Abstract
It is not known how plants synthesize the p-aminobenzoate (PABA) moiety of folates. In Escherichia coli, PABA is made from chorismate in two steps. First, the PabA and PabB proteins interact to catalyze transfer of the amide nitrogen of glutamine to chorismate, forming 4-amino-4-deoxychorismate (ADC). The PabC protein then mediates elimination of pyruvate and aromatization to give PABA. Fungi, actinomycetes, and Plasmodium spp. also synthesize PABA but have proteins comprising fused domains homologous to PabA and PabB. These bipartite proteins are commonly called “PABA synthases,” although it is unclear whether they produce PABA or ADC. Genomic approaches identified Arabidopsis and tomato cDNAs encoding bipartite proteins containing fused PabA and PabB domains, plus a putative chloroplast targeting peptide. These cDNAs encode functional enzymes, as demonstrated by complementation of an E. coli pabA pabB double mutant and a yeast PABA-synthase deletant. The partially purified recombinant Arabidopsis protein did not produce PABA unless the E. coli PabC enzyme was added, indicating that it forms ADC, not PABA. The enzyme behaved as a monomer in size-exclusion chromatography and was not inhibited by physiological concentrations of PABA, its glucose ester, or folates. When the putative targeting peptide was fused to GFP and expressed in protoplasts, the fusion protein appeared only in chloroplasts, indicating that PABA synthesis is plastidial. In the pericarp of tomato fruit, the PabA-PabB mRNA level fell drastically as ripening advanced, but there was no fall in total PABA content, which stayed between 0.7 and 2.3 nmol·g-1 fresh weight.
Tetrahydrofolate and its derivatives, collectively termed folates, are essential cofactors for one-carbon transfer reactions and thus are required for synthesis of methionine, purines, and thymidylate, and for interconversion of glycine and serine (1, 2). Plants, fungi, and many microorganisms synthesize folates de novo, but humans and other higher animals do not and therefore require a dietary supply (1, 3, 4). Because folate deficiency is a worldwide health problem and plant foods are major sources of folates for humans, there is much interest in engineering plants to enhance folate content (1, 5). To do this, the plant folate synthesis pathway must first be understood.
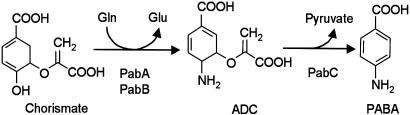
Folates are made up of pterin, p-aminobenzoate (PABA), and glutamate moieties. In plants, it is now clear that the pterin moiety is formed from GTP, most probably in the cytosol (6), then coupled to PABA in mitochondria (7), and that the subsequent glutamylation and reduction steps are also mitochondrial (7, 8). The origin of the PABA moiety in plants, however, remains unknown. In Escherichia coli, PABA is made from chorismate in two steps (Fig. 1). In the first step, the PabA and PabB proteins cooperate to replace the hydroxyl group of chorismate with an amino group, yielding 4-amino-4-deoxychorismate (ADC) (4, 9). PabA acts as a glutamine amidotransferase, supplying an amino group to PabB, which carries out the amination reaction. Together, PabA and PabB are termed ADC synthase (ADCS) (4). PabB can also convert chorismate to ADC in the absence of PabA if unphysiologically high concentrations of NH3 are given in place of glutamine (4). The second step of PABA synthesis in E. coli entails elimination of pyruvate and aromatization of the ADC ring to give PABA, catalyzed by ADC lyase (PabC) (4, 10).
Fig. 1.
The PABA biosynthesis pathway in E. coli. PabA and PabB associate to form the ADCS complex; PabC (ADC lyase) is a separate enzyme (4).
No biochemical work has been done on PABA synthesis in fungi or other lower eukaryotes such as Plasmodium spp., but genomic and genetic data show that these organisms and also actinomycetes have bipartite proteins with domains homologous to PabA and PabB (11-14). These hybrid proteins are commonly named “PABA synthase,” reflecting an assumption that they are sufficient for PABA synthesis. Although there are no data to show that they indeed produce PABA and not just ADC, this seems a valid possibility. Firstly, there are no obvious ADC lyase orthologs in yeast, actinomycete, or Plasmodium genomes. Secondly, PABA synthases contain an extra region between the PabA and PabB domains that could potentially carry a novel ADC lyase activity. Lastly, PabA and PabB are, respectively, homologous to TrpG and TrpE, which convert chorismate all of the way to the PABA analog anthranilate (o-aminobenzoate) without need of a lyase (9, 15). It thus remains to be established whether PABA synthases are bifunctional (i.e., strictly equivalent to PabA and PabB) or trifunctional, having in addition ADC lyase activity.
In this study, we identified tomato and Arabidopsis cDNAs encoding hybrid PabA-PabB proteins and showed these proteins to be ADCSs that are plastid-targeted. Because tomato fruit is our system for folate engineering (6) and nothing was known about PABA synthesis in fruits, we also analyzed ADCS expression in relation to PABA levels during tomato fruit ripening.
Materials and Methods
Plants Material. Tomato (Lycopersicon esculentum Mill. cv. MicroTom) plants were grown in a greenhouse (maximum temperature 27°C) in potting mix with standard fertilizer and pesticide treatments. Fruit and leaves were harvested in June of 2003, frozen in liquid N2, and stored at -80°C.
cDNA Clones and Expression Constructs. Arabidopsis cDNA clone RAFL09-32-D04 coding for AtADCS was isolated at the RIKEN Genomic Sciences Center (16). 5′-Truncated tomato expressed sequence tags (TIGR contig TC108445) were used to design a primer (5′-CCTTTGGGGTGGAACCACGA-3′), for RACE PCR, which yielded the missing 5′ sequence. Full-length cDNAs encoding LeADCS were then obtained from fruit mRNA by reverse transcription and PCR amplification with the primers 5′-ATTTCTGCACCAAGCGTTTT-3′ (forward) and 5′-AAAATGAAACGTGGAATCATCA-3′ (reverse). To express AtADCS and LeADCS in yeast or E. coli, targeting regions were removed and replaced by initiation codons by using suitable PCR primers; the changes were V85 → M for AtADCS and V84 → M for LeADCS. The vector for yeast expression was pVT103-U (17); the truncated ADCS cDNAs were ligated between its BamHI and PvuII sites and electroporated into E. coli DH10B cells. Yeast transformation and retransformation were carried out as described (6). The E. coli vector for complementation experiments was pLOI707HE, which allows tight control of gene expression by isopropyl β-d-1-thiogalactopyranoside (IPTG) (18). The truncated ADCS cDNAs were inserted between the NotI and SstI sites and introduced into E. coli DH10B. For overexpression, the truncated AtADCS sequence was inserted between the NcoI and XhoI sites of pET-28a (Novagen) and introduced first into E. coli DH10B, then into BL21-CodonPlus (DE3)-RIL cells (Stratagene). E. coli PabC cloned in pJMG30 (10), PabA in pSZD51 (19), and PabB in pSZD52 (19) were likewise introduced into BL21-CodonPlus (DE3)-RIL cells.
Functional Complementation. Yeast strains BY4741 (Mata his3-1 leu2-0 met15-0 ura3-0 YNR033w::kanMX4) and 971/6c (Mata ade2-1 his3-11,15 leu2-3,112 ura3-1 can1) were obtained from EUROSCARF (Frankfurt) and M. L. Agostini Carbone (Università di Milano), respectively. Yeast cells transformed with pVT103-U constructs were cultured at 30°C in appropriately supplemented synthetic minimal medium, prepared as specified in the 1984 Difco manual, except that folic acid was omitted, and PABA was included at 0.2 μg·ml-1or omitted. E. coli BN1163 (pabA1, pabB::Kan, rpsL704, ilvG-, rfb-50, rph-1) harboring pLOI707HE constructs was cultured at 37°C in M9 synthetic minimal medium containing 50 μg·ml-1 kanamycin, 10 μg·ml-1 tetracycline, and 100 μM IPTG, plus or minus 0.5 μg·ml-1 PABA.
Recombinant Protein Production and Analysis. E. coli cells were grown at 37°C in LB medium until A600 was ≈1, at which point IPTG was added (final concentrations were 100 μM for At-ADCS and 500 μM for PabA, -B, and -C), and incubation was continued for 3 h at 30°C (AtADCS) or 37°C (PabA, -B, and -C). Subsequent operations were at 4°C. Pelleted cells from 50- to 500-ml cultures were resuspended in 1-4 ml of 0.1 M Tris·HCl (pH 7.5)/10 mM 2-mercaptoethanol and shaken with 0.1-mm zirconia-silica beads in a MiniBeadbeater (Biospec Products, Bartlesville, OK) at 5,000 rpm for 6 × 20 s. The extracts were centrifuged (15,000 × g, 20 min) and desalted on PD-10 columns (Amersham Biosciences) equilibrated in 0.1 M Tris-HCl (pH 7.5)/10 mM 2-mercaptoethanol/10% (vol/vol) glycerol. Desalted extracts were routinely frozen in liquid N2 and stored at -80°C; this preserved enzyme activity. AtADCS was partially purified, and its molecular mass was estimated by using a Waters 626 HPLC system equipped with a Superdex 200 HR 10/30 column (Amersham Biosciences). Desalted extract (0.2 ml) was applied to the column, which was equilibrated and eluted with 0.1 M Tris·HCl (pH 7.5)/10 mM 2-mercaptoethanol. Carbonic anhydrase, BSA, β-amylase, and apoferritin were used as standards. Protein was estimated by Bradford's method (20), using BSA as the standard.
Enzyme Assays. Assays of PABA synthesis activities were modifications of published procedures (19). Briefly, standard assays (100 μl) contained 50 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 10 mM DTT, 5 mM l-glutamine, 100 μM chorismate (glutamine-dependent assays) or 40 mM triethanolamine (pH 8.5), 26 mM (NH4)2SO4, 8 mM MgCl2, 4 mM DTT, and 80 μM chorismate (NH3-dependent assays) and were run at 37°C for 30-120 min. Desalted PabC extract (7 μg of protein) was added when indicated. Reactions were stopped with 20 μl of 75% (vol/vol) acetic acid, incubated on ice for 1 h, and centrifuged (15,000 × g, 4°C, 20 min). Supernatants (60 μl) were injected onto a Supelco Discovery C18 column (5 μm, 250 × 4.6 mm) and eluted isocratically with 0.5% acetic acid containing 20% (vol/vol) methanol at a flow rate of 1 ml·min-1. The PABA peak was detected by fluorescence (290-nm excitation, 340-nm emission) and quantified relative to standards.
Transient Expression of GFP Fusion Protein in Arabidopsis Protoplasts. The N-terminal region of AtADCS (MNFSFC... GFVRT; residues 3-87) was amplified by PCR with the primers 5′-GAGAGTCGACATGA ATTTT TCGT T T TGT TCA AC-3′ and 5′-GAGACCATGGAAGTCCTCACAAAACCAAGCTTC-3′. [The sequence context of the codon for the methionine at position 3 is closer to the plant translation initiation consensus (21) than that of the first methionine codon.] The amplificate was digested with SalI and NcoI and cloned in frame upstream of the GFP gene in the 35Ω-sGFP(S65T) plasmid (22). Arabidopsis protoplasts were prepared from cell suspension cultures and transformed with the AtADCS construct or the empty vector as described (8, 23). Samples were analyzed by confocal laser scanning microscopy with a Leica TCS-SP2 operating system. GFP and chlorophyll fluorescence were excited (488 and 633 nm, respectively) and collected sequentially. Fluorescence emission was collected from 500 to 535 nm for GFP and 643 to 720 nm for chlorophyll.
PABA Determination. Total PABA (i.e., free PABA plus PABA glucose ester) was isolated from fruit pericarp tissue by methanol extraction, cation exchange chromatography, and ethyl acetate partitioning and quantified by HPLC with fluorescence detection as described (24).
Real-Time Quantitative RT-PCR. Total RNA was isolated from fruit and leaf samples (1-2 g) as described (6) and DNase-treated (DNA-free, Ambion, Austin, TX). Real-time quantitative RTPCR was performed on 250 ng of total RNA in 25-μl reactions by using TaqMan One-Step RT-PCR Master Mix Reagents and a GeneAmp 5700 sequence-detection system (Applied Biosystems). The primers and TaqMan probe were as follows: forward primer, 5′-GGA ATGACCT TGGGCGTGTA-3′; reverse primer, 5′-TGCATAGGATTCAATTTCCATGA-3′; probe, 5′-TGAGACTGGCTCTGTTCATGTCCCACA-3′, with the fluorescent reporter dye 6-carboxyfluorescein and the quencher 6-carboxytetramethylrhodamine. Controls without reverse transcriptase were run to check that the amplifications were not due to contaminating genomic DNA. [3H]UTP standard RNA was prepared from LeADCS cDNA by using the MAXIscript in vitro transcription kit (Ambion). mRNA integrity was checked by semiquantitative RT-PCR analysis of the Never-ripe (Nr) transcript (25). PCR (35 cycles) was conducted on cDNA prepared from 250 ng of total RNA by using the Nr-specific primers 5′-GCAGACGATTTATTCAACTT-3′ and 5′-TTACAGACTTCTTTGATAGC-3′. Controls without reverse transcription gave no amplification product.
Results
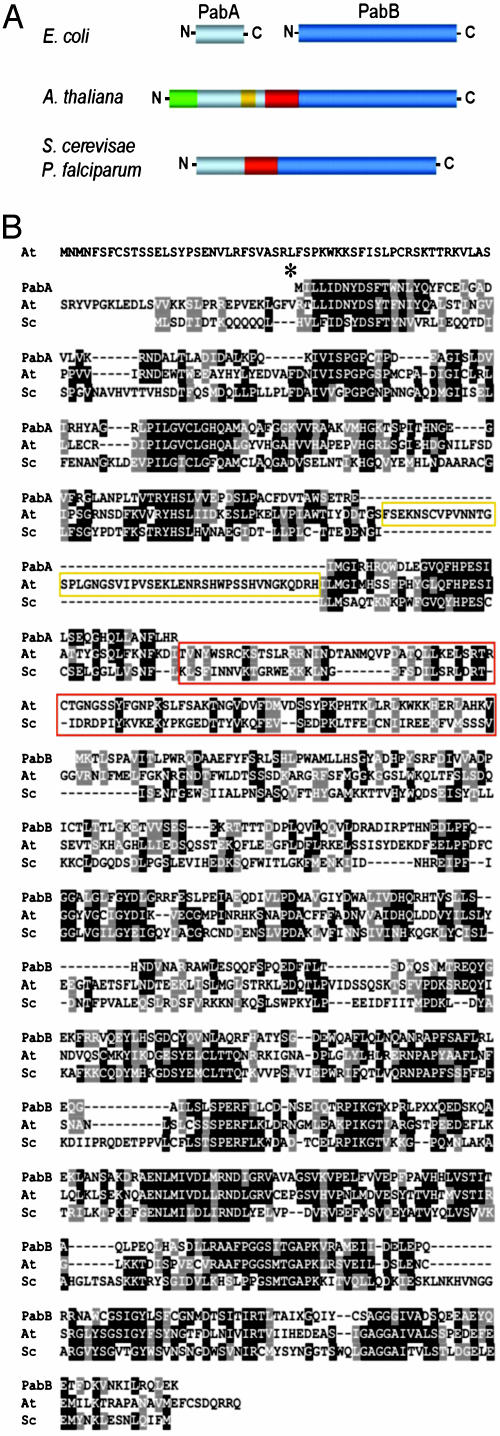
Plants Have Genes Specifying PabA-PabB Hybrid Proteins. blast searches of GenBank and TIGR databases with the protein sequences of E. coli PabA and PabB detected a single Arabidopsis gene (At 2 g 28880) and a cognate cDNA (RAFL 09-32-D04) encoding a 103-kDa polypeptide with PabA- and PabB-like domains. Truncated tomato expressed sequence tags specifying a similar protein were also found; 5′ RACE was used to obtain the missing sequence, and complete tomato cDNAs encoding a 102-kDa protein were then isolated by RT-PCR. The deduced Arabidopsis protein is shown in Fig. 2 and is aligned with its tomato counterpart in Fig. 7, which is published as supporting information on the PNAS web site. Both plant proteins comprise tandem domains that share 33-37% identity with E. coli PabA and PabB, and 26% identity with yeast PABA synthase (Figs. 2 A and 7). The plant proteins share 61% overall identity with each other. As in yeast and other PABA synthases, the PabA and PabB domains of the plant proteins are separated by a long linker (≈90 residues) whose sequence is not conserved but is rich in basic amino acids (Figs. 2B and 7). In contrast to yeast PABA synthase, the plant proteins have a ≈45-residue insertion near the end of the PabA domain. Like the interdomain linker, this extra sequence is not conserved between Arabidopsis and tomato but is basic in character. The plant proteins also have nonconserved N-terminal extensions of ≈85 residues that display the features of chloroplast targeting peptides (Figs. 2 and 7).
Fig. 2.
Primary structure of the deduced Arabidopsis PabA-PabB polypeptide. (A) Scheme comparing the organization of the Arabidopsis polypeptide with that of eukaryotic PABA synthases and with E. coli PabA and PabB. Interdomain linker, red; insertion in PabA domain, orange; putative chloroplast targeting sequence, green. (B) Sequence alignment of E. coli PabA (GenBank accession no. AAG58468) and PabB (AGEC1), yeast PABA synthase (Sc, NP_014431), and the Arabidopsis polypeptide (NP_850127). Identical residues are shaded in black, and similar residues are shaded in gray. Dashes are gaps introduced to maximize alignment. The residue (V85) at which the protein was truncated in expression constructs is marked by an asterisk. The interdomain linker is boxed in red, and the insertion in the PabA domain, in orange.
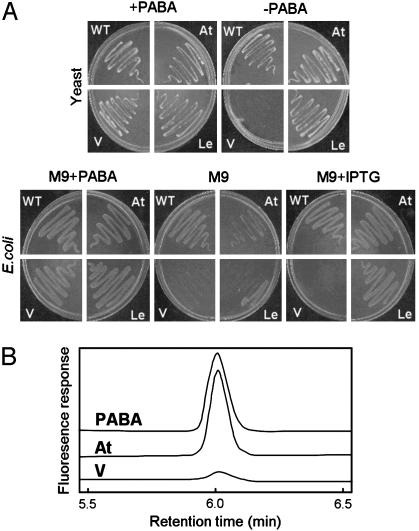
Plant PabA-PabB cDNAs Complement E. coli and Yeast PABA Synthesis Mutants. To determine whether the Arabidopsis and tomato cDNAs encode functional enzymes, the sequences predicted to specify the mature proteins (Figs. 2B and 7) were cloned into E. coli expression vector pLOI707HE and yeast expression vector pVT103-U. pLOI707HE allows tightly IPTG-controlled gene expression in diverse host strains (18). The resulting constructs were transformed into an E. coli pabA pabB double mutant and a yeast PABA synthase deletant, both of which are PABA auxotrophs (Fig. 3A). The Arabidopsis and tomato constructs yielded PABA-independent transformants of the E. coli and yeast mutants with high frequency, and their growth was similar to that of wild-type strains (Fig. 3A). No complementation was seen with vectors alone (Fig. 3A). In E. coli harboring plant cDNA constructs, complementation depended strictly on addition of the IPTG inducer, indicating that the plant cDNAs were responsible for the effect (Fig. 3A). Retransformation of the yeast mutant with rescued plasmid containing the Arabidopsis cDNA restored PABA prototrophy, further confirming that complementation was due to the plant cDNA (data not shown). Taken together, these data indicate that plant PabA-PabB proteins have PabA plus PabB activity (i.e., that they are functional ADCSs) but say nothing about their PabC (ADC lyase) activity.
Fig. 3.
Plant PabA-PabB proteins encode functional enzymes. (A) Complementation of a yeast PABA synthase mutant (Upper) and an E. coli pabA pabB double mutant (Lower). Similar numbers of cells of wild-type (WT) yeast (strain 971/6c) or E. coli (strain K12) and mutants transformed with vector alone (V) or containing Arabidopsis (At) or tomato (Le) cDNAs were plated on minimal media plus or minus PABA and, for E. coli, plus or minus IPTG (100 μM). Plates were incubated for 5 days at 30°C (yeast) or 2 days at 37°C (E. coli). (B) HPLC fluorescence assays of ADCS activity in desalted extracts of E. coli overexpressing the Arabidopsis PabA-PabB protein (At) or the vector alone (V). Assays contained 5 mM l-glutamine, 100 μM chorismate, and excess E. coli PabC. The uppermost trace is authentic PABA.
Arabidopsis PabA-PabB Has ADC Synthase but Not Lyase Activity. To investigate ADCS and lyase activities directly, the mature Arabidopsis PabA-PabB protein was overexpressed in E. coli by using the pET-28a plasmid and the host strain BL21-CodonPlus (DE3)-RIL (which is wild type with respect to pabA, pabB, and pabC). ADCS activity was first assayed in desalted extracts, by using glutamine as amino donor and adding excess E. coli PabC to convert ADC to PABA, which was quantified by HPLC. In this coupled assay, vector-alone cell extracts produced traces of PABA (Fig. 3B), attributable to the endogenous PabA and PabB activities of the host. Extracts of cells overexpressing the Arabidopsis PabA-PabB protein gave 20-fold more PABA (Fig. 3B), providing initial biochemical evidence for ADCS activity.
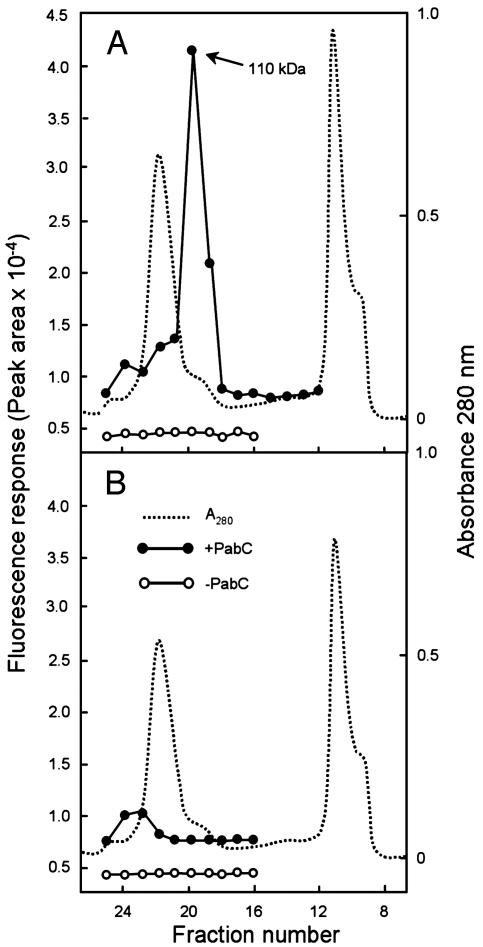
To corroborate this result and to test for ADC lyase activity, we used size-exclusion chromatography to separate the recombinant Arabidopsis PabA-PabB protein from the E. coli host's PabC enzyme, which is smaller (≈50 kDa; ref. 19). By using glutamine as amino donor, the column fractions were assayed for PABA formation, plus or minus addition of E. coli PabC. When PabC was added, there was a large peak of PABA-forming activity at ≈110 kDa (fractions 19-20), which is close to the predicted mass of the monomeric recombinant enzyme (93 kDa) (Fig. 4A). This peak disappeared when PabC was omitted (Fig. 4A) and was not given by extracts of cells transformed with vector alone (Fig. 4B). The minor peak of PABA formation in fractions 23-24 is attributable to endogenous activity from the host cells (Fig. 4 A and B). These data show that the Arabidopsis PabA-PabB protein has ADCS activity but lacks ADC lyase activity. The Arabidopsis and tomato proteins may thus be termed ADCSs (AtADCS and LeADCS, respectively). The AtADCS peak of Fig. 4A was unstable, losing all activity within 12 h at 4°C.
Fig. 4.
The Arabidopsis PabA-PabB protein has ADCS but not ADC lyase activity. Extracts of E. coli cells overexpressing the Arabidopsis PabA-PabB protein (A) or harboring the vector alone (B) were fractionated by size exclusion chromatography. Fractions were assayed for protein (absorbance at 280 mm) and for PABA synthesis from chorismate (100 μM) and l-glutamine (5 mM) in the presence and absence of excess E. coli PabC protein. For clarity, the baselines for the activities in the presence of PabC have been offset in A and B by +0.5 × 10-4 fluorescence units. The molecular mass of the Arabidopsis PabA-PabB protein was determined to be 110 ± 15 kDa (mean ± SE of three experiments).
Amino Donors and Inhibitors of Arabidopsis ADC Synthase. When testing amino donors, we compared the Arabidopsis enzyme with the E. coli enzyme, using assay conditions developed for the latter (19). Like PabA, AtADCS did not use asparagine (Table 1). AtADCS was able to use NH3, like PabB, but apparently more efficiently because the NH3-dependent rate was 50% of the glutamine-dependent rate for AtADCS but only 9% for PabB (Table 1). AtADCS was not inhibited by physiological concentrations of PABA (0.5 μM), PABA glucose ester (6 μM), or folates (10 μM tetrahydrofolate, 5-methyltetrahydrofolate, or 5-formyltetrahydrofolate pentaglutamate), nor was it sensitive to the PABA analogs PABA ethyl ester, p-acetamidobenzoate, p-hydroxybenzoate, or anthranilate (50 μM) (data not shown).
Table 1. Activities of Arabidopsis and E. coli ADC synthases with various amino donors.
| Enzyme activity, nmol·h-1·mg-1 protein
|
||
|---|---|---|
| Amino donor | AtADCS | PabA + PabB |
| l-Glutamine 5 mM | 4.0 ± 0.3 | 35.0 ± 3.0 |
| l-Asparagine 5 mM | <0.01 | <0.01 |
| [NH4+ + NH3] 52 mM | 2.0 ± 0.2 | 2.8 ± 0.4 |
Assay conditions were based on those developed for the E. coli enzyme (19), as described in Materials and Methods. All assays contained excess PabC to convert ADC to PABA, which was determined by HPLC fluorescence. The AtADCS preparation was partially purified by size exclusion chromatography (Fig. 4A). E. coli PabA and PabB preparations were desalted extracts of cells overexpressing each protein. Data are means ± SE of five replicates.
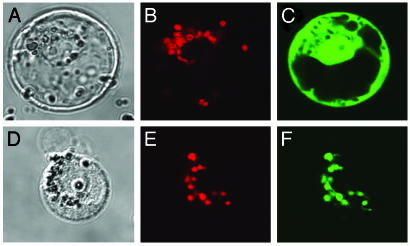
Subcellular Localization of Arabidopsis ADC Synthase. To analyze the subcellular localization of AtADCS in vivo, its N-terminal region (85 residues) was fused to the GFP marker protein. Expression of this fusion protein in Arabidopsis protoplasts resulted in a punctate pattern of green fluorescence that colocalized with the red autofluorescence of chlorophyll (Fig. 5 E and F). In contrast, control protoplasts expressing GFP alone showed green fluorescence throughout the cytoplasm and nucleus (Fig. 5C). These data demonstrate that AtADCS contains a functional chloroplast targeting peptide, suggesting that plant ADCS is a plastidial enzyme.
Fig. 5.
Expression in Arabidopsis protoplasts of GFP fused to the N-terminal region (residues 3-87) of the Arabidopsis PabA-PabB protein and of GFP alone. Images are optical photomicrographs (A and D), chlorophyll fluorescence (B and E, red pseudocolor), and GFP fluorescence (C and F, green pseudocolor). A-C correspond to the GFP control, and D-F correspond to the fusion protein.
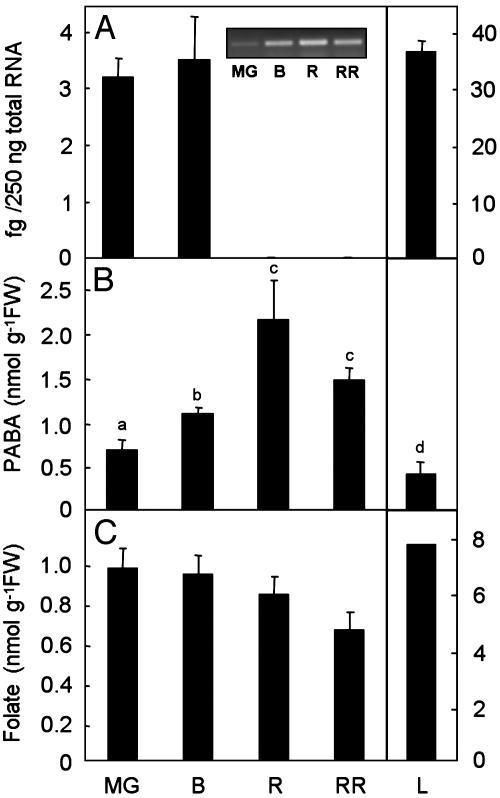
ADC Synthase mRNA Expression During Tomato Fruit Development. Because nothing is known about PABA synthesis in plants, we measured LeADCS transcript expression and total PABA pools in leaves and in fruits harvested at four stages. LeADCS mRNA levels in unripe fruits were initially ≈10% of that in leaves but dropped dramatically to beneath the detection limit after the breaker stage (Fig. 6A). The detection of the Nr transcript, which is induced in ripening fruit (25), showed that this fall was not due to general mRNA degradation as ripening advanced (Fig. 6A Inset). Concentrations of PABA in the fruit increased progressively as the fruit ripened from the mature green to the red and red-ripe stages (Fig. 6B). As previously observed (24), the majority of PABA (80-90%) was present as its glucose ester (data not shown).
Fig. 6.
Levels of LeADCS mRNA, PABA, and folate in tomato fruits and in leaves. (A) LeADCS mRNA was quantified by real-time RT-PCR on 250 ng of total RNA from pericarp tissue of fruits at mature green (MG), breaker (B), ripe (R), and red-ripe (RR) stages, or from young leaves (L). Data are means ± SE of three determinations. Note the 10-fold difference in scale between the fruit and leaf mRNA data. (Inset) Semiquantitative RT-PCR analysis of the Neverripe transcript, as a quality control for mRNA integrity. (B) Total PABA levels in pericarp tissue and young leaves. Values with different letter annotation are significantly different (P < 0.01, least significant difference) by ANOVA. Data are means ± SE of three replicates. (C) Folate contents of fruits (6) and leaves (3), taken from the literature.
Discussion
We have identified Arabidopsis and tomato cDNAs encoding ADCS, the first of two enzymes in the PABA branch of the folate synthesis pathway. Both plant proteins are bipartite, with tandem domains homologous to the PabA and PabB subunits of the E. coli enzyme, and functional complementation tests as well as enzyme assays show that they have both PabA and PabB activities. Similar bipartite proteins exist in other PABA-producing organisms, including fungi, actinomyces, and Plasmodium (11-14). However, none of these has been biochemically characterized, and it is particularly unclear whether they have ADC lyase in addition to ADCS activity. Compounding the confusion is the near-universal use in publications and databases of the term “PABA synthase” to describe deduced PabA-PabB hybrid proteins, for it implies that they mediate the lyase reaction. Our finding that the Arabidopsis ADCS has no lyase activity provides, to our knowledge, the first evidence on this point for any PabA-PabB protein. Besides invalidating the name “PABA synthase,” this evidence points to the existence of a yet-to-be-discovered ADC lyase in plants (and probably also in other organisms with PabA-PabB proteins).
The Arabidopsis and tomato ADCSs have predicted plastid transit peptides, and the putative targeting sequence from Arabidopsis was able to direct a passenger protein into chloroplasts. ADCSs are therefore most probably plastid enzymes, as previously hypothesized (26). This location is consistent with the evidence that the chorismate substrate for ADCS is produced in plastids (27). Moreover, because the intermediate ADC is very labile (28), the plastid location of ADCS suggests that plant ADC lyase may also be plastidial. Above all, our evidence that the PABA moiety of folate is made in plastids underscores the unique spatial organization of plant folate synthesis, in which three subcellular compartments participate (7, 26).
The properties of AtADCS (minus its predicted targeting sequence) are generally similar to those of the E. coli PabA-PabB complex. First, the plant enzyme has an absolute preference for glutamine over asparagine, a preference that is noteworthy given that asparagine can be more abundant than glutamine in leaves and chloroplasts (29-31). Second, AtADCS retains a capacity to use NH3 as amino donor, showing that the structure of the bipartite protein is not such that only NH3 generated in situ by the PabA domain can be channeled to the active site of the PabB domain. Third, as for the E. coli enzyme, there is no indication that AtADCS is subject to feedback inhibition by physiological levels of PABA or folates, nor is the glucose ester of PABA inhibitory, this conjugate being the predominant form of PABA in plants (24). Such lack of inhibition contrasts starkly with the extreme end-product sensitivity of the related enzyme, anthranilate synthase (32). How flux to PABA is regulated at the metabolic level thus remains a mystery in plants, as in bacteria (9).
The expression of LeADCS in immature fruits implies that they synthesize at least part of their own supply of PABA and therefore may not need to import it from leaves. In this connection, it is interesting that the relative levels of LeADCS mRNA in leaves and unripe fruits roughly match their relative folate contents, leaves having ≈10-fold more transcript and 8-fold more folate than fruits (Fig. 6 A and C). The disappearance of the LeADCS mRNA during ripening suggests that fruits eventually lose their ability to synthesize PABA, in much the same way as they lose the capacity to produce the pterin moiety of folate due to a collapse in the expression of GTP cyclohydrolase I (6). It therefore seems likely that folate synthesis in ripening fruit is limited to some extent by falling enzyme activity in both the pterin and the PABA branches of the folate synthesis pathway. There is a modest upward trend in the fruit PABA pool during ripening, presumably due mainly to continued activity of the ADCS enzyme. However, the PABA pool in fruit remains at all times fairly small, so that even if all of it were converted to folate, the folate content of the fruit would still be less than half that in leaves. Thus, because the plant ADCS seems not to be feedback regulated by PABA or folate, boosting ADCS activity merits exploration as part of a rational strategy to increase fruit folate level by metabolic engineering.
Supplementary Material
Acknowledgments
We thank Ruth White for help with sequencing and sequence analysis, and Dr. Didier Grundwald for expertise in confocal microscopy. This work was supported in part by the Florida Agricultural Experiment Station, an endowment from the C. V. Griffin, Sr., Foundation, and grants from the National Science Foundation to (A.D.H. and J.F.G.). This article is Journal Series No. R-09811.
Abbreviations: ADC, 4-amino-4-deoxychorismate; ADCS, ADC synthase; IPTG, isopropyl β-d-1-thiogalactopyranoside; PABA, p-aminobenzoate.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY096797.1 and AY425708).
References
- 1.Scott, J. M., Rébeillé, F. & Fletcher, J. (2000) J. Sci. Food Agric. 80, 795-824. [Google Scholar]
- 2.Appling, D. R. (1991) FASEB J. 5, 2645-2651. [DOI] [PubMed] [Google Scholar]
- 3.Cossins, E. A. (2000) Can. J. Bot. 78, 691-708. [Google Scholar]
- 4.Green, J. M., Nichols, B. P. & Matthews, R. G. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), pp. 665-673.
- 5.DellaPenna, D. (1999) Science 285, 375-379. [DOI] [PubMed] [Google Scholar]
- 6.Basset, G., Quinlivan, E. P., Ziemak, M. J., Diaz De La Garza, R., Fischer, M., Schiffmann, S., Bacher, A., Gregory, J. F., III, & Hanson, A. D. (2002) Proc. Natl. Acad. Sci. USA 99, 12489-12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rébeillé, F. & Douce, R. (1999) in Regulation of Primary Metabolic Pathways in Plants, eds. Kruger, N. T., Hill, S. A. & Ratcliffe, R. G. (Kluwer, Dordrecht, The Netherlands), pp. 53-99.
- 8.Ravanel, S., Cherest, H., Jabrin, S., Grunwald, D., Surdin-Kerjan, Y., Douce, R. & Rébeillé, F. (2001) Proc. Natl. Acad. Sci. USA 98, 15360-15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viswanathan, V. K., Green, J. M. & Nichols, B. P. (1995) J. Bacteriol. 177, 5918-5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, J. M., Merkel, W. K. & Nichols, B. P. (1992) J. Bacteriol. 174, 5317-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edman, J. C., Goldstein, A. L. & Erbe, J. G. (1993) Yeast 9, 669-675. [DOI] [PubMed] [Google Scholar]
- 12.Criado, L. M., Martin, J. F. & Gil, J. A. (1993) Gene 126, 135-139. [DOI] [PubMed] [Google Scholar]
- 13.James, T. Y., Boulianne, R. P., Bottoli, A. P., Granado, J. D., Aebi, M. & Kües, U. (2002) J. Basic Microbiol. 42, 91-103. [DOI] [PubMed] [Google Scholar]
- 14.Triglia, T. & Cowman, A. F. (1999) Exp. Parasitol. 92, 154-158. [DOI] [PubMed] [Google Scholar]
- 15.Morollo, A. A. & Bauerle, R. (1993) Proc. Natl. Acad. Sci. USA 90, 9983-9987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seki, M., Narusaka, M., Kamiya, A., Ishida, J., Satou, M., Sakurai, T., Nakajima, M., Enju, A., Akiyama, K., Oono, Y., et al. (2002) Science 296, 141-145. [DOI] [PubMed] [Google Scholar]
- 17.Vernet, T., Dignard, D. & Thomas, D. Y. (1987) Gene 52, 225-233. [DOI] [PubMed] [Google Scholar]
- 18.Arfman, N., Worrell, V. & Ingram, L. O. (1992) J. Bacteriol. 174, 7370-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols, B. P., Seibold, A. M. & Doktor, S. Z. (1989) J. Biol. Chem. 264, 8597-8601. [PubMed] [Google Scholar]
- 20.Bradford, M. M. (1976) Anal. Biochem. 72, 248-254. [DOI] [PubMed] [Google Scholar]
- 21.Koziel, M. G., Carozzi, N. B. & Desai, N. (1996) Plant Mol. Biol. 32, 393-405. [DOI] [PubMed] [Google Scholar]
- 22.Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H. & Sheen, J. (1996) Curr. Biol. 6, 325-330. [DOI] [PubMed] [Google Scholar]
- 23.Abel, S. & Theologis, A. (1994) Plant J. 5, 421-427. [DOI] [PubMed] [Google Scholar]
- 24.Quinlivan, E. P., Roje, S., Basset, G., Shachar-Hill, Y., Gregory, J. F., III, & Hanson, A. D. (2003) J. Biol. Chem. 278, 20731-20737. [DOI] [PubMed] [Google Scholar]
- 25.Wilkinson, J. Q., Lanahan, M. B., Yen, H. C., Giovannoni, J. J. & Klee, H. J. (1995) Science 270, 1807-1809. [DOI] [PubMed] [Google Scholar]
- 26.Hanson, A. D. & Gregory, J. F., III (2002) Curr. Opin. Plant Biol. 5, 244-249. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann, K. M. (1995) Plant Cell 7, 907-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tewari, Y. B., Jensen, P. Y., Kishore, N., Mayhew, M. P., Parsons, J. F., Eisenstein, E. & Goldberg, R. N. (2002) Biophys. Chem. 96, 33-51. [DOI] [PubMed] [Google Scholar]
- 29.Mills, W. R. & Joy, K. W. (1980) Planta 148, 75-83. [DOI] [PubMed] [Google Scholar]
- 30.Riens, B., Lohaus, G., Heineke, D. & Heldt, H. W. (1991) Plant Physiol. 97, 227-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winter, H., Robinson, D. G. & Heldt, H. W. (1993) Planta 191, 180-190. [Google Scholar]
- 32.Radwanski, E. R. & Last, R. L. (1995) Plant Cell 7, 921-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.