was removed still exhibited rapid cleavage, providing evidence that the extraneous 5′ sequence was not responsible for rapid cleavage (mutants RS19Δ5′ and RS19Δ5′α; Figs. 1C and 2C and Table 1; others not shown).
A portion of the 3′ primer binding site of RS19 (pink nucleotide in Fig. 1C) is partially complementary to the sequence upstream of the cleavage site, including part of the linker sequence (orange nucleotide in Fig. 1C), forming a helix analogous to helix Ia of the natural ribozyme, although longer and containing several non-Watson–Crick pairs. As a consequence of this helix Ia, the internal loop that contains the cleavage site is enlarged by four additional nucleotides on the 3′ side (5′CUAA3′; pink box in Fig. 1C). Disruption of helix Ia, by deletion of the nucleotides comprising its 3′ side and the extra CUAA sequence, decreased kobs by ≈10-fold, to 0.33 s-1 (mutant RS19ΔIa, Figs. 1C and 2C and Table 1). Similarly, a 5′ permutation with a long linker between stem loop I and helix II but lacking helix Ia (mutant AN16ΔIa, Fig. 1B) cleaved at a similar rate (kobs = 0.31 s-1; Fig. 2B and Table 1). These data suggest that observed cleavage rates in the range of 0.3 s-1 are the fastest that can be obtained simply by having a long linker between stem loop I and the rest of the ribozyme. Mutants in which helix Ia is restored with different complementary sequences comprising 6–12 bp restored rapid cleavage in 3′ permutations (RS19Δ5′α, RS19β, RS19γ, and RS19Δ5′γ with kobs = 2.0, 1.7, 1.8, and 2.9 s-1, respectively; Figs. 1C and 2C and Table 1) and a 5′ permutation (AN16εL, kobs = 2.2 s-1; Figs. 1B and 2B). These data provide evidence that helix Ia contributes to very rapid cleavage.
The Natural Sequence of the Cleavage Site Loop Favors Ligation. Restoring helix Ia but not the extra CUAA sequence in the cleavage site loop also restored rapid cleavage in both 3′ (RS19ΔL: kobs = 1.9 s-1; Figs. 1C and 2C and Table 1) and 5′ (AN16ε: kobs = 1.9 s-1; Figs. 1B and 2B) permutation mutants. However, unlike RS19, which cleaved rapidly and essentially to completion, AN16ε and RS19ΔL showed a burst of rapid cleavage but a low apparent extent (fmax); only 15% or 11%, respectively, of these RNAs appeared to cleave (Figs. 2B and C and Table 1). Burst or biphasic kinetics are often interpreted to be caused by the presence of two subpopulations of RNA with different kinetics (26); however, chase experiments (see below) showed that the low extent of cleavage was not caused by a large fraction of slowly cleaving or inactive RNA, but by an equilibrium between Pre RNA and Rz·S, in which equilibrium strongly favors the ligated state. The burst phase is followed by a reversible dissociation of Rz·S into free Rz + S (Fig. 3A) that is much slower and not detectable at the RNA concentration and time scale used in Fig. 2 (but is detectable in the longer time course shown in Fig. 3C, ○). The burst kinetics of the AN16ε and RS19ΔL mutants provide evidence that the natural sequence of the VS cleavage site loop favors ligation.
Fitting the burst phase of the RS19ΔL cleavage data to a simple approach-to-equilibrium kinetic model that includes rate constants only for cleavage (k1) and ligation (k-1), suggests that the kobs of 1.9 s-1 for RS19ΔL is comprised of an apparent k1 of 0.21 s-1 and k-1 of 1.7 s-1. Similar results were obtained for AN16ε (Table 1). The somewhat unintuitive notion that kobs = k1 + k-1 stems from the fact that as the ratio of k1/k-1 decreases, so does the apparent extent of cleavage. From this finding it follows that the half-maximal extent of cleavage also decreases and is therefore attained in shorter time than would be required if cleavage were to go to completion. Consequently, kobs may appear to be large because of a large ligation rate constant, k-1, even though the cleavage rate constant, k1, itself is rather small. It is interesting to note that k1 for RS19ΔL and AN16ε is similar to the kobs of AN16ΔIa (0.31 s-1) and RS19ΔIa (0.33 s-1), both of which cleave almost to completion, and may reflect the cleavage rate constant attainable simply by lengthening the linker between stem loop I and helix II. These data suggest that the effect of having a stable helix Ia in a RNA that contains the natural cleavage site loop is to increase the ligation rate constant, resulting in a lowering of the apparent extent of cleavage.
To show that most of the RS19ΔL RNA that was uncleaved at the end of the burst phase was indeed active, we allowed the self-cleavage reaction to proceed well past the burst phase and then added an excess of an unlabeled S analog [Ch, Fig. 3B; determined by titration experiments (data not shown) to be at saturating concentration] to outcompete reassociation of Rz and S. The Ch was designed to form a helix Ia with Rz that was even more stable than in RS19, comprising 12 bp, such that its dissociation would be insignificant over the time course of the current experiments. Also, Ch contained eight additional nucleotides at its 3′ end to provide a size difference that would allow electrophoretic separation of the product of Rz and Ch ligation (RzCh) from the original or religated Pre (Fig. 3C), thereby allowing estimation of the apparent rate of formation of RzCh (see below). Fig. 3C (•) shows that upon addition of Ch most of the remaining RS19ΔL Pre was observed to cleave, providing evidence that the remaining Pre is indeed catalytically active and that the low extent of cleavage in the absence of Ch reflects an equilibrium that favors ligation.
The apparent rate constant for the disappearance of Pre after addition of Ch (≈0.020 s-1 from a simple first-order analysis of the data in Fig. 3C) may reflect dissociation of Rz·S into Rz + S (k2) or other slow steps that occur after the chemical step. The apparent rate constant for the formation of RzCh is slightly slower still, 0.0061 s-1, after a noticeable lag of several seconds. Control experiments in which ligation was initiated by mixing purified Rz and saturating concentrations of Ch showed that binding and ligation of Ch are much faster than this; formation of RzCh occurred in these experiments with an apparent rate constant of 0.4 s-1, reaching an equilibrium in which ≈40% of Rz was ligated (data not shown). More detailed kinetic analyses suggest that the lag observed during formation of RzCh in the cleavage-Ch experiment described above can be accounted for by two consecutive slow steps with rate constants similar to each other between the chemical step and formation of RzCh (R.Z. and R.A.C., unpublished data).
Experiments in which Ch was added before starting the cleavage reaction showed that the presence of Ch had no effect on the rapid burst phase of the RS19ΔL cleavage reaction (Fig. 3D and Table 1), as expected if this phase represents approach to equilibrium. These data provide further evidence that RS19ΔL cleaves very rapidly to an equilibrium that favors ligation, and the extent of observed cleavage also remains very low because the binding equilibrium between Rz + S and Rz·S favors Rz·S at the concentrations of RNA used.
The fast cleavage phase of RS19, which encompasses the whole time course and proceeds almost to complete cleavage, was also unaffected by the presence of Ch (Fig. 3E), providing further evidence that the equilibrium strongly favors cleavage in this RNA. Ligated RzCh appeared after a lag of several seconds with an apparent first-order ligation rate constant of 0.0077 s-1, which may reflect the rate-limiting dissociation of the RS19 Rz·S complex. Thus, even though RS19 is almost completely cleaved in <1 s, the Ch experiment shows that several more seconds are required before enough of Rz·S dissociates to allow detectable ligation of released Rz to Ch. These observations emphasize that even though cis-cleavage reactions are often described as “single turnover,” they do not necessarily reflect only a single catalytic event. In situations where the religation rate constant is similar to or greater than the rate constant for product dissociation, many cycles of cleavage and ligation may occur before the products dissociate, even in an RNA like RS19 where equilibrium favors cleavage.
Extra Nucleotides in the Cleavage Site Loop Shift the Equilibrium in Favor of Cleavage. Introducing the extra CUAA sequence into the cleavage site loop of RNAs with a stable helix Ia and a long linker still allowed rapid cleavage; furthermore, it also restored the high final extent of cleavage originally observed with RS19. For example, insertion of the CUAA sequence into the 5′ permutation AN16ε, making mutant AN16εL (Fig. 1B), produced little difference in the apparent cleavage rate constant (kobs = 1.9 vs. 2.2 s-1, respectively), but AN16εL cleaved almost to completion (Fig. 2B and Table 1). 3′ Permutations containing the CUAA sequence and various lengths and sequences of helix Ia also exhibited rapid cleavage with kobs ranging from 1.7 to 2.9 s-1 and high final extents of cleavage (mutants RS19Δ5′α, RS19β, RS19γ, and RS19Δ5′γ; Fig. 1C and 2C and Table 1). The approach to equilibrium kinetic analysis of RS19, as described above for RS19ΔL, suggests an apparent k1 of ≥2.7 s-1 and k-1 of ≤0.32 s-1; the other mutants that contained the extra CUAA sequence also exhibited a high k1 and low k-1 (Table 1). Thus, compared to RS19ΔL and AN16ε, which have the natural sequence in the cleavage site loop, the additional CUAA sequence in the cleavage site loop shifts the equilibrium in favor of cleavage by both increasing the apparent cleavage rate constant and decreasing the apparent ligation rate constant.
Even in the absence of a linker between helices Ia and II, insertion of the CUAA sequence into the cleavage site loop increased the apparent cleavage rate constant of G11 from 0.0021 to 0.14 s-1 (mutant G11L; Figs. 1B and 2B and Table 1). Thus, the extra loop nucleotides contribute to increased apparent cleavage rate independently of, and partially additively with, the effect of the linker between helices Ia and II.
A series of mutants (RS19L1 to RS19L5) with varying numbers and identities of extra nucleotides in the cleavage loop showed that RNAs containing CAAA, AUAA, or AAAA showed the same self-cleavage kinetics as the CUAA sequence in RS19 (Table 1). Increasing the size of the loop by 1 nt (CUAAA) also had little effect on cleavage kinetics. Decreasing the number of extra loop nucleotides to three or two resulted in progressively slower cleavage. A further decrease to a single extra nucleotide in the loop (mutant RS19L1) or removal of all extra nucleotides (mutant RS19ΔL, described above), changed the shape of the cleavage curve substantially, such that these latter two RNAs exhibited a rapid burst of cleavage but only to a low apparent extent. Ch experiments like those described above for RS19ΔL showed that the uncleaved portion of RS19L1 RNA also reflects a cleavage–ligation equilibrium that favors ligation (Table 1 and data not shown). The RS19L1 mutant was notable in having the fastest apparent rate constant observed for any VS ribozyme to date (10 s-1), resulting from a cleavage rate constant as rapid as that of RS19 (2.6 s-1), and the fastest apparent ligation rate constant of any VS ribozyme (7.4 s-1) (Table 1).
An Even Faster On-Pathway Process. Closer examination of the early portion of the self-cleavage time courses of RS19 (Fig. 4) and other fast-cleaving RNAs (data not shown) revealed that the first-order analyses described above are an oversimplification. During the first 0.1 s, in which the first 15–20% of RS19 Pre cleaved, a lag phase is clearly evident (Fig. 4). Fitting the entire cleavage curve to a variety of hypothetical kinetic schemes revealed an excellent fit to the following model:
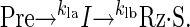 |
When a reaction proceeds through an on-pathway intermediate (I), and k1a and k1b are within about an order of magnitude of each other, each is partially rate limiting, and the observed reaction progress curve exhibits the shape shown in Fig. 4 (3, 27). Because the proportion of RNA in the I state cannot be measured directly, two mathematical solutions are obtained in which the magnitudes of the two rate constants are the same in both solutions, but in one the faster rate constant is k1a, whereas in the other the faster rate constant is k1b. By fitting the RS19 self-cleavage data to this model, k1a and k1b were estimated to be 15.4 ± 3.6 and 4.0 ± 0.40 s-1. Even the slower of these rate constants is fast in comparison with previously studied ribozymes, and the faster rate constant implies that some process on the reaction pathway occurs at the remarkably rapid rate of almost 1,000 min-1.
Implications for Ribozyme Biology. Previous experiments with full-length VS RNA and deletion and substitution mutants had shown that the VS ribozyme is capable of both cleavage and ligation (20, 28, 29). The experiments described in this article have identified sequence and structural features of the cleavage site loop of the VS ribozyme that substantially affect the apparent cleavage and ligation rate constants and the equilibrium between cleavage and ligation. The observation that the natural sequence around the cleavage/ligation site strongly favors ligation may explain why the majority of VS RNA monomers isolated from mitochondria are in the circular, ligated state (11). The minus strand of the satellite RNA of tobacco ringspot virus, which contains the hairpin ribozyme, also ligates efficiently (30), and equilibrium favors ligation in model hairpin ribozymes in which product release is minimized (31). Equilibrium of minimal hammerhead ribozymes can be shifted in favor of ligation by a covalent crosslink that constrains the mobility of two helices (32). Recent work shows that natural hammerhead ribozymes may be similarly constrained by a loop–loop interaction outside of the catalytic core (33, 34). Some of these larger hammerhead ribozymes cleave to an apparent low final extent (34); perhaps, like the RS19ΔL version of the VS ribozyme (above), the low extent of cleavage may indicate that these ribozymes actually favor ligation. Because the circular forms of these RNAs are thought to be templates for rolling circle replication that would be essential for propagation of the RNAs, it may be that ligation rate constants that exceed those for cleavage have been evolved by natural selection to ensure that a substantial fraction of these RNAs is in the circular form.
The VS ribozyme variants described here show that ribozymes using the chemical pathway that produces 2′3′ cyclic phosphate and 5′ hydroxyl termini are capable of rate enhancements much larger than previously appreciated and provide an experimental system for the direct investigation of these fast processes.
Acknowledgments
This work was funded by the Canadian Institutes of Health Research, the Canada Foundation for Innovation, and the Canada Research Chairs Program.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: VS, Varkud satellite; Ch, chase; Pre, precursor; Rz·S, cleaved ribozyme–substrate complex.
References
- 1.Campbell, F. E., Jr., Cassano, A. G., Anderson, V. E. & Harris, M. E. (2002) J. Mol. Biol. 317, 21-40. [DOI] [PubMed] [Google Scholar]
- 2.Katoh, H., Yoshinaga, M., Yanagita, T., Ohgi, K., Irie, M., Beintema, J. J. & Meinsma, D. (1986) Biochim. Biophys. Acta 873, 367-371. [DOI] [PubMed] [Google Scholar]
- 3.Beebe, J. A. & Fierke, C. A. (1994) Biochemistry 33, 10294-10304. [DOI] [PubMed] [Google Scholar]
- 4.Herschlag, D. & Cech, T. R. (1990) Biochemistry 29, 10159-10171. [DOI] [PubMed] [Google Scholar]
- 5.Glasner, M. E., Bergman, N. H. & Bartel, D. P. (2002) Biochemistry 41, 8103-8112. [DOI] [PubMed] [Google Scholar]
- 6.Tang, J. & Breaker, R. R. (2000) Proc. Natl. Acad. Sci. USA 97, 5784-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breaker, R. R., Emilsson, G. M., Lazarev, D., Nakamura, S., Puskarz, I. J., Roth, A. & Sudarsan, N. (2003) RNA 9, 949-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narlikar, G. J. & Herschlag, D. (1997) Annu. Rev. Biochem. 66, 19-59. [DOI] [PubMed] [Google Scholar]
- 9.Emilsson, G. M., Nakamura, S., N., Roth, A. & Breaker, R. R. (2003) RNA 9, 907-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bevilacqua, P. C. (2003) Biochemistry 42, 2259-2265. [DOI] [PubMed] [Google Scholar]
- 11.Saville, B. J. & Collins, R. A. (1990) Cell 61, 685-696. [DOI] [PubMed] [Google Scholar]
- 12.Sigurdsson, S. T., Thomson, J. B. & Eckstein, F. (1998) in RNA Structure and Function, eds. Simons, R. W. & Grunberg-Manago, M. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 339-376.
- 13.Butcher, S. E. (2001) Curr. Opin. Struct. Biol. 11, 315-320. [DOI] [PubMed] [Google Scholar]
- 14.Beattie, T. L., Olive, J. E. & Collins, R. A. (1995) Proc. Natl. Acad. Sci. USA 92, 4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins, R. A. (2002) Biochem. Soc. Trans. 30, 1122-1126. [DOI] [PubMed] [Google Scholar]
- 16.Lafontaine, D. A., Norman, D. G. & Lilley, D. M. (2002) Biochem. Soc. Trans. 30, 1170-1175. [DOI] [PubMed] [Google Scholar]
- 17.Jones, F. D. & Strobel, S. A. (2003) Biochemistry 42, 4265-4276. [DOI] [PubMed] [Google Scholar]
- 18.Hiley, S. L., Sood, V. D., Fan, J. & Collins, R. A. (2002) EMBO J. 21, 4691-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, H. C., De Abreu, D. M., Tillier, E. R., Saville, B. J., Olive, J. E. & Collins, R. A. (1993) J. Mol. Biol. 232, 351-361. [DOI] [PubMed] [Google Scholar]
- 20.Andersen, A. A. & Collins, R. A. (2000) Mol. Cell 5, 469-478. [DOI] [PubMed] [Google Scholar]
- 21.Milligan, J. F., Groebe, D. R., Witherell, G. W. & Uhlenbeck, O. C. (1987) Nucleic Acids Res. 15, 8783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins, R. A. & Olive, J. E. (1993) Biochemistry 32, 2795-2799. [DOI] [PubMed] [Google Scholar]
- 23.Hiley, S. L. & Collins, R. A. (2001) EMBO J. 20, 5461-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzmic, P. (1996) Anal. Biochem. 237, 260-273. [DOI] [PubMed] [Google Scholar]
- 25.Li, Y. & Breaker, R. R. (1999) J. Am. Chem. Soc. 121, 5364-5372. [Google Scholar]
- 26.Esteban, J. A., Walter, N. G., Kotzorek, G., Heckman, J. E. & Burke, J. M. (1998) Proc. Natl. Acad. Sci. USA 95, 6091-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, K. A. (1992) in The Enzymes, ed. Sigman, D.S. (Academic, San Diego), Vol. XX, pp. 1-61. [Google Scholar]
- 28.Jones, F. D., Ryder, S. P. & Strobel, S. A. (2001) Nucleic Acids Res. 29, 5115-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saville, B. J. & Collins, R. A. (1991) Proc. Natl. Acad. Sci. USA 88, 8826-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buzayan, J. M., Gerlach, W. L. & Bruening, G. (1986) Nature 323, 349-352. [Google Scholar]
- 31.Nesbitt, S. M., Erlacher, H. A. & Fedor, M. J. (1999) J. Mol. Biol. 286, 1009-1024. [DOI] [PubMed] [Google Scholar]
- 32.Stage-Zimmermann, T. K. & Uhlenbeck, O. C. (2001) Nat. Struct. Biol 8, 863-867. [DOI] [PubMed] [Google Scholar]
- 33.De la Pena, M., Gago, S. & Flores, R. (2003) EMBO J. 22, 5561-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khvorova, A., Lescoute, A., Westhof, E. & Jayasena, S. (2003) Nat. Struct. Biol. 10, 708-712. [DOI] [PubMed] [Google Scholar]