Abstract
Fluorescence in situ hybridization (FISH) is an effective method for the physical mapping of genes and repetitive DNA sequences on chromosomes. Physical mapping of unique nucleotide sequences on specific rice chromosome regions was performed using a combination of chromosome identification and highly sensitive FISH. Increases in the detection sensitivity of smaller DNA sequences and improvements in spatial resolution have ushered in a new phase in FISH technology. Thus, it is now possible to perform in situ hybridization on somatic chromosomes, pachytene chromosomes, and even on extended DNA fibers (EDFs). Pachytene-FISH allows the integration of genetic linkage maps and quantitative chromosome maps. Visualization methods using FISH can reveal the spatial organization of the centromere, heterochromatin/euchromatin, and the terminal structures of rice chromosomes. Furthermore, EDF-FISH and the DNA combing technique can resolve a spatial distance of 1 kb between adjacent DNA sequences, and the detection of even a 300-bp target is now feasible. The copy numbers of various repetitive sequences and the sizes of various DNA molecules were quantitatively measured using the molecular combing technique. This review describes the significance of these advances in molecular cytology in rice and discusses future applications in plant studies using visualization techniques.
Keywords: Oryza sativa L., chromosome, fluorescence in situ hybridization (FISH), extended DNA fibers (EDFs), genomic in situ hybridization (GISH)
Introduction
In situ hybridization (ISH) is a useful method to visualize the localization of nucleotide sequences on chromosomes, nuclei, and tissues. The principle of ISH is to hybridize labeled nucleotide sequences (or probes) as reporter molecules directly onto complementary DNA or RNA sequences on slide glass. Following hybridization, the reporter molecule in the hybridized DNA is detected by antibodies or affinity chemicals labeled with fluorescent molecules, to be visualized under a microscope, for example. This ISH using fluorescence to detect DNA probes is referred to as fluorescence in situ hybridization (FISH). Technical details on using this technique in both animals and plants have been published in many good manuals.1),2) The visualization of DNA probes using various techniques has been performed in recent decades and has been applied in the study of many plants of agricultural importance as well as in plant genome research. FISH has been extensively used in important members of the plant kingdom, such as rice, wheat, maize, tomato, Brassica, and Arabidopsis.3)–8) Technical advances in cytology and FISH applications, such as DNA mapping on chromosomes, nuclei, and DNA fibers, have dramatically progressed to allow the detection of DNA sequences of fewer than a few kilobase pairs. FISH mapping of single genes enables a direct comparison of the physical location of a gene and its position on the linkage map.9) Currently, the functional aspects of histone protein modifications are being analyzed. Histone modifications, such as methylation and acetylation, are known to be involved in the epigenetic regulation of gene expression and are thus related to functional regions of chromosomes, aiding in the differentiation of heterochromatin and euchromatin. Technological advancements have been utilized in the development of a number of genetic methods, each with advantages and drawbacks. In this review, we discuss the effects and versatility of these methods in plant research, as the development of visualization methods is considered a significant milestone in molecular cytology.
Detection of ribosomal RNA genes by in situ hybridization
Efforts to visualize specific DNA sequences directly on chromosomes had been pursued for years, but good methods for identifying chromosomes or detecting genes on a chromosome first became available at the beginning of the 1980s. The first reproducible result was the success of Fukui in physically locating 18S-5.8S-25S ribosomal RNA gene (45S rDNA) loci at the end of a pair of chromosomes using 125iodine-labeled rRNA probes.9) Following hybridization with a radioactive ribosomal RNA probe, the radioactive hybridizing signals were detected using a photographic emulsion layered over the surface of the samples. Ten years later, ISH using haptenes such as biotin for a reporter molecule was developed, and soon it was found that these methods had many advantages over the earlier radioactive methods. One of several remarkable achievements that employed non-radioactive labeling systems, such as a colorimetric procedure using an enzymatic reaction, was the detection of 45S rDNA sites on rice chromosomes.10) Although the stability and safety of the detection procedures were high, the low spatial resolution, the probe size limitation, and the limited number of probes that could be used in single ISH experiment remained problems.
Based on these technological achievements, fluorescence in situ hybridization (FISH) technology was developed. The FISH technique for human chromosomes was developed first,11),12) followed by a method for the wheat genome.13),14) The significant advantages of FISH included good sensitivity and high spatial resolution, the ability to simultaneously detect several probes using different fluorochrome/color combinations, and versatility in three-dimensional analyses. The development of the FISH methodology allowed dramatic advances in molecular cytological studies even in rice, which has very small chromosomes. For example, 45S rDNA loci were detected in different rice species, and variability in the number of 45S rDNA loci among several rice species was demonstrated.15) A FISH study was also performed using two-color fluorescence to simultaneously detect 5S and 45S rDNAs loci.16) Both rDNA loci were visualized on the chromosomes of six species of the genus Oryza, and the rice chromosomes harboring the rDNA loci were identified based on their condensation patterns.10) The positions of the rDNAs were shown to be different in the different rice species.17) The 45S rDNA locus of japonica rice is localized on chromosome 9. Indica rice has the two loci on chromosomes 9 and 10. O. officinalis and O. eichingeri both have the three loci on chromosomes 4, 7, and 11 (Fig. 1, Table 1). Recent studies have demonstrated that the number of rDNA loci varies from one to eight among wild rice species.18) This variability is likely caused by either chromosome exchange or translocation and/or by a ribosomal RNA gene during the evolution of rice wild species. Variations in the location of the rDNA loci might indicate the transpositional nature of the rDNAs, as has been suggested in the genera Triticeae and Allium.19),20) Recently, flow cytometry and sorting studies of plant chromosomes have been carried out using suspensions of intact chromosomes, and individual chromosomes have been discriminated in cereals.21),22) 5S rDNA, 17S rDNA, and centromeric DNA have been used as probes in FISH for rye and barley chromosomes in suspension to label the specific chromosomes. Bright signals were detected at the specific regions of interest on the chromosomes. These results indicate that this method would be useful for the selection and sorting of plant chromosomes.
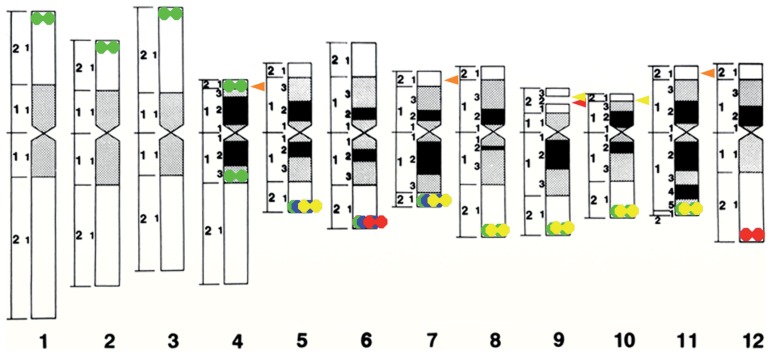
Fig. 1.
Distribution patterns of the genome-specific repetitive sequence TrsA and rDNA sites on a rice chromosome idiogram. Black and gray boxes: heavily and moderately condensed regions, respectively, based on the condensation patterns determined by CHIAS3. Doublet circles: Trs sites—two in O. sativa ssp. japonica (red), six in O. sativa ssp. indica (yellow), three in O. glaberrima (blue) and 12 in O. meridionalis (green). Most of those sites are located on the distal ends of long arms. Arrowhead: rDNA site—one in O. sativa ssp. japonica (red), two in O. sativa ssp. indica (yellow) and three each in O. officinalis and O. eichingeri (orange).
Table 1.
Chromosomal locations of 45S rDNA, 5S rDNA and TrsA of rice species
| Respective chromosomes | ||||
|---|---|---|---|---|
|
|
||||
| Genome | Species | 45SrDNA Ribosomal RNA gene |
5SrDNA Ribosomal RNA gene |
TrsA Tandem repeat A |
| AA | O. sativa ssp. japonica | 9 | 11 | 6, 12 |
| AA | O. sativa ssp. indica | 9, 10 | 11 | 5, 7, 8, 9, 10, 11 |
| AA | O. glaberrima | 9 | 5, 6, 7 | |
| AA | O. meridionalis | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 | ||
| BB | O. punctata, diploid* | 9 | 11 | |
| CC | O. officinalis | 4, 7, 11 | 11 | |
| CC | O. eichingeri | 4, 7, 11 | 11 | |
| EE | O. australinensis | 9 | 7 | |
| FF | O. brachyantha | 9 | 7 | |
Oryza punctata (diploid) has BB genome, while the tetraploid form of this species possesses the genome formula, BBCC.86)
Variability of chromosomal construction by repetitive sequences in Oryza species
Tandem repeat sequences and retroelements constitute a large fraction of the genomic DNA of plants. Rice, 2n = 24 with a genome size of 390 Mbp, has the smallest genome size among the major cereals,23) smaller than the other important cereals such as sorghum, maize, barley, and wheat (with genome sizes of 735, 2,600, 5,400, and 16,900 Mbp, respectively). The rice genome is an important reference point for all cereal genomes because it shows co-linearity with other agriculturally important crops.24) The rice genome contains 28,000 genes,25) and it also contains abundant repeated sequences. In plant genomes, repetitive DNAs are known to contribute considerably to the chromosome structure, including the heterochromatin, centromere, and telomere. FISH is very useful in identifying the genome-wide distribution pattern of different types of repetitive sequences in rice.
TrsA26) and Os4827) were the first tandem repeat sequences identified in rice. TrsA repeats have a 355-bp unit length and are present at 2,000–6,000 copies in japonica rice with the A genome. TrsA repeats occupy at least 10% of the total genome in indica rice. TrsA was localized to the short arm of chromosome 6 (6S) and the long arm of chromosome 12 (12L) (Fig. 1, Table 1). TrsA sites have also been detected in the subtelomeric regions in both japonica and indica rice chromosomes.28) TrsA copies are located at the distal end of chromosomes 6 and 12 in japonica rice and chromosomes 5, 7, 8, 9, 10, and 11 in indica rice.29) Although two pairs of TrsA were visually detected in japonica rice by FISH, six chromosome ends (3L, 5L, 6S, 8L, 9L, 12L) have been reported to contain TrsA.30) This suggests that four of the chromosomal ends (3L, 5L, 8L, 9L) are low-copy sites of TrsA that are undetectable by FISH. Wild rice species with the A genome shows variable numbers of TrsA copies and chromosomal positions; for example, chromosomes 5, 6, and 7 in O. glaberrima and all chromosomes but chromosome 12 in O. meridionalis contain TrsA copies (Table 1). Heterochromatic chromomere regions at the distal ends of O. meridionalis chromosomes consist of TrsA copies. Thus, TrsA likely contributes to the chromosome terminal structures and genome sizes of various rice species.31)
FISH studies have revealed different classes of retrotransposons that contribute to the chromosomal organization. The retrotransposable elements named LTR (long terminal repeats), such as Ty-1 copia-type and Ty-3 gypsy-type, are widespread in plant genomes.32) These elements also contribute to the large variations in genome size among diploid rice species.33) For example, O. australiensis (E genome) has the largest and O. brachyantha (F genome) the smallest genome size in rice. O. sativa (A genome), including all cultivated species, has an intermediate genome size. O. brachyantha, with a genome size of 346 Mb/1C, has a limited number of repetitive DNA sequences specific to the F genome.34) In comparison, O. australiensis, with a genome size of 946 Mb/1C, shows an overall amplification of the genome-specific DNA sequence RIRE1 throughout its chromosomes. RIRE1, a Ty-1 copia-type retroelement, is detected throughout the chromosomes of O. australiensis except the nucleolar organizing region (NOR) and centromeric regions. Other plant retroelements, such as BARE1 of barley, have high homology to RIRE1 and have been reported to show a chromosomal distribution pattern similar to that of RIRE1.35) The amplification of such copia-type retroelements causes variations in chromosome morphology and genome size among species, even within the same genus. The genetic markers of the wild-type rice O. punctata (BB genome) and O. officinalis (CC genome) have synthetic arrangements similar to the corresponding chromosomes of O. sativa.36) The positions of individual markers on the corresponding chromosomes among the three different rice species with A, B, and C genomes were quite similar.
In plant genomes, centromeres contain several kinds of retrotransposons and satellite repeats with 100–200 bp units of the short motif. The centromeric satellite sequences are the most rapidly evolved in the genome. The Ty-1 copia-type transposable element is considered the main tandem array that composes the pericentromeric regions in Arabidopsis thaliana.37) Moreover, the maize centromere is composed of fragments of the Ty-3 gypsy-type retrotransposons; CentA (a 156-bp satellite repeat) and CentC (a 156-bp repeat) are present in the centromeric regions.38) An RIRE7 with a tandem repeat sequence, named TrsD, is homologous to the tandem repeat sequences RCS2 and CentC, which were previously identified in the centromeric regions of rice and maize chromosomes, respectively.38)–40) Interestingly, the RIRE7 sequence is homologous to several DNA segments present in the centromeric regions of cereal chromosomes. FISH analysis using a BAC clone containing both RIRE7 and TrsD sequences also revealed their presence in the pericentromeric regions of the pachytene chromosomes in O. sativa cv. Nipponbare. High copy numbers of Ty-3 gypsy-type have been confirmed in many plant species, including rice, Arabidopsis, maize, barley, and wheat.37),38),41)–43) The gag-polymerase region of the Ty-3 gypsy-type retrotransposon, appearing as RIRE7, CRM, and LjRE2 repeats in rice, maize, and Lotus, respectively, has been detected in the pericentromeric regions of pachytene chromosomes by FISH.38)–41),43)–45) The centromeric sequences and structures are well understood in rice.45)–47)
The relationship between chromosomal function and repeat sequences in the centromere has been demonstrated by the concentration of histone variants. CentO, which is the sequence corresponding to the spindle-binding region of the kinetocore, provides evidence to confirm the relationship between chromosome function and the DNA sequence.48) Ty-3 gypsy-type centromere specific retrotransposons containing CentO are highly enriched in chromosome domains containing the centromere-specific histone H3 variant (CENH3).48) The CENH3 binding regions are thought to be the functional regions of the centromere. CENH3 genes have been found in all eukaryotes investigated, including humans (CENP-A)49) and A. thaliana (HTR12).50) Nagaki et al. have determined that centromeric sequences of approximately 750 kb bind to the rice CENH3.51) In the case of rice with 390 Mb genome size, which has no distinct heterochromatic chromocenters like Arabidopsis in nuclei,52) but has conspicuous heterochromatic regions in pachytene chromosomes, the signals of histone H3K9me2 were dispersed all over the nuclei with strong spot-like signals and enriched at heterochromatic regions in pachytene chromosomes.53) It is also worth noticing that histone H3K9me2 especially marked condensed regions within each chromomere. Pericentromeric regions composed of highly condensed chromomeres showed stronger and larger signals, that suggests chromatin condensation might directly or indirectly correlate with histone H3K9me2 in rice, too. In the centromeric regions responsible for kinetochore assembly and microtubule attachment, where a specialized histone H3 variant called CENH3 (CENP-A) replaces the canonical histone,52) H3K9me2 was not detected. We suppose that rice histone H3K9me2 can occur at condensed regions within each chromomere in varying degrees, and the degree of H3K9me2 is proportional to the degree of chromatin condensation. Moreover, it can be said that the pattern of H3K9me2 in rice pachytene chromosome might be consistent with uneven transcriptional activity along chromosomes.
Physical mapping of agricultural important genes on rice chromosomes
Molecular cytology techniques enabled the detection of unique rice genomic DNA sequences related to important agricultural traits. A yeast artificial chromosome (YAC) covering the gall midge resistance gene (Gm2) was mapped to chromosome 1 of the resistant variety of rice.54),55) Jiang et al.56) and Nakamura et al.54),57) visualized bacterial artificial chromosome (BAC) clones closely linked to a rice bacterial blight resistance locus, Xa21, and to the rice blast resistance gene Pi-b, respectively, both on chromosome 2 (Fig. 2). Finally, the improved detection sensitivity enabled the visualization of a small RFLP marker of only 1.29 kb.54) A single RFLP marker of 1.29 kb in size was mapped to the distal end of the long arm of rice chromosome 4 (chromosomal address: 4q2.1). Image analysis of the FISH results allowed the refinement of the localization of the molecular markers on the quantitative rice chromosome map. The combination of chromosome identification, physical mapping, and image analysis gives the most accurate physical mapping of genes. For example, a 180 kb BAC clone was FISH-mapped at 96.2% distance from the end of the short arm of chromosome 2 (Fig. 2g). The position of this BAC clone was localized at 98.7% (190.4 cM) from the end of the same chromosome arm on the genetic map.54) Thus, only a small difference was found between the estimated positions of this BAC clone on the physical and genetic map.
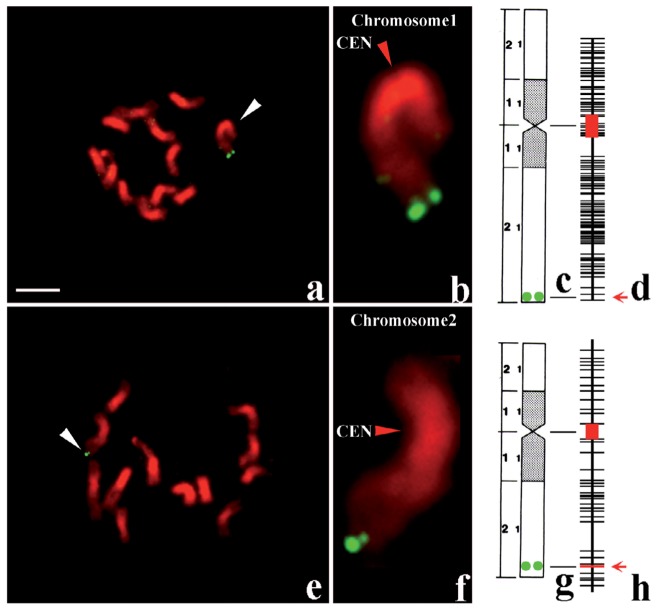
Fig. 2.
Physical mapping by FISH of a YAC clone (400 kb) carrying the gall midge resistance gene, Gm2, and a BAC clone (180 kb) containing a rice blast resistance gene, Pi-b, on rice haploid chromosomes.54) a–d: Mapping of the Gm2 carrying YAC clone that appeared as green fluorescent doublets on the probed chromosome 1. e–h: Mapping of the Pi-b carrying BAC clone that appeared as the green fluorescent doublets on the probed chromosome 2. a and e: An entire somatic haploid metaphase plate with 12 chromosomes, of which probed chromosome is indicated by a white arrowhead. b and f: Enlarged image of the respective signal-tagged chromosome, of which centromere is shown by red arrowhead. c and g: Cytological maps of chromosome 1 and 2, carrying Gm2 and Pi-b gene, respectively. Green doublet circles show the site of the probed YAC or BAC clone. d and h: Genetic maps of chromosome 1 and 2, of which centromeric regions are indicated by red boxes, and the sites of probed YAC and BAC clones by red arrows.
High resolution analysis for the integration of genetic, physical and cytological chromosome maps
Genetic linkage maps and physical maps are both beneficial to determine the positions of genes and specific DNA sequences in plant genomes. However, linkage maps based on recombination frequencies do not directly reflect physical distances, because genetic recombination does not occur at random along chromosomes. Therefore, the integration of linkage maps and physical maps is essential for the accurate positioning of genes and specific DNA sequences.58)–60) A standardized rice karyotype was constructed using pachytene chromosomes of O. sativa to facilitate rice chromosome identification.61) This karyotype consists of landmarks in cultivated and wild rice species by FISH using centromere specific and chromosomal arm specific BAC clones.62),63)
Further integration among the linkage map, somatic prometaphase map, and pachytene map, based on the positions of common BAC/PAC clones by FISH,53) was demonstrated. An idiogram depicting the distribution of heterochromatin in the rice pachytene chromosome was developed based on the patterns of 4’,6-diamidino-2-phenylindole (DAPI) and propidium iodide (PI) staining. In comparing the three maps, discrepancies between the positions of DNA markers on the linkage map and the FISH signal positions on pachytene chromosomes map were detected.64) In addition, a difference among the three maps in the position of NOR on chromosome 9 was shown. In the linkage map, NOR was not mapped, suggesting that the NOR region has no effective genetic marker. As previous studies demonstrated on chromosome 4, consistencies were found between the condensation patterns of somatic prometaphase chromosomes and the chromomeric characters in pachytene chromosomes. This result suggests that chromosomes might condense during mitosis and meiosis through the same mechanism.65)
Characterization of genome organization of hybrids and polyploids by GISH
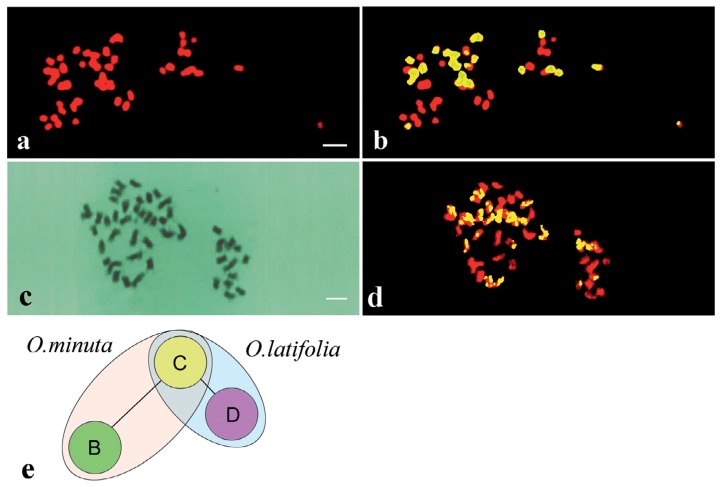
Chromosome painting by genomic in situ hybridization (GISH), a modified version of FISH technology, is also versatile tool in the field of plant cytology and cytogenetics.66) GISH allows the visualization and comparison of chromosomes and genomes of different materials, enabling one to characterize them as polyploids, F1 hybrids and their progenies, partial allopolyploids, polyhaploids, or recombinant lines.67)–71) Fig. 3 shows the distinction between two kinds of genomes in two wild tetraploids, O. minuta (BBCC, Fig. 3a and b) and O. latifolia (CCDD, Fig. 3c and d). The genetic distances among the three genomes in the two species can be resolved (Fig. 3e). Using the C genome as a pivotal genome by GISH, the genetic distance of the B genome from C is larger than that of the D genome from C. Recombination events can be diagnosed using GISH, which allows new insights into chromosome structure differentiation between the three genomes.
Fig. 3.
Genomic in situ hybridization (GISH) of two amphidiploids, O. minuta (BBCC, 2n = 48) and O. latifolia (CCDD, 2n = 48).68) a and b: Chromosomes of O. minuta. (a) Counterstained with propidium iodide, and (b) composite chromosome images of the counterstaining (red) and C genome signal image (yellow). c and d: GISH of O. latifolia (CCDD, 2n = 48) chromosomes. (c) Giemsa stained chromosomes before GISH and (d) a composite chromosome image of counterstaining (red) and signal image (yellow). e: Schematic representation of the genetic relationships between the B, C, and D genomes. Distances between the genomes are in arbitrary units. Bar indicates 10 μm.
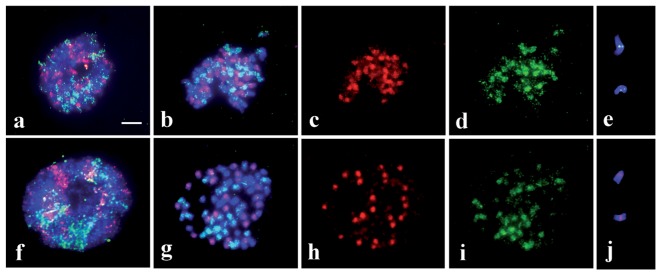
Shishido et al.69) have shown the introgression of chromosome fragments among different genomes in somatic hybrids of rice species. Somatic hybrids between O. sativa (AA, 2n = 24) and O. punctata tetraploid (BBCC, 2n = 48) were produced by somatic cell fusion. Multicolor GISH (McGISH) with two genomic probes, one for the A genome and the other for the C genome, and DAPI staining of DNA were performed. Although the expected hexaploid somatic hybrid (2n = 6x = 72) should have the A, B, and C genomes with 72 chromosomes, the somatic hybrids showed aneuploidy with 65–72 chromosomes (Fig. 4). GISH revealed that A genome chromosomes were never eliminated, whereas frequent loss of B genome chromosomes and an occasional loss of C genome chromosomes were observed (Fig. 4a, 4b, 4f, 4g). GISH revealed the chromosome constitution of all aneuploids, demonstrating its important role as a tool for genome monitoring in plant breeding. Shishido et al.72) have reported highly reproducible, complete protocols for chromosome painting by GISH in rice chromosomes. GISH is also an effective method for basic genetic research and practical breeding in tomato and eggplant species.73),74) However, the feasibility of chromosome painting by GISH for hybrids consisting of different genomes depends on the diversity of the different contributor genomes. For example, the use of GISH in Brassica allopolyploids consisting of a combination of the A, B, and C genomes could not discriminate the three genomes because the repeated sequences are highly homologous among the three genomes.75) However, recently, total genomic DNA from B. oleracea (CC genome, 2n = 18) was hybridized to mitotic or meiotic chromosomes of B. napus (AACC, 2n = 38) in the presence of blocking DNA from B. rapa (AA, 2n = 20), and nine pairs of the C genome chromosomes were clearly detected.76)
Fig. 4.
Multicolor genomic in situ hybridization (McGISH) of somatic hybrids having the A, B, and C genomes with the different combinations of the genomic probes.69) Upper panel: McFISH results with two probes of A (red) and B (green) genomes with the counterstained C genome (blue) with 4’,6-diamidino-2-phenylindole (DAPI). Lower panel: McFISH results with two probes of A (red) and C (green) genomes with the counterstaining B genome with DAPI (blue). a and f: McGISH of the whole nucleus, in which A genome is colored in red, B genome in green or blue, and C genome in blue or green. b and g: Identification of the chromosomes of three genomes by their fluorescent color, those of the A, B and C genome chromosomes being identified with red, green or blue, and blue or green fluorescence, respectively. c and h: Identification of the A genome chromosomes with red fluorescence. d and i: Identification of the B and C genome chromosomes with green fluorescence. e and j: Insertion of the B genome chromosome fragments to the C genome chromosomes and that of the A genome chromosome fragments to the B genome chromosomes, respectively. Bar indicates 5 μm.
High spatial resolution for visualizing DNA domains and sequences using extended DNA fiber FISH
In plants, FISH resolution is 5–10 Mb using well-spread metaphase spread chromosomes, 1.2 Mb in pachytene chromosomes, and 100 kb for interphase nuclei.77) These values depend on the degree of chromatin compactness and also on the genome size, phase of nuclear division, cell type, and degree of heterochromatinization. Two target sequences located in close vicinity would show overlapping FISH signals on chromosomes and even in nuclei.
Fiber DNA technology provided a breakthrough for estimating the distance between TrsA and telomere sequences at a chromosomal end. Extended DNA fibers (EDFs) were isolated from rice nuclei according to the modified methods of Fransz et al.78) Calculations showed that 1 μm of signal track on a DNA fiber corresponded to 3.27 kb of actual nucleotide length, indicating that EDF-FISH provides a much higher spatial resolution power than does any other FISH method. Resolution ranging from 2 to 3.29 kb can be obtained on fully extended chromatin. In fact, the detection sensitivity can be further increased to detect a probe of only 300 bp in size by using EDF-FISH. This approach allows the accurate estimation of the number of copies of a repetitive sequence and of the physical length of target nucleotide sequences.79)
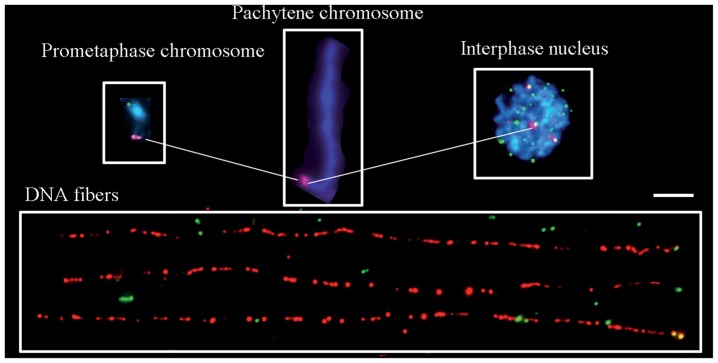
Fig. 5 shows multicolor FISH (McFISH)80) using telomere- and TrsA specific DNA probes on rice somatic cells, pachytene chromosomes, and EDFs. TrsA’s were localized to the terminal regions of the long arms of two japonica rice chromosomes.28) The simultaneous detection of TrsA and telomere sequences by dual-color FISH experiments on chromosomes even allowed detection of overlapping signals (Fig. 5). TrsA and telomeric signals overlapped partially in nuclei as well. This result demonstrates that the two DNA sequences are located within a distance of less than 1 Mb. The fluorescent signals on the EDFs showed a “beads-on-a-string” pattern, as shown in previous reports (Fig. 5). EDF-FISH also revealed the approximate copy numbers of telomeric and TrsA sequences, based on their previously reported unit sizes of 7 and 355 bp, respectively. The lengths of the FISH signal tracks of the telomere (green) and TrsA (red) were measured using the chromosome image analyzing system CHIAS3, and their copy numbers were estimated. The results of TrsA and telomere FISH on rice chromosomes and extended DNA fibers are as follows. 1) All chromosome ends possess telomere repeats measuring roughly 3–4 kb. 2) The subtelomeric TrsA is located only at the ends of the long arms of chromosomes 6 and 12 and is 82 and 241 kb long, respectively, corresponding to 231 and 682 copies. 3) The telomere and TrsA repeats are separated by less than a few thousand intervening nucleotide sequences.
Fig. 5.
High resolution FISH of rice somatic chromosomes, pachytene chromosomes, the nucleus, and extended DNA fibers. Upper left: DAPI- stained mitotic prometaphase chromosome 12 with telomere signals (green) and the subtelomeric tandem repeat TrsA (red). Upper middle: A pachytene bivalent of the same chromosomes with TrsA (red), in which heterochromatin and euchromatin are well differentiated. Upper right: Interphase nucleus, in which telomere sequences, TTTAGGGs and TrsA are colored in green and red, respectively. Bottom: Extended DNA fibers (EDFs) measuring 74 μm with 242 kb TrsA’s (red) and telomere sequence (green). EDFs resemble “beads-on-a-string”. All figures are at the same magnification. Bar indicates 5 μm.
Quantifying the length of purified DNA clones
Measurement of amounts of DNA molecules is essential for many molecular biological experiments. Ordinary methods for measuring the amount of DNA molecules include gel electrophoresis, sometimes followed by Southern blot analysis. An alternative and simple technique, a modified DNA combing protocol, has also been reported.81) This technique allows DNA molecules to be stretched on aminosilane-coating glass slides, requires only small amounts of DNA (10–20 ng), and can be performed rapidly with a fluorescence microscope to measure DNA lengths. This procedure allows measuring the length of BAC, PAC, cosmid, and plasmid DNAs with sizes ranging from fewer than 10 kb to 300 kb. DNA molecules appear as relaxed circles, supercoiled fibers, and/or strands with a nick. Fiber lengths are measured using a fluorescence microscope equipped with a CCD camera, and sizing is done by calculating the number of pixels between two points on the digital image. In this method, generating the pixel-to-kilobase conversion is essential and can be done by determining the length of known DNA fibers (such as lambda DNA) under the same magnification. A 100 kb DNA molecule corresponds to 250 pixels under the condition that a 10 μm long DNA molecule corresponds to 73.5 pixels using the ×100 objective lens microscope, when the DNA molecule is fully stretched to its theoretical size.
Size measurement of DNA molecules by visualization with combing FISH
DNA combing is a useful technique for high-resolution measurements in plants.82) Digitally measured distances can also be transformed into kilo-bases of DNA using the length of a BAC clone of known length along with the length of a standard. The lengths of plant DNA fragments as small as 2 kb have been directly measured on circular BAC molecules using this method.
DNA combing in conjunction with FISH has enabled the high-resolution visual mapping of a multiple gene cluster on a large DNA fragment. Cheng et al.83) calibrated the average DNA extension of seven sequenced BAC clones using the fiber-FISH technique in rice and obtained a value of 3.21 kb/μm. Fiber-FISH results obtained from a BAC contig spanning 1 Mb of DNA strongly matched sequencing data, demonstrating the utility of this technique in cytological mapping. Other research has confirmed the insertion of organellar DNA on the long arm of rice chromosome 10.84) Sequence of the inserted chloroplast DNA is nearly identical to regions of the rice chloroplast genome sequence, suggesting that the chloroplast DNA was transferred recently. In Brassica species, self-incompatibility is regulated by a single S locus with multiple alleles that span several hundreds of kilobases and contain several genes, including SLG and SRK.85) A 76-kb fragment in a P1-derived artificial chromosome (PAC) clone containing the SLG9 and SRK 9 genes was used to directly visualize the S locus. Using DNA combing and FISH, Suzuki et al. demonstrated that the positions of the fluorescent signals of SLG9 and SRK 9 on the clone are consistent with their positions on the restriction map.85) Combing-FISH has superior resolving power and can be used to determine the precise lengths of repetitive sequences. Genes mapping performed visually by high-resolution FISH is an important technique in genome research.
Conclusions
Rice genetics and chromosome research has a long history.86),87) FISH has much been improved since its first development some 20 years ago. FISH is currently one of the major techniques in plant cytology and biology. The increased sensitivity of the technique and its ability to detect gene locations provide a powerful research tool in genetic studies. The detection sensitivities of FISH are well improved that >10 kb DNA detection on the chromosome and >0.3 kb on the extended DNA fibers. Gene mapping of rice genomes is becoming ever more important in rice genetics and breeding. The accumulation of sequence data and gene functional data would further support rice breeding efforts.
The improved sensitivity of FISH is leading FISH technology to the next step, detecting DNA-protein interactions with the use of chromatin immunoprecipitation (ChIP) and immunostaining. Histone acetylation occurs at N-terminal lysines of H3 and H4. Three-dimensional observation revealed that barley shows typical heterochromatin configuration in interphase nucleus.88) These techniques should greatly aid in studies of genetics and also epigenetics. Fluorescence techniques using green and/or red fluorescence protein, like GFP and DsRed, respectively, could also be applied to study living cell dynamics in plants.89)
Acknowledgements
The authors thank Prof. Dr. Hans J. de Jong, Wageningen University, for his invaluable technical advice and suggestions in the course of these studies.
Profile
Nobuko Ohmido graduated the master degree of Agriculture at Kobe University in 1989 and took a doctor degree of Agriculture supervised by Professor Emeritus Toshiro Kinoshita at Hokkaido University in 1994. She started research career in the field of plant breeding technology in 1991 at Hokuriku National Agricultural Experiment Station, Ministry of Agriculture, Forestry and Fishers. She performed the pioneer of rice and other crops chromosome researches using fluorescence in situ hybridization. Cause of these performances, she received the following awards; the Society of Chromosome Research Award from The society of chromosome research in 2001, Young Scientist Award from The society of Japanese Science of Breeding in 2002, Technology Award from Japanese Society of Plant Cell and Molecular Biology in 2007. She moved to Faculty of Human Development (present name; Graduate School of Human Development and Environment), Kobe University as an associate professor in 2004. The research is “Analysis and usage of genome and chromosome in plant science”. Current studies are the genome and chromosome researches of rice, bean, vegetable, forage crops, and a new bio-fuel plant for the co-existence and co-prosperity with food production and environment of plants. The research purpose is to develop new plant science technologies to resolve difficulties of environmental programs and to effectively utilize the plant’s specific functions for the facilitation of human life.

References
- 1).Gill B.S., Friebe B. (1998) Plant cytogenetics at the dawn of the 21st century. Curr. Opin. Plant Biol. 1, 109–115 [DOI] [PubMed] [Google Scholar]
- 2).Mukai Y. (1996) In situ hybridization. InPlant Chromosomes Laboratory Methods (eds. Fukui K., Nakayama S.). CRC Press, New York, pp. 155–170 [Google Scholar]
- 3).Mukai Y. (2005) Perspectives in molecular cytogenetics of wheat. Frontiers of Wheat Bioscience. Memorial Issue, Wheat Inf. Serv. 100, 17–31 [Google Scholar]
- 4).de Jong H. (2003) Visualizing DNA domains and sequences by microscopy: a fifty-year history of molecular cytogenetics. Genome 46, 943–946 [DOI] [PubMed] [Google Scholar]
- 5).Lysak M.A., Pecinka A., Schubert I. (2003) Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Res. 11, 195–204 [DOI] [PubMed] [Google Scholar]
- 6).Jiang J., Gill B.S. (2006) Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49, 1057–1068 [DOI] [PubMed] [Google Scholar]
- 7).Lysak M., Fransz P., Schubert I. (2006) Cytogenetic analyses of Arabidopsis. Methods Mol. Biol. 323, 173–186 [DOI] [PubMed] [Google Scholar]
- 8).Harper L.C., Cande W.Z. (2000) Mapping a new frontier: development of integrated cytogenetic maps in plants. Funct. Integr. Genomics 1, 89–98 [DOI] [PubMed] [Google Scholar]
- 9).Fukui K. (1990) Localization of rRNA genes on rice chromosomes. Rice Biotech. Quart. 1, 18–19 [Google Scholar]
- 10).Fukui K., Iijima K. (1991) Somatic chromosome map of rice by imaging methods. Theor. Appl. Genet. 81, 589–596 [DOI] [PubMed] [Google Scholar]
- 11).Langer P.R., Waldrop A.A., Ward D.C. (1981) Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc. Natl. Acad. Sci. USA 78, 6633–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Pardue M.L., Gall J.G. (1970) Chromosomal localization of mouse satellite DNA. Science 168, 1356–1358 [DOI] [PubMed] [Google Scholar]
- 13).Schwarzacher T., Leitch A.R., Bennett M.D., Heslop-Harrison J.S. (1989) In situ hybridization of parental genomes in a wide hybrid. Ann. Bot. (London) 64, 315–324 [Google Scholar]
- 14).Yamamoto M., Mukai Y. (1989) Application of fluorescence in situ hybridization to molecular cytogenetics of wheat. Wheat Inf. Serv. 69, 30–32 [Google Scholar]
- 15).Fukui K., Ohmido N., Khush G.S. (1994) Variability in rDNA loci in genus Oryza detected through fluorescence in situ hybridization. Theor. Appl. Genet. 87, 893–899 [DOI] [PubMed] [Google Scholar]
- 16).Ohmido N., Fukui K. (1995) Cytological studies of African cultivated rice, Oryza glaberrima. Theor. Appl. Genet. 91, 212–217 [DOI] [PubMed] [Google Scholar]
- 17).Shishido R., Sano Y., Fukui K. (2000) Ribosomal DNAs: an exception to the conservation of gene order in rice genomes. Mol. Gen. Genet. 263, 586–591 [DOI] [PubMed] [Google Scholar]
- 18).Chung M.C., Lee Y.I., Cheng Y.Y., Chou Y.J., Lu C.F. (2008) Chromosomal polymorphism of ribosomal genes in the genus Oryza. Theor. Appl. Genet. 116, 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Dubcovsky J., Dvorak J. (1995) Ribosomal RNA 3 multigene loci: nomads of the Triticeae genomes. Genetics 140, 1367–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Schubert I., Wobus U. (1985) In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92, 143–148 [Google Scholar]
- 21).Dolezel J., Kubalakov 3 a M., Bartos J., Macas J., 3 (2004) Flow cytogenetics and plant genome mapping. Chromosome Res. 12, 77–91 [DOI] [PubMed] [Google Scholar]
- 22).Ma Y., Lee J.H., Li L.C., Uchiyama S., Ohmido N., Fukui K. (2005) Fluorescent labeling of plant chromosomes in suspension by FISH. Genes Genet. Syst. 80, 35–39 [DOI] [PubMed] [Google Scholar]
- 23).International Rice Genome Sequence Project: http://rgp.dna.affrc.go.jp/E/IRGSP/index.html
- 24).Moore G., Foote T., Helentjaris T., Devos K., Kurata N., Gale M. (1995) Was there a single ancestral cereal chromosome? Trends Genet. 11, 81–82 [DOI] [PubMed] [Google Scholar]
- 25).National Institute of Agribiological Sciences Rice Full-Length cDNA Project Team (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301, 376–379 [DOI] [PubMed] [Google Scholar]
- 26).Ohtsubo H., Umeda M., Ohtsubo E. (1991) Organization of DNA sequences highly repeated in tandem in rice genomes. Jpn. J. Genet. 66, 241–254 [DOI] [PubMed] [Google Scholar]
- 27).Wu H.K., Chung M.C., Wu T.Y., Ning C.N., Wu R. (1991) Localization of specific repetitive DNA sequences in individual rice chromosomes. Chromosoma 100, 330–338 [DOI] [PubMed] [Google Scholar]
- 28).Ohmido N., Fukui K. (1997) Visual verification of close disposition between a rice A genome-specific DNA sequence (TrsA) and the telomere sequences. Plant Mol. Biol. 35, 963–968 [DOI] [PubMed] [Google Scholar]
- 29).Ohmido N., Kijima K., Ashikawa I., de Jong H.J., Fukui K. (2001) Visualization of the terminal structure of rice chromosomes 6 and 12 using multicolor FISH to chromosomes and extended DNA fibers. Plant Mol. Biol. 47, 413–421 [DOI] [PubMed] [Google Scholar]
- 30).Mizuno H., Wu J., Kanamori H., Fujisawa M., Namiki N., Saji S, et al. (2006) Sequencing and characterization of telomere and subtelomere regions on rice chromosomes 1S, 2S, 2L, 6L, 7S, 7L, and 8S. Plant J. 46, 206–217 [DOI] [PubMed] [Google Scholar]
- 31).Ohmido N., Fukui K. (2004) Recent advances in FISH analysis of plant chromosomes. Recent Res. Devel. Biochem. 5, 267–279 [Google Scholar]
- 32).Heslop-Harrison J.S., Brandes A., Taketa S., Schmidt T., Vershinin A.V., Alkhimova E.G., et al. (1997) The chromosomal distributions of Ty1-copia group retrotransposable elements in higher plants and their implications for genome evolution. Genetica 100, 197–204 [PubMed] [Google Scholar]
- 33).Nakajima R., Noma K., Ohtsubo H., Ohtsubo E. (1996) Identification and characterization of two tandem repeat sequences (TrsB and TrsC) and a retrotransposon (RIRE1) as genome-general sequences in rice. Genes Genet. Syst. 71, 373–382 [DOI] [PubMed] [Google Scholar]
- 34).Uozu S., Ikehashi H., Ohmido N., Ohtsubo H., Ohtsubo E., Fukui K. (1997) Repetitive sequences: cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol. Biol. 35, 791–799 [DOI] [PubMed] [Google Scholar]
- 35).Suoniemi A., Anamthawat-Jonsson K., Arna T., 3, Schulman A.H. (1996) Retrotransposon BARE-1 is a major, dispersed component of the barley (Hordeum vulgare L.) genome. Plant Mol. Biol. 30, 1321–1329 [DOI] [PubMed] [Google Scholar]
- 36).Tang X., Bao W., Zhang W., Cheng Z. (2007) Identification of chromosomes from multiple rice genomes using a universal molecular cytogenetic system. J. Integr. Plant Biol. 49, 953–960 [Google Scholar]
- 37).Heslop-Harrison J.S., Brandes A., Schwarzacher T. (2003) Tandemly repeated DNA sequences and centromeric chromosomal regions of Arabidopsis species. Chromosome Res. 11, 241–253 [DOI] [PubMed] [Google Scholar]
- 38).Ananiev E.V., Phillips R.L., Rines H.W. (1998) Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95, 13073–13078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Dong F., Miller J.T., Jackson S.A., Wang G.L., Ronald P.C., Jiang J. (1998) Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc. Natl. Acad. Sci. USA 95, 8135–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Kumekawa N., Ohmido N., Fukui K., Ohtsubo E., Ohtsubo H. (2001) A new gypsy-type retro-transposon, RIRE7: preferential insertion into the tandem repeat sequence TrsD in pericentromeric heterochromatin regions of rice chromosomes. Mol. Gen. Genomics 265, 480–488 [DOI] [PubMed] [Google Scholar]
- 41).Ma J., Wing R.A., Bennetzen J.L., Jackson S.A. (2007) Plant centromere organization: a dynamic structure with conserved functions. Trends Genet. 23, 134–139 [DOI] [PubMed] [Google Scholar]
- 42).Fukui K.N., Suzuki G., Lagudah E.S., Rahman R., Appels R., Yamamoto M., Mukai Y. (2001) Physical arrangement of retrotransposon-related repeats in centromeric regions of wheat. Plant Cell Physiol. 42, 189–196 [DOI] [PubMed] [Google Scholar]
- 43).Cheng Z.J., Murata M. (2003) A centromeric tandem repeat family originating from a part of Ty3/gypsy-retroelement in wheat and its relatives. Genetics 164, 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Ohmido N., Ishimaru A., Kato S., Sato S., Tabata S, Fukui K. (2009) Integration of cytogenetic and genetic linkage maps of Lotus japonicus, a model plant for legumes. Chromosome Res. (in press) [DOI] [PubMed] [Google Scholar]
- 45).Zhang Y., Huang Y., Zhang L., Li Y., Lu T., Lu Y., et al. (2004) Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 32, 2023–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Cheng Z., Dong F., Langdon T., Ouyang S., Buell C.R., Gu M., et al. (2002) Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14, 1691–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Zhang W., Yi C., Bao W., Liu B., Cui J., Yu H., et al. (2005) The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata. Plant Physiol. 139, 306–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Jiang J., Birchle J.A., Parrott W.A., Dawe R.K. (2003) A molecular view of plant centromeres. Trends Plant Sci. 8, 570–575 [DOI] [PubMed] [Google Scholar]
- 49).Palmer D.K., O’Day K., Trong H.L., Charbonneau H., Margolis R.L. (1991) Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA 88, 3734–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Talbert P.B., Masuelli R., Tyagi A.P., Comai L., Henikoff S. (2002) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14, 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Nagaki K., Neumann P., Zhang D., Ouyang S., Buell C.R., Cheng Z., et al. (2005) Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol. Biol. Evol. 22, 845–855 [DOI] [PubMed] [Google Scholar]
- 52).Houben A., Demidov D., Gernand D., Meister A., Leach C.R., Schubert I. (2003) Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J. 33, 967–973 [DOI] [PubMed] [Google Scholar]
- 53).Iwata I. (2008) Cytological research of rice chromosomes. Kobe University Graduation thesis, pp. 1–51 [Google Scholar]
- 54).Ohmido N., Akiyama Y., Fukui K. (1998) Physical mapping of unique nucleotide sequences on identified rice chromosomes. Plant Mol. Biol. 38, 1043–1052 [DOI] [PubMed] [Google Scholar]
- 55).Rajyashiri K.R., Nair S., Ohmido N., Fukui K., Kurata N., Sasaki T., et al. (1998) Isolation and FISH mapping of Yeast Artificial Chromosomes (YACs) encompassing an allele of the Gm2 gene for gall midge resistance in rice. Theor. Appl. Genet. 97, 507–514 [Google Scholar]
- 56).Jiang J., Gill B.S., Wang G.L., Ronald P.C., Ward D.C. (1995) Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 92, 4487–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Nakamura S., Asakawa S., Ohmido N., Fukui K., Shimizu N., Kawasaki S. (1997) Constitution of an 800-kb contig in the near-centromeric region of the rice blast resistance gene Pi-ta2 using a highly representative rice BAC library. Mol. Gen. Genet. 254, 611–620 [DOI] [PubMed] [Google Scholar]
- 58).Szinay D., Chang S.B., Khrustaleva L., Peters S., Schijlen E., Bai Y., et al. (2008) High-resolution chromosome mapping of BACs using multi-colour FISH and pooled-BAC FISH as a backbone for sequencing tomato chromosome 6. Plant J. 56, 627–637 [DOI] [PubMed] [Google Scholar]
- 59).Wang C.J., Harper L., Cande W.Z. (2006) High-resolution single-copy fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 18, 529–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).Cheng Z., Buell C.R., Wing R.A., Gu M., Jiang J. (2001) Toward a cytological characterization of the rice genome. Genome Res. 11, 2133–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).Tang X., Bao W., Zhang W., Cheng Z. (2007) Identification of chromosomes from multiple rice genomes using a universal molecular cytogenetic marker system. J. Integr. Plant Biol. 49, 953–960 [Google Scholar]
- 62).Wu J., Mizuno H., Hayashi-Tsugane M., Ito Y., Chiden Y., Fujisawa M., et al. (2003) Physical maps and recombination frequency of six rice chromosomes. Plant J. 36, 720–730 [DOI] [PubMed] [Google Scholar]
- 63).Kato S., Ohmido N., Fukui K. (2003) Development of a quantitative pachytene chromosome map in Oryza sativa by imaging methods. Genes Genet. Syst. 78, 155–161 [DOI] [PubMed] [Google Scholar]
- 64).Feng Q., Zhang Y., Hao P., Wang S., Fu G., Huang Y., et al. (2002) Sequence and analysis of rice chromosome 4. Nature 420, 316–320 [DOI] [PubMed] [Google Scholar]
- 65).Schubert I., Fransz P.F., Fuchs J., de Jong J.H. (2001) Chromosome painting in plants. Methods Cell Sci. 23, 57–69 [PubMed] [Google Scholar]
- 66).Mukai Y., Gill B.S. (1991) Detection of barley chromatin added to wheat by genomic in situ hybridization. Genome 34, 448–452 [Google Scholar]
- 67).Mukai Y., Nakahara Y., Yamamoto M. (1993) Simultaneous discrimination on the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 36, 489–494 [DOI] [PubMed] [Google Scholar]
- 68).Fukui K., Shishido R., Kinoshita T. (1997) Identification of the rice D-genome chromosomes by genomic in situ hybridization. Theor. Appl. Genet. 95, 1239–1245 [Google Scholar]
- 69).Shishido R., Apisitwanich S., Ohmido N., Okinaka Y., Mori K., Fukui K. (1998) Detection of specific chromosome reduction in rice somatic hybrids with the A, B, and C genomes by multi-color genomic in situ hybridization. Theor. Appl. Genet. 97, 1013–1018 [Google Scholar]
- 70).Tan G., Jin H., Li G., He R., Zhu L., He G. (2005) Production and characterization of a complete set of individual chromosome additions from Oryza officinalis to Oryza sativa using RFLP and GISH analyses. Theor. Appl. Genet. 111, 1585–1595 [DOI] [PubMed] [Google Scholar]
- 71).Yan H., Liu G., Cheng Z., Min S., Zhu L. (2001) Characterization of euploid backcross progenies derived from interspecific hybrids between Oryza sativa and O. eichingeri by restriction fragment length polymorphism (RFLP) analysis and genomic in situ hybridization (GISH). Genome 44, 86–95 [DOI] [PubMed] [Google Scholar]
- 72).Shishido R., Ohmido N., Fukui K. (2001) Chromosome painting as a tool for rice genetics and breeding. Methods Cell Sci. 23, 125–132 [PubMed] [Google Scholar]
- 73).Escalante A., Imanishi S., Hossain M., Ohmido N., Fukui K. (1998) RFLP analysis and genomic in situ hybridization (GISH) in somatic hybrids and their progenies between Lycopersicon esculentum and Solanum lycopersicoides. Theor. Appl. Genet. 96, 719–726 [Google Scholar]
- 74).Iwamoto Y., Hirai M., Ohmido N., Fukui K., Ezura H. (2007) Fertile somatic hybrids between Solanum integrifolium and S. sanitwongsei (syn. S. kurzii) as candidates for bacterial wilt-resistant rootstock of eggplant. Plant Biotech. 24, 179–184 [Google Scholar]
- 75).Snowdon R.J., Kohler W., Friedt W., Kohler A. (1997) Genomic in situ hybridization in Brassica amphidiploids and interspecific hybrids. Theor. Appl. Genet. 95, 1320–1324 [Google Scholar]
- 76).Howell E.C., Kearsey M.J., Jones G.H., King G.J., Armstrong S.J. (2008) A and C genome distinction and chromosome identification in Brassica napus by sequential fluorescence in situ hybridization and genomic in situ hybridization. Genetics 180, 1849–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77).de Jong H.J., Fransz P., Zabel P. (1999) High-Resolution FISH in plants: techniques and applications. Trends Plant Sci. 4, 258–263 [DOI] [PubMed] [Google Scholar]
- 78).Fransz P., Alonso-Blanco C., Liharska T., Peeters A.J.M., Zabel P., de Jong J.H. (1996) High-resolution physical mapping in Arabidopsis thaliana and tomato by fluorescence in situ hybridization to extended DNA fibers. Plant J. 9, 421–430 [DOI] [PubMed] [Google Scholar]
- 79).Ohmido N., Kijima K., Akiyama Y., de Jong J.H., Fukui K. (2000) Quantification of total genomic DNA and selected repetitive sequences reveals concurrent changes in different DNA families in indica and japonica rice. Mol. Gen. Genet. 263, 388–394 [DOI] [PubMed] [Google Scholar]
- 80).Mukai Y. (1996) Multicolor fluorescence in situ hybridization: a new tool for genome analysis. InMethods of Genome Analysis in Plants (ed. Jauhar P.P.). CRC Press, New York, pp. 181–192 [Google Scholar]
- 81).Henegariu O., Grober L., Haskins W., Bowers P.N., State M.W., Ohmido N., et al. (2001) Rapid DNA fiber technique for size measurements of linear and circular DNA probes. Biotechniques 31, 246–250 [DOI] [PubMed] [Google Scholar]
- 82).Jackson S.A., Wang M.L., Goodman H.M., Jiang J. (1998) Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41, 566–572 [PubMed] [Google Scholar]
- 83).Cheng Z., Buell C.R., Wing R.A., Jiang J. (2002) Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes, and extended DNA fibers. Chromosome Res. 10, 379–387 [DOI] [PubMed] [Google Scholar]
- 84).Yuan Q., Hill J., Hsiao J., Moffat K., Ouyang S., Cheng Z., et al. (2002) Genome sequencing of a 239-kb region of rice chromosome 10L reveals a high frequency of gene duplication and a large chloroplast DNA insertion. Mol. Genet. Genomics 267, 713–720 [DOI] [PubMed] [Google Scholar]
- 85).Suzuki G., Kai N., Hirose T., Fukui K., Nishio T., Takayama S., et al. (1999) Genomic organization of the S locus: identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics 153, 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86).Morishima H. (1984) Wild plants and domestication. InBiology of Rice (eds. Tsunoda S., Takahashi N.). Japan Sci. Soc. Press, Amsterdam, pp. 3–30 [Google Scholar]
- 87).Ohmido N., Fukui K., Kinoshita T. (2005) Advances in rice chromosomes research. Proc. Jpn. Acad., Ser. B 81, 382–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88).Wako T., Fukuda M., Furushima-Shimogawara R., Belyaev N.D., Fukui K. (2002) Cell cycle-dependent and lysine residue-specific dynamic changes of histone H4 acetylation in barley. Plant Mol. Biol. 49, 645–653 [DOI] [PubMed] [Google Scholar]
- 89).Ohmido N., Wako T., Fukui K. (2008) Nuclear organization analyzed by visualization. The Nucleus 50, 473–489 [Google Scholar]