Abstract
KaiA is a two-domain circadian clock protein in cyanobacteria, acting as the positive element in a feedback loop that sustains the oscillation. The structure of the N-terminal domain of KaiA is that of a pseudo-receiver, similar to those of bacterial response regulators, which likely interacts with components of the clock-resetting pathway. The C-terminal domain of KaiA is highly conserved among cyanobacteria and enhances the autokinase activity of KaiC. Here we present the NMR structure of the C-terminal domain of KaiA from the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. This domain adopts a novel all α-helical homodimeric structure. Several mutations known to affect the period of the circadian oscillator are shown to be located at an exposed groove near the dimer interface. This NMR structure and a 21-Å-resolution electron microscopy structure of the hexameric KaiC particle allow us to postulate a mode of KaiA–KaiC interaction, in which KaiA binds a linker region connecting two globular KaiC domains.
Circadian timekeeping oscillators and their accompanying machinery comprise widespread regulatory mechanisms that control the ≈24-h temporal pattern of diverse physiological processes, ranging from leaf movement to transcription regulation (1, 2). Circadian oscillators have been identified in virtually all eucaryotic groups, including insects, fungi, plants, and mammals (3), and in cyanobacteria (4) and share mechanistic similarities, such as positive and negative transcription–translation feedback loops (3). The cyanobacterial clock is the simplest and potentially oldest (5) identified thus far, composed of three interacting clock proteins, KaiA, KaiB, and KaiC, (4, 6) that form heteromultimeric complexes in vivo of sizes that oscillate in a circadian manner (7). Transcription from practically all promoter regions in Synechococcus elongatus PCC 7942 is under circadian control, and transcription levels oscillate in phase or in antiphase with transcription from the kai locus (4, 8). Indeed, even promoters from other organisms, such as Escherichia coli, become rhythmically expressed in S. elongatus (8).
We previously reported that KaiA enhances the autokinase activity of KaiC in vitro (9). The KaiC phosphorylation state changes in vivo with a circadian pattern (10, 11), and ATP-binding by KaiC promotes formation of a hexameric KaiC particle (12, 13), which is presumed to be the functional form of the protein. Evidence suggests that KaiC binds forked DNA when in the hexameric form (13). KaiC preparations have been subjected to electron microscopic studies by two independent groups resulting in two structures (12, 13). KaiB is known to attenuate the KaiA-enhanced KaiC autokinase activity (9, 10, 14); however, no other information regarding its biochemical function is presently available.
KaiA is a two-domain protein. We previously showed that the C-terminal KaiA domain alone is sufficient in vitro to enhance the autokinase activity of KaiC (9), whereas the N-terminal domain has no effect in that assay. The N-terminal domain of KaiA adopts the fold of a canonical response regulator receiver domain (9, 15), although the primary sequence is dissimilar to that of receivers, and it lacks the conserved aspartyl residue necessary for phosphorylation. Similar to other pseudo-receiver domains (16), KaiA activation most likely involves direct protein–protein interactions of the N-terminal domain (with an as-yet-unidentified protein) that result in functional modulation of the C-terminal effector domain. We postulated that KaiA is a two-domain response regulator that acts as a timing input device to control the phase of the circadian period by modulating the KaiC phosphorylation state (9).
Here we report the solution structure of the ≈12-kDa KaiC-interacting C-terminal KaiA domain (residues 180–283, henceforth named ThKaiA180C) from the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1 (17), which grows naturally at temperatures >50°C. The superior thermal stability of this domain compared with the equivalent domain of KaiA from S. elongatus (Fig. 5, which is published as supporting information on the PNAS web site) enabled us to solve the structure at the physiological temperature of 50°C. We also determined the structure of S. elongatus KaiC by electron microscopy (EM) and single-particle reconstruction to a resolution of 21 Å. The structures raise interesting proposals for the mode of KaiA–KaiC interaction.
Materials and Methods
Protein Preparation. Samples of S. elongatus KaiC and full-length KaiA were prepared as described (9). Expression and purification of ThKaiA180C are described elsewhere (18). NMR samples consisted of ≈1.3 mM protein in a buffer containing 20 mM NaCl and 20 mM Na2HPO4, pH 7.07, at 23°C. Heterodimeric (13C/12C) samples of ThKaiA180C were prepared by mixing equimolar amounts of oxidized 13C/15N-enriched and -unenriched protein in 6 M GdHCl/50 mM NaCl/20 mM Tris·HCl, pH 7.0, with the addition of 100 mM β-mercaptoethanol. Complete reduction of the intermolecular disulfide bond was monitored by SDS/PAGE. The protein was reconstituted by slow dilution with a 20-fold excess volume of 50 mM NaCl/20 mM Tris·HCl, pH 7.0, buffer and exchanged to the final NMR buffer and reoxidized. The final protein concentration of the heterodimeric samples was a 1.0 mM ThKaiA180C dimer (0.5 mM heterodimer plus 0.25 mM fully enriched homodimer plus 0.25 mM unenriched homodimer). Protein concentration was determined by absorbance at 280 nm by using a corrected extinction coefficient as suggested by Pace et al. (19).
NMR Experiments. Sequential backbone and sidechain assignments of ThKaiA180C have been reported (18). All experiments were performed at 50°C with Varian Inova 600- and 500-MHz spectrometers equipped with triple axis gradient probes. The isomerization state of all proline residues was determined as trans (20). Stereospecific assignments of methylene and methyl protons and χ1 and leucine χ2 angle determinations were preformed as described (9). Interproton nuclear Overhauser effect (NOE) distance restraints were obtained by 13C/13C-edited 4D NOESY (100-ms mixing time), 15N-edited 3D NOESY (100-ms mixing time), and 13C-edited/12C-filtered 3D NOESY (21) (120-ms mixing time) spectra. Hydrogen bond restraints were applied based on hydrogen exchange protection data collected by NMR and the existence of expected regular secondary structure NOEs (20). Backbone dynamics measurements (15N T1, 15N T2, and 15N{-1H} NOE) were performed and analyzed as described (22). Spectrometer temperature was calibrated with a methanol sample.
NMR Structure Calculations. ϕ and Ψ dihedral angle values were derived from TALOS (23). Only residues for which all 10 predictions lie in the same region of the Ramachandran plot were used. The XPLOR-NIH software package (24) was used for all stages of NMR structure calculations. A distance-geometry simulating annealing protocol in Cartesian space was used to generate an initial set of structures from NOE and dihedral angle data. After inspection, the structures were refined by a simulated slow-cooling process and a final minimization. During refinement, additional restraints were used, including a conformational database potential (25), direct refinement against 13Cα and 13Cβ chemical shifts (26), and a radius of gyration restraint (27). The latter was applied independently to each monomeric subunit for residues of the globular core (inclusive, residues 186–204, 215–243, and 257–275) with a calculated value of 10.94 Å for the radius of gyration. Noncrystallographic symmetry was enforced between the two monomeric subunits throughout the calculation because only one set of resonances was observed. However, to better sample possible conformations of the intermolecular disulfide bond, C272 was excluded from this symmetry term. The family of structures consists of 25 low-energy structures that satisfy all experimental restraints (Table 1, which is published as supporting information on the PNAS web site).
EM. N-terminal His-tagged KaiC (0.1 mg/ml) in 300 mM NaCl/40 mM Tris·HCl, pH 7.5/1 mM ATP was negatively stained with 1% (wt/vol) uranyl acetate on a glow-discharged carbon grid. Images were recorded in a JEOL 1200 transmission electron microscope operating at 100 kV and at a calibrated magnification of ×38,900. Micrographs were digitized with a Leafscan 45 microdensitometer (Leaf Systems, Southborough, MA) at 20-μm increments corresponding to 5.14 Å per pixel at the specimen level. Single-particle analysis was performed by using the EMAN software package (28). About 1,000 particle images were used for the final reconstruction. The volume of the particle was rendered based on the calculated molecular mass of 344.5 kDa and an average density of 1.37 Da/Å3. The resolution of the 3D reconstruction was estimated to be ≈21 Å by Fourier shell correlation (Fig. 6B, which is published as supporting information on the PNAS web site).
Figure Preparation. Figures were prepared by SPOCK (29). The three N-terminal non-KaiA-derived residues of ThKaiA180C are not shown in the figures.
Results
The ThKaiA180C Domain. KaiA is the cyanobacterial clock protein for which the greatest divergence in sequence exists among different cyanobacterial groups (9, 30). This divergence appears primarily in the N-terminal pseudo-receiver domain; the C-terminal domain is highly conserved among all KaiA proteins known to date (Fig. 7, which is published as supporting information on the PNAS web site). For instance, the S. elongatus and T. elongatus N- and C-terminal domains share 30% and 62% identical residues, respectively. Furthermore, there are KaiA proteins that lack the N-terminal domain altogether (Anabaena sp. PCC 7120) or have only a truncated version (Nostoc punctiforme). The highly conserved C-terminal domain does not show significant sequence similarity to any other protein or derived domain from any organism studied. This simultaneous high degree of conservation in cyanobacteria and absence in other organisms suggests that the C-terminal domain of KaiA serves a function specific to this system.
The C-terminal domain of the S. elongatus KaiA was unfavorable for NMR structure determination because of limited aliphatic proton chemical shift dispersion and fast exchange of amide protons. In addition, thermal denaturation experiments showed little cooperativity of unfolding (Fig. 5). In contrast, the C-terminal domain of KaiA from T. elongatus has greater chemical shift dispersion and displays excellent cooperativity (Fig. 5). Thus, we proceeded to solve the structure of the T. elongatus KaiA C-terminal domain, which by sequence identity should be virtually equivalent to that of S. elongatus.
We previously showed that the N-terminal domain of S. elongatus KaiA is monomeric in solution even at high concentrations (9). Full-length S. elongatus KaiA, however, elutes off a calibrated gel filtration column at a molecular mass higher than the monomeric 32 kDa (data not shown). Analytical ultracentrifugation equilibrium of full-length KaiA showed a single species with molecular mass consistent with a dimer (Fig. 8, which is published as supporting information on the PNAS web site). Additionally, NMR relaxation measurements performed at 50°C on ThKaiA180C yielded average 15N T2 values for ordered residues (15N{-1H} NOE values >0.6; Fig. 9C, which is published as supporting information on the PNAS web site) of ≈70 ms and a calculated total correlation time, τc, of ≈9.4 ns (Fig. 9B), which are consistent with a molecular species of ≈24 kDa, corresponding to the ThKaiA180C homodimer. Only a single set of resonances was observed for each residue, which indicates that ThKaiA180C exists as a symmetric homodimer on the NMR timescale.
Structure Determination. The solution structure of ThKaiA180C was calculated from a total of 2,207 experimental restraints per monomeric subunit (Table 1). There were 50 unambiguous intersubunit distance restraints, most identified through a 13C-edited/12C-filtered 3D NOESY spectrum (21) collected in a equimolar mixture of fully 13C/15N-enriched and -unenriched protein. An additional 87 restraints were identified from the 13C/13C-edited 4D NOESY and 15N-edited 3D NOESY spectra as having inter-/intrasubunit ambiguity and were treated as such. We previously reported that the single cysteine residue of the monomeric subunit, C272, forms an intersubunit disulfide bond with C272′ (prime denotes the second subunit) (18), which was accounted for in the structure calculation. The precision of the final family of structures was calculated against the average structure for all ordered residues and is 0.31 ± 0.06 Å for the backbone and 0.75 ± 0.06 Å for all heavy atoms of a monomeric subunit. For the dimer, this precision is 0.42 ± 0.13 Å for the backbone and 0.81 ± 0.08 Å for all heavy atoms (Fig. 1B and Table 1).
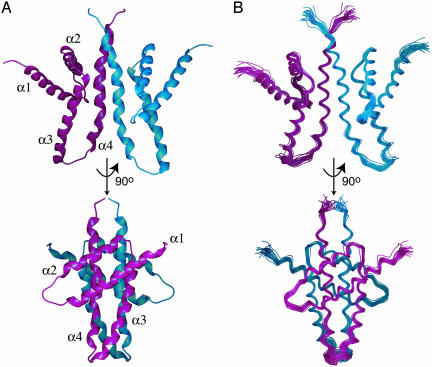
Fig. 1.
The solution structure of ThKaiA180C. Shown here are a schematic representation of the average minimized structure (PDB ID code 1Q6A) (A) and the overlaid backbone of a 25-structure ensemble (PDB ID code 1Q6B) (B) of ThKaiA180C at two mutually orthogonal views. The four helices composing the monomer structure are identified. The two monomeric subunits are discriminated by color (magenta and blue). Note the wide angle formed between the packing helical pairs, which places ThKaiA180C in the so-called X class of four-helix bundles (35).
Description of the ThKaiA180C structure. The monomeric subunit. ThKaiA180C is composed of four α-helices (henceforth α1, α2, α3, and α4) organized in two antiparallel helical pairs (α1 with α2 and α3 with α4) that pack against each other at a ≈54° angle (Fig. 1 A). The loops connecting helices α1 with α2 and α3 with α4 experience significant internal motion as indicated both by the lower number of long-range 1H–1H NOEs and heteronuclear 15N{-1H} NOE values <0.6. In contrast, the loop connecting α2 with α3 has an average 15N{-1H} NOE value of 0.75 and shows contacts both in the same monomeric subunit and across the dimer interface (see below).
The hydrophobic core of the monomeric subunit is mostly contained in the region where the four helices pack against each other. It is composed of 11 hydrophobic residues whose sidechains are completely buried, three of which are aromatic (Fig. 7). An additional 11 nonpolar residues pack around this core, thereby increasing the total hydrophobic area buried. The hydrogen bonding network is mostly restricted to residues that are close in the primary sequence (e.g., the i to i+4 hydrogen bonds of α-helical segments). Only four hydrogen bonds were inferred from the structure to occur between residues far apart in the sequence. These hydrogen bonds are between α1 and α2 (O′ of L189 and Nζ of K221), between α1 and α3 (Oη of Y197 and Nδ1 of H236) and between α1 and α4 (Oη of Y197 and N of L264, and between Oη of Y204 and the Oδ1 of D266). Stability and fold specificity is further enhanced through a number of intra- and interhelical salt-bridges (e.g., between residues R187 and E234 of α1 and α3). Sequence alignment between the C-terminal domains of KaiA from T. elongatus and S. elongatus shows that all buried residues are identical or conservatively substituted (Fig. 7). Similarly, three of the four long-range hydrogen bonds inferred from the structure involve residues conserved between these two organisms. In contrast, many of the favorable electrostatic interactions in T. elongatus KaiA are eliminated in S. elongatus through substitutions of charged residues with uncharged polar ones. This difference could account for the significant reduction in stability and cooperativity of unfolding observed in the C-terminal domain of S. elongatus KaiA (Fig. 5).
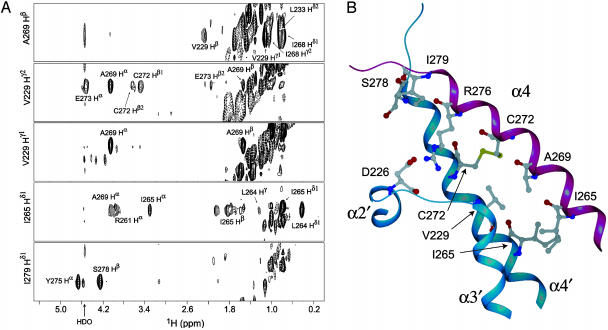
The dimer interface. The dimer interface of ThKaiA180C (Fig. 2B) involves almost the entire α4, the N-terminal part of α3, and the loop connecting α2 with α3. Formation of the dimer buries >900 Å2 of accessible surface area. Residues I265 to A269 of α4 pack against residues I265′ to A269′ of α4′ and the N-terminal part of α3′. Contacts form between residues I265 and I265′, I265 and L264′, and A269 and V229′. Fig. 2 A shows strips from the 13C-edited/12C-filtered 3D NOESY spectrum, where some of these contacts are identified.
Fig. 2.
(A) Selected 13C–1H strips from the 13C-edited/12C-filtered 3D NOESY spectrum (21) identifying NOEs at the dimer interface of ThKaiA180C. The spectrum was acquired in a heterodimeric sample of unenriched and 13C/15N-enriched protein. (B) The dimer interface of ThKaiA180C. Shown here is α4 of one subunit in magenta, and in blue the C-terminal region of α2′, the loop between α2′ and α3′, and the N-terminal region of α3′ and α4′ are shown. Residues identified as being part of the dimer interface by the 13C-edited/12C-filtered 3D NOESY and the intersubunit disulfide bond and salt-bridge are also shown.
Beyond A269 the dimer interface resembles that of a coiled-coil between α4 and α4′ extending from C272 to I279. The first interaction in this region is an intersubunit disulfide bond connecting residues C272 and C272′. It is worth noting that C272 is conserved in all but one of the known KaiA proteins (Fig. 7). Because no noncrystallographic symmetry requirement was applied on C272, the calculated disulfide geometry was an asymmetric intermediate of the left-hand-spiral and right-handhook disulfide conformations (31), both in terms of average Cα separation and sidechain dihedral angles adopted. The Hβ resonances of C272 appear significantly broader in all of the NMR spectra collected, likely because of localized motions on a μs-to-ms timescale.
The α4–α4′ coiled-coil is further stabilized by an intersubunit salt-bridge between residues R276 and D226′. In addition, Oε2 of E273 was inferred to participate in a bifurcated hydrogen bond with Oγ of S228′ and N of V229′. However, these hydrogen bonds do not occur in all of the structures of the ensemble; therefore, their presence is not certain. Although E273 is conserved among all KaiA molecules known, S228 is unique to T. elongatus KaiA.
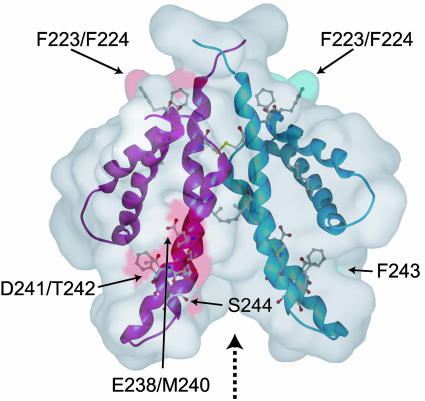
Possible Function of ThKaiA180C. Dimer formation creates a groove between α3–α4 and α3′–α4′ (dashed arrow in Fig. 3), thereby exposing several residues to solvent. Most are polar (D241, T242, S244, K245, K248, S253, E254, D255, and R261) and a couple are hydrophobic (L249 and L258). A number of substitutions in KaiA that cause altered circadian phenotype in S. elongatus map at this exposed region, and others map either at or near the hydrophobic core or the dimer interface (Fig. 3) (32). The substitutions at the hydrophobic core and dimer interface likely alter normal circadian rhythmicity by destabilizing the structural integrity of the C-terminal domain of KaiA. We postulate that the solvent-exposed groove directly interacts with KaiC, and substitutions within that region weaken the interaction between the two proteins, thereby altering the clock mechanism.
Fig. 3.
Accessible surface area representation of the ThKaiA180C structure and residue substitutions producing an altered circadian oscillator phenotype identified in S. elongatus KaiA as mapped at this domain. The accessible surface area for each substituted residue is colored in magenta or blue for the two monomeric subunits. The groove formed between α3 and α4 and α3′ and α4′ is indicated by a dashed arrow. Several of the circadian phenotype substitutions map at this groove.
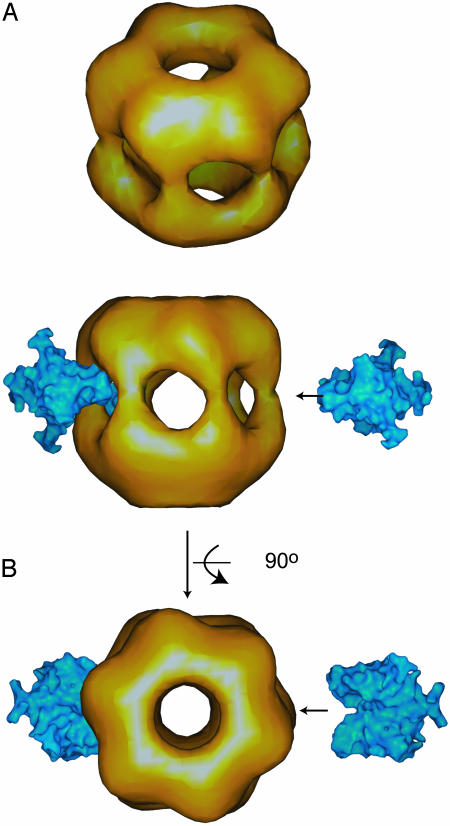
EM of KaiC. We calculated a 21-Å-resolution hexameric KaiC structure from EM data collected on negatively stained S. elongatus KaiC by the single particles method (Fig. 6). KaiC appears to form a dumbbell-shaped symmetric hexamer with a ≈50-Å radius and a 100-Å height (Fig. 4A), in agreement with what has been previously reported (12, 13). KaiC has two domains (CI, residues 1–250, and CII, residues 252–519) (6), both of which interact with KaiA in vitro. Our electron microscope reconstruction of the KaiC particle clearly shows two globular domains connected by an ≈10-Å linker region.
Fig. 4.
(A) Twenty-one-angstrom-resolution EM structure of the hexameric S. elongatus KaiC particle. The structure was derived from analysis of ≈1,000 single hexameric KaiC particle images. The ≈29-Å opening at the top of the structure and the separation between the two globular KaiC domains (putative CI and CII) can be seen. (B) Our proposed model of KaiA–KaiC interaction is shown. Depicted here is one C-terminal KaiA dimer in a molecular surface area representation modeled as “bound” to KaiC at the interface linker between the putative CI and CII domains. A second C-terminal KaiA dimer is also shown, and an arrow indicates the proposed approach.
Openings on both ends of the hexameric structure (≈29-Å top opening diameter, ≈23-Å bottom opening diameter) can be seen in our reconstruction of the S. elongatus KaiC particle, in agreement with data from Mori et al. (13). However, an electron microscope reconstruction by Hayashi et al. (12) of the T. elongatus hexameric KaiC features a hole only at the top of the particle. The latter structure also does not display holes at the side of the particle, although significant depressions can be seen. The KaiC sequence in these two strains is highly conserved (81% identical residues, 92% similar residues); therefore, we do not expect that the differences observed in the electron microscope reconstructions reflect real differences in the protein structures but, rather, a difference in the analysis and interpretation of the data. The presence of two openings in our structure is consistent with experimental data that show binding of forked DNA by KaiC and those author's proposal of the structure (13).
Discussion
The overall fold of the ThKaiA180C monomeric unit can be described as a right-turning, four-helix bundle (33) with similar topology to that of previously studied proteins, such as cytochrome b562 (34). However, the wide (54° on average) angle between the helical pairs places ThKaiA180C in the so-called X class of four-helix bundles as defined by Harris et al. (35). Note that this wide angle of helix packing is close to what Chothia et al. (36) predicted as a possible preferred angle in their “ridges-into-grooves” model. Examples of this X class are found as smaller parts in larger proteins, such as helices α1, α9, α10, and α11 of 3-isopropylmalate dehydrogenase (37). ThKaiA180C is a self-contained, X-class, four-helix bundle; structure-based search algorithms (38–40) did not find any other stand-alone proteins of this class in the Protein Data Bank. This finding is in contrast to the structure of the N-terminal KaiA domain, which was found to adopt a very common α/β fold (9). In addition, the quaternary homodimeric structure of ThKaiA180C is unusual, because dimerization involves a combination of coiled-coil and helix-in-cleft interactions.
Because the C-terminal domain of KaiA has a groove between the two monomeric subunits and many circadian phenotype substitutions map in that region, we propose that this groove of KaiA interacts with KaiC by binding the narrow linker connecting the two globular KaiC domains (putative CI and CII). This possible arrangement is shown in Fig. 4B, with one KaiA C-terminal domain bound to KaiC in the manner we propose and a second KaiA C-terminal domain indicating the proposed mode of approach. It is worth noting that our proposal is consistent with previous studies that identified putative KaiA-binding domains in KaiC close to the CI–CII interface (41) and provides a structural form-fitting basis for the mode of interaction.
In vivo studies have shown the stoichiometry of KaiA to KaiC to be ≈1:20 (14) (corresponding to a stoichiometry of 1 KaiA dimer per 6.7 KaiC hexamers). Although this would seem to indicate that, on average, KaiA–KaiC complexes involve a single KaiA dimer, it is still possible that the active species of this complex has a higher KaiA:KaiC stoichiometry. Therefore, the KaiC phosphorylation state may be controlled simply through variation of the number of KaiA dimers bound to the hexameric KaiC particle or their complete absence. Similarly, amino acid substitutions in KaiA with altered circadian phenotype could attenuate KaiA stimulation of the KaiC autokinase activity, either through disruption of the KaiA structure or by weakening the KaiA–KaiC interaction. Notably, the phenotypes are longer free-running periods or arrhythmia (32), which suggests that a reduced ability of KaiA to enhance the KaiC phosphorylation rate directly lengthens the circadian period.
The size of the clock protein complexes varies in vivo during the circadian cycle (7). KaiA alternates between a complex of molecular mass approximately equal to that of the KaiA dimer during the subjective day and a complex with a mass >500 kDa during the subjective night. We expect that this latter complex contains one or more KaiA dimers bound to the hexameric KaiC particle in addition to other clock components, such as KaiB and SasA. Therefore, a possible mechanism of circadian oscillator phase resetting emerges, in which protein–protein interactions at the N-terminal pseudo-receiver domain are followed by signal propagation to the C-terminal KaiC-interacting domain of KaiA (9). The conformational change at the C-terminal domain would affect the strength of the KaiA–KaiC interaction, thereby altering the number of KaiA dimers bound to the KaiC particle and, thus, the phase of the oscillator.
Supplementary Material
Acknowledgments
We thank Dr. Satoshi Tabata (Kazusa DNA Research Institute, Chiba, Japan) for providing T. elongatus DNA, Dr. Karl Koshlap for technical assistance with the NMR spectrometers, and Shannon Canales for helpful comments on the manuscript. Funding was provided to A.C.L. and S.S.G. by National Institutes of Health Grants GM064576 and GM062419, respectively. A.H. thanks the Office of the Vice President for Research for financial support. The NMR instrumentation in the Biomolecular NMR Laboratory at Texas A&M University was supported by National Science Foundation Grant DBI-9970232.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NOE, nuclear Overhauser effect; EM, electron microscopy.
Data Deposition: The solution structure has been deposited in the Protein Data Bank, www.rcsb.org (PBD ID codes 1Q6A and 1Q6B for the average minimized structure and the 25-structure ensemble, respectively).
References
- 1.Bünning, E. (1973) The Physiological Clock: Circadian Rhythms and Biological Chronometry (Springer, New York), 3rd. Ed.
- 2.Young, M. W. & Kay, S. A. (2001) Nat. Rev. Genet. 2, 702-715. [DOI] [PubMed] [Google Scholar]
- 3.Dunlap, J. C. (1999) Cell 96, 271-290. [DOI] [PubMed] [Google Scholar]
- 4.Ishiura, M., Kutsuna, S., Aoki, S., Iwasaki, H., Andersson, C. R., Tanabe, A., Golden, S. S., Johnson, C. H. & Kondo, T. (1998) Science 281, 1519-1523. [DOI] [PubMed] [Google Scholar]
- 5.Dvornyk, V., Vinogradova, O. & Nevo, E. (2003) Proc. Natl. Acad. Sci. USA 100, 2495-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwasaki, H., Taniguchi, Y., Ishiura, M. & Kondo, T. (1999) EMBO J. 18, 1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kageyama, H., Kondo, T. & Iwasaki, H. (2002) J. Biol. Chem. 277, 7157-7164. [DOI] [PubMed] [Google Scholar]
- 8.Liu, Y., Tsinoremas, N. F., Johnson, C. H., Lebedeva, N. V., Golden, S. S., Ishiura, M. & Kondo, T. (1995) Genes Dev. 9, 1469-1478. [DOI] [PubMed] [Google Scholar]
- 9.Williams, S. B., Vakonakis, I., Golden, S. S. & LiWang, A. C. (2002) Proc. Natl. Acad. Sci. USA 99, 15357-15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu, Y., Mori, T. & Johnson, C. H. (2003) EMBO J. 22, 2117-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki, H., Nishiwaki, T., Kitayama, Y., Nakajima, M. & Kondo, T. (2002) Proc. Natl. Acad. Sci. USA 99, 15788-15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi, F., Suzuki, H., Iwase, R., Uzumaki, T., Miyake, A., Shen, J. R., Imada, K., Furukawa, Y., Yonekura, K., Namba, K. & Ishiura, M. (2003) Genes Cells 8, 287-296. [DOI] [PubMed] [Google Scholar]
- 13.Mori, T., Saveliev, S. V., Xu, Y., Stafford, W. F., Cox, M. M., Inman, R. B. & Johnson, C. H. (2002) Proc. Natl. Acad. Sci. USA 99, 17203-17208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitayama, Y., Iwasaki, H., Nishiwaki, T. & Kondo, T. (2003) EMBO J. 22, 2127-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volz, K. (1993) Biochemistry 32, 11741-11753. [DOI] [PubMed] [Google Scholar]
- 16.O'Hara, B. P., Norman, R. A., Wan, P. T. C., Roe, S. M., Barrett, T. E., Drew, R. E. & Pearl, L. H. (1999) EMBO J. 18, 5175-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaoka, T., Satoh, K., Katoh, S. (1978) Plant Cell Physiol. 19, 943-954. [Google Scholar]
- 18.Vakonakis, I., LiWang, A. C. (2004) J. Biomol. NMR, in press.
- 19.Pace, C. N., Vajdos, F., Fee, L., Grimsley, G. & Gray, T. (1995) Protein Sci. 4, 2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wüthrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 21.Lee, W., Revington, M. J., Arrowsmith, C. & Kay, L. E. (1994) FEBS Lett. 350, 87-90. [DOI] [PubMed] [Google Scholar]
- 22.LiWang, A. C., Cao, J. J., Zheng, H., Lu, Z., Peiper, S. C. & LiWang, P. J. (1999) Biochemistry 38, 442-453. [DOI] [PubMed] [Google Scholar]
- 23.Cornilescu, G., Delaglio, F. & Bax, A. (1999) J. Biomol. NMR 13, 289-302. [DOI] [PubMed] [Google Scholar]
- 24.Schwieters, C. D., Kuszewski, J. J., Tjandra, N. & Marius Clore, G. (2003) J. Magn. Reson. 160, 65-73. [DOI] [PubMed] [Google Scholar]
- 25.Kuszewski, J., Gronenborn, A. M. & Clore, G. M. (1996) Protein Sci. 5, 1067-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuszewski, J., Qin, J., Gronenborn, A. M. & Clore, G. M. (1995) J. Magn. Reson. B 106, 92-96. [DOI] [PubMed] [Google Scholar]
- 27.Kuszewski, J., Gronenborn, A. M. & Clore, G. M. (1999) J. Am. Chem. Soc. 121, 2337-2338. [Google Scholar]
- 28.Ludtke, S. J., Baldwin, P. R. & Chiu, W. (1999) J. Struct. Biol. 128, 82-97. [DOI] [PubMed] [Google Scholar]
- 29.Christopher, J. A. (1999) spock (Center for Macromolecular Design, Texas A&M Univ.), Version 1.0b.
- 30.Lorne, J., Scheffer, J., Lee, A., Painter, M. & Miao, V. P. (2000) FEMS Microbiol. Lett. 189, 129-133. [DOI] [PubMed] [Google Scholar]
- 31.Richardson, J. S. (1981) Adv. Protein Chem. 34, 167-339. [DOI] [PubMed] [Google Scholar]
- 32.Nishimura, H., Nakahira, Y., Imai, K., Tsuruhara, A., Kondo, H., Hayashi, H., Hirai, M., Saito, H. & Kondo, T. (2002) Microbiology 148, 2903-2909. [DOI] [PubMed] [Google Scholar]
- 33.Kamtekar, S. & Hecht, M. H. (1995) FASEB J. 9, 1013-1022. [DOI] [PubMed] [Google Scholar]
- 34.Lederer, F., Glatigny, A., Bethge, P. H., Bellamy, H. D. & Matthew, F. S. (1981) J. Mol. Biol. 148, 427-448. [DOI] [PubMed] [Google Scholar]
- 35.Harris, N. L., Presnell, S. R. & Cohen, F. E. (1994) J. Mol. Biol. 236, 1356-1368. [DOI] [PubMed] [Google Scholar]
- 36.Chothia, C., Levitt, M. & Richardson, D. (1981) J. Mol. Biol. 145, 215-250. [DOI] [PubMed] [Google Scholar]
- 37.Imada, K., Sato, M., Tanaka, N., Katsube, Y., Matsuura, Y. & Oshima, T. (1991) J. Mol. Biol. 222, 725-738. [DOI] [PubMed] [Google Scholar]
- 38.Shindyalov, I. N. & Bourne, P. E. (1998) Protein Eng. 11, 739-747. [DOI] [PubMed] [Google Scholar]
- 39.Madej, T., Gibrat, J. F. & Bryant, S. H. (1995) Proteins 23, 356-369. [DOI] [PubMed] [Google Scholar]
- 40.Holm, L. & Sander, C. (1996) Science 273, 595-603. [DOI] [PubMed] [Google Scholar]
- 41.Taniguchi, Y., Yamaguchi, A., Hijikata, A., Iwasaki, H., Kamagata, K., Ishiura, M., Go, M. & Kondo, T. (2001) FEBS Lett. 496, 86-90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.