Abstract
The actin cytoskeleton drives cell locomotion and tissue remodeling. The invention of live-cell fluorescence single-molecule imaging opened a window for direct viewing of the actin remodeling processes in the cell. Since then, a number of unanticipated molecular functions have been revealed. One is the mechanism of F-actin network breakdown. In lamellipodia, one third of newly polymerized F-actin disassembles within 10 seconds. This fast F-actin turnover is facilitated by the filament severing/disrupting activity involving cofilin and AIP1. Astoundingly fast dissociation kinetics of the barbed end interactors including capping protein suggests that F-actin turnover might proceed through repetitive disruption/reassembly of the filament near the barbed end. The picture of actin polymerization is also being revealed. At the leading edge of the cell, Arp2/3 complex is highly activated in a narrow edge region. In contrast, mDia1 and its related Formin homology proteins display a long-distance directional molecular movement using their processive actin capping ability. Recently, these two independently-developed projects converged into a discovery of the spatiotemporal coupling between mDia1-mediated filament nucleation and actin disassembly. Presumably, the local concentration fluctuation of G-actin regulates the actin nucleation efficiency of specific actin nucleators including mDia1. Pharmacological perturbation and quantitative molecular behavior analysis synergize to reveal hidden molecular linkages in the actin turnover cycle and cell signaling.
Keywords: single-molecule imaging, actin turnover, mDia1, pharmacokinetic simulation, G-actin
1. Introduction
The actin cytoskeleton is the major component beneath the surface of eukaryotic cells. Remodeling of the actin cytoskeleton is directly coupled to cell shape changes. To accomplish body functions such as morphogenesis, migration of immune cells, formation of the neural network, endocytosis, exocytosis and cytokinesis, cells must reorganize cell surface structures through mobilization of the actin remodeling machinery. The actin cytoskeleton not only facilitates transformation of cell structures but also generates forces that drive translocation of the whole cell body and even tissue remodeling.
Recent development of live-cell fluorescence single-molecule imaging has given us a window for direct viewing of cellular biochemical reactions.1) Currently, the prevailing method to examine molecular functions is to knock out or knock down gene expression and to observe resultant phenotypes. For example, my research group has recently carried out comprehensive screening to elucidate the role of Rho family GTPases in the PDGF-induced chemotaxis of fibroblast cells. This study identified Cdc42, Rac1 and RhoG as crucial regulators of fibroblast chemo-taxis. Knock down of three GTPases had severe and distinct morphological effects on chemotactic cells. Interestingly, impaired chemotaxis in all situations arises from the reduced cell migration speed but not loss in the directionality.2) As seen in this example, genetic manipulation provides a powerful means to elucidate molecular functions. However, due to its slowness, genetic manipulation does not always clarify the primary site of action of the molecule. Success in elucidating mechanistic aspects of complex biological systems such as actin-based motility relies on the methods enabling real time monitoring of the process.
In 2002, my study with Tim Mitchison convincingly demonstrated that intracellular molecules can be visualized at the single-molecule level.3) It was fortunate that my research subject was actin filament dynamics. The assembly to disassembly step of cytoskeletal proteins is a suitable target for fluorescence single-molecule observation. With the aid of the low-noise charge-coupled devise (CCD) cameras, it is possible to visualize a single fluorechrome attached to filamentous actin (F-actin). Signals from a non-diffusing fluorechrome add up in a small spot on the CCD chip with the exposure time of ~2 seconds. In contrast, signals from a freely diffusing fluorechrome are blurred on the image. Thus, cytoskeletal proteins which cease random walk for a certain period can be visualized as single-molecules in a state-specific manner (Fig. 1).
Fig. 1.
Overview of fluorescence single-molecule imaging. The upper panels show conventional live-cell imaging of EGFP-actin in a Xenopus fibroblast cell. A part of the timelapse images are paneled on the right. The retrograde flow of bulk F-actin structures can be visualized although movement of individual F-actin subunits and local F-actin assembly and disassembly can not be assessed by the conventional imaging of GFP probes. The lower panels show fluorescence single-molecule imaging employing the same EGFP-actin probe. The density of labeled probes is much less than that in the upper example. Low noise CCD cameras can capture the images of individual cell structure-anchored fluorechromes with long exposure times, typically 0.2 to 2 seconds. Signals from soluble probes are blurred. By this way, the association-dissociation kinetics of cytoskeletal proteins can be monitored with high resolution. Scale bars, 5 μm.
Using this approach, my research has elucidated detailed F-actin turnover kinetics at the leading edge of fibroblasts3) as well as F-actin association kinetics of major actin regulators.4),5) It is now possible to compare kinetics data obtained in living cells directly with data from in vitro actin biochemistry. In a number of cases, single-molecule observation has revealed unexpectedly fast actin remodeling processes. In the former part of this review, I overview the molecular kinetics involved in breakdown of the actin network.
In the latter, I describe the feedback F-actin restoration mechanism regulated by monomeric actin (G-actin). In the cell, actin exists in two forms, monomeric G-actin and filamentous F-actin (Fig. 2). F-actin turnover is very dynamic. In cultured cells, the conversion between G- and F-actin occurs in every a half to a few minutes.3),6)–8) G-actin and F-actin exist roughly at the 1:1 ratio. The cellular concentration of G-actin may exceed a hundred micromolar. Similarly abundant G-actin sequestering proteins prevent G-actin from uncontrolled polymerization. The large mass of the G-actin pool and its sequestering proteins has kept researchers’ eyes off their regulatory roles. However, two independently-developed projects in my research recently converged into a discovery of the dynamic association of filament nucleation to actin disassembly mechanisms in the cell.9) I discuss the spatiotemporal coupling between actin disassembly and reassembly through the concentration fluctuation and heterogeneity of free G-actin.
Fig. 2.
Homeostasis between G-actin and F-actin. In typical cultured cells, conversion between monomeric ‘G-actin’ and filamentous ‘F-actin’ occurs in every a half to a few minutes. G-actin and F-actin exist roughly at the 1:1 ratio. G-actin sequestering proteins bind G-actin and prevent uncontrolled actin nucleation by lowering the concentration of ‘free’ G-actin. The concentration of total G-actin may exceed 100 μM while the concentration of free G-actin is kept low at a range of 0.1–1 μM.9),170) Because of the large size of the G-actin pool, little has been investigated about the role of the G-actin pool in actin remodeling. However, my two independently-developed researches have converged into an intriguing finding that the actin disassembly and F-actin nucleation activities are tightly coupled in times and places within the cell.9)
2. Force generation by actin system in non-muscle cells
To generate the force to drive cell locomotion, the actin cytoskeleton operates two major force-generating systems.10) One uses the actin polymerization force and the other is the actomyosin contraction mechanism. The leading edge of motile cells forms two morphologically distinct pseudopods called filopodia and lamellipodia. In both, actin forms polarized arrays of filaments directing fast-growing barbed ends towards the cell edge.11),12) In lamellipodia, F-actin free from barbed end inhibitors such as capping protein exists in abundance.4),13)–15) Many free barbed ends are presumably in contact with the leading edge plasma membrane. Due to the flexible nature of either the plasma membrane16) or the lateral bending of actin filaments,17) G-actin is allowed to occasionally assemble onto the barbed end in contact with the plasma membrane. Such insertion events lead to generation of force pushing the cell edge forward. This mechano-chemical coupling has been demonstrated using pharmacological intervention of actin elongation18) and theorized in the context of the Brownian ratchet mechanism.19)
In the more central region of the cell at the back of lamellipodia, actin forms an array of long, often anti-parallel filaments. This type of F-actin is connected to the extracellular matrix and the neighboring cells, through actin stress fibers to focal adhesions and circumference bundles to adherence junctions, respectively. The conventional type-II myosin associating with such actin arrays generates contractile forces. Cells exhibiting the amoeboid mode of migration use the actomyosin mechanism.20) Actomyosin also plays a crucial role in migration of epithelial sheets during embryo morphogenesis.21) In addition, blebbing, the third form of cell edge protrusion, is driven by actomyosin.22),23) The local cortical rupture as a consequence of increased actomyosin tension24) or uneven transient pressure formed by a poroelastic cytoplasm25) may lead to the formation of blebs. Blebbing is frequently observed in cells exhibiting amoeboid migration and cytokinesis accompanied by an increase in cortical tension.
Here I propose another potent regulatory step conducted by actin polymerization that is the formation of structure-bridging actin filaments. Thus far, much of our attention to the role of actin polymerization has been paid in cell edge protrusion. However, in the central region, F-actin turnover is ubiquitously found.7),26) Anti-parallel F-actin structures such as actin stress fibers and the cytokinetic contractile ring provide the scaffold for conventional myosin. The amount of myosin-generated force and its transmission should be modulated by the density and the polarity of surrounding actin filaments. Therefore, assembly of cell structure-bridging F-actin structures would be a vital step to control the force generation for cell locomotion and tissue remodeling.
Under many conditions, rapid remodeling of actin stress fibers is observed in cultured cells. Extra-cellular ligands such as lysophosphatidic acid stimulate the rapid formation of actin stress fibers. Upon wounding the chick embryonic skin, thick actin cables are formed at the front of the marginal epithelial cells within 5 min.27) Rho, a small GTPase, is a molecular switch in the formation of actin stress fibers28) and the contractile ring during cytokinesis.29),30) In mid 90’s, my work in graduate school identified mDia1, a member of formin homology proteins (Formins) as an effector of Rho.31) Further analysis revealed that its FH1–FH2 unit structure, which is conserved among Formins, is capable of inducing the formation of long thin actin fibers.32) These actin fibers often bridge across the diameter of the cell. This activity cooperates with ROCK, a Rho effector kinase that upregulates actomyosin contractility,33)–37) to induce morphologically-varied actin fibers. It is noteworthy that overexpression of active mDia1 induces the bleb formation in a fraction of cells,32) suggesting a close link between mDia1 and actomyosin-derived cortical tension. Thus, cell morphology-based analysis provided a number of insights into the function of mDia1. At the same time, I realized the difficulty in capturing the precise site of action of this potent actin fiber inducer because of the complex nature of cellular F-actin remodeling. Direct viewing of the molecular behavior of mDia1 and its related molecules has now turned out to be a powerful tool to clarify how structure-bridging actin arrays are generated.
3. F-actin turnover measurement: different interpretations
Before the invention of live-cell fluorescence single-molecule observation,3) cellular F-actin turnover had been characterized mainly by two methods. One observes primary incorporation sites of newly-administrated actin probes. Researchers fixed cells 20–30 seconds after microinjection of labeled actin and observed distribution of the probe incorporation either by electron microscopy14) or by fluorescence microscopy.13) These studies observed that the extreme leading edge of the cell assembles F-actin at the fastest rate.
The other approach uses photoactivation of fluorescence (PAF) of caged fluorescent actin6),8) or fluorescence recovery after photobleaching (FRAP) of fluorescently-labeled actin.7),38),39) These methods simultaneously visualize movement and disintegration of the cytoskeletal network. In an early study, Yu-li Wang38) discovered the continuous centripetal movement of the lamellipodium actin network which is called the retrograde actin flow. This finding illuminated the main body of the long known centripetal movement of the cell peripheral architecture.40),41) This centripetal actin flow is thought to serve for adhesion-induced cell edge protrusion, as proposed in a hypothetical model called the ‘clutch model’.42) In this model, strengthening linkage between cell adhesion molecules and the retrograde actin flow reduces the flow speed. If the filament growth rate is unaltered at the leading edge, the reduction in the actin flow rate may lead to cell edge extension. Recently, the interaction either between Shootin-1 and L1-CAM43) or between N-cadherin and F-actin44) has been proposed to constitute the molecular ‘clutch’ to promote neurite outgrowth. Since this massive actin flow is found in most of cultured cells, there must be more adhesion molecules that function in a similar manner. Notably, a similar flow of E-cadherin complexes between two adjacent cells is also found at the cell-to-cell junction.45) The relationship between this ‘physical’ clutch mechanism and the adhesion-induced ‘chemical’ cell signaling remains to be clarified.
With regard to F-actin turnover, an important notion was brought by PAF experiments using caged fluorescent actin. Based on the F-actin disintegration faster than the filament density decay expected from the retrograde flow rate, Theriot and Mitchison proposed that actin frequently polymerizes away from the leading edge.6) This proposal contradicted the treadmilling model which had prevailed in the research field. Actin forms a polarized filament with the barbed end growing and the pointed end shrinking at steady state,46) which is referred to as treadmilling. At the leading edge, the majority of actin filaments direct their fast-growing barbed ends toward the cell edge.11) F-actin assembles the fastest at the leading edge13),14) and then flows inward.18),38) From these observations, the treadmilling model, in which actin polymerizes exclusively at the leading edge while disassembling at the back of lamellipodia, was widely accepted. The proposal by Theriot and Mitchison provoked debate over the treadmilling and their nucleation release model.6),47) I reinvestigated this issue using fluorescence single-molecule observation with Mitchison, and found that about one third of newly assembled F-actin subunits had a short lifetime less than 10 seconds in lamellipodia of Xenopus XTC fibroblasts.3) Since it takes 2–5 minutes for the actin network to travel through the entire width of a lamellipodium, the results support Theriot and Mitchison’s conclusion. The fast disassembling population of F-actin was also observed by a computer assisted fluorescence speckle analysis of X-rhodamine-labeled actin.26)
FRAP experiments often show slow recovery kinetics of F-actin in lamellipodia. A recent study39) strongly argues that FRAP data showing slow recovery of the photobleached actin are inconsistent with single-molecule observation results showing fast disassembling population.3) This discrepancy might arise in part from the intrinsic problems in FRAP and PAF experiments.
First, single-molecule analysis detects fast F-actin species repeatedly within the observed time window. By contrast, FRAP and PAF analyses measure the recovery kinetics of the label existing at a given moment. Hence direct comparison between single-molecule lifetime distribution and the FRAP/PAF decay rate may not be valid. Reinterpreted to the FRAP/PAF situation by weighing lifetime of each population, the single-molecule lifetime data yield that at a given moment, nearly a half of F-actin subunits have lifetime of >100 seconds. F-actin forms high order structures and its stability is enhanced by association with tropomyosin,48),49) fascin8),50),51) and so on.
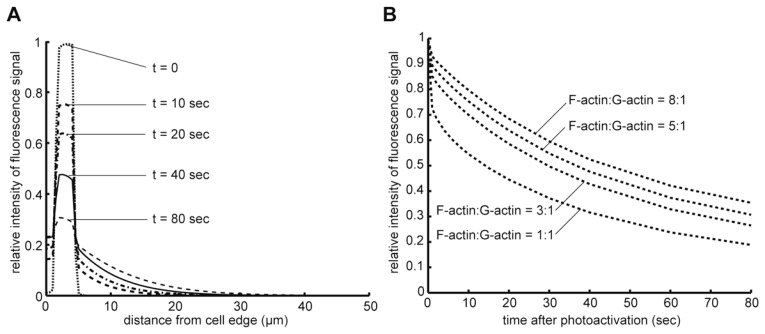
Second, as shown by mathematical simulation,52) reincorporation of dissociated labels into the marked zone in FRAP and PAF experiments may retard the decay rate substantially. In lamellipodia, F-actin exists at ≈1000 μM53) and therefore the ratio of F- and G-actin could become as high as 5~10:1. In such situations, the reincorporation mechanism retards the apparent FRAP decay ~2 times slower than true actin disassembly as duration in the G-actin state becomes very short in the actin turnover cycle (Fig. 3). These two problems need special attention when FRAP/PAF analysis is applied for molecules which rapidly switch between fast and slow diffusion states.
Fig. 3.
Simulation of PAF (photoactivation of fluorescence) experiments using the Tardy model: a delay in the recovery kinetics due to local reincorporation. Tardy et al.52) mathematically analyzed actin-based PAF and FRAP experiments incorporating the exchange between F-actin and G-actin. Although the model does not consider uneven distribution of F-actin and the retrograde actin flow, the early decay or recovery kinetics in the local cell environment can be estimated by this model. (A) The time-resolved distribution of PAF signals in a 50 μm wide cell. The photoactivation label is placed between 1.5 μm and 4.5 μm away from the cell edge. Diffusion constant of G-actin, the turnover rate from F-actin to G-actin and the ratio between F- and G-actin are 4.0 × 10−8 cm2/s,7) 0.03 s−1 and 5:1, respectively. Although the overall ratio between F- and G-actin is about 1:1, F-actin exists at ~1000 μM in lamellipodia,53) which should lead to a local high F-/G-actin concentration ratio. (B) Dependency of PAF signal decay on the ratio between F- and G-actin. The conditions are the same as above except for the F-/G-actin ratios. Note that the simulated decay rate of PAF labels becomes substantially slower (apparent T1/2 is approximately 30~50 sec) than the true F-actin disassembly rate (T1/2 = 23 sec).
In addition, as I discuss in the following sections, actin disassembly may proceed in part through release of actin oligomers.4) This mechanism, which is now under investigation in my research group, may further slow actin diffusing out of the FRAP and PAF areas.
4. Single-molecule kinetics of F-actin turnover and actin regulators in lamellipodia
I summarize single-molecule kinetics of actin and its regulators in a single defined cell system, XTC Xenopus fibroblast cells. These cells are suitable for live cell imaging as they grow at room temperature in ambient atmosphere. A condition where cells form a wide flat lamellipodia was developed.3) Since thickness of lamellipodia is less than 200 nm,53) it is possible to observe molecules in the entire lamellipodium using conventional epifluorescence microscopy. This method can also be applied to other mammalian cell types with modification.43) I have found that several types of mammalian cells including mouse NIH3T3 cells and rat 3Y1 cells tend to lose activity rapidly under strong xenon illumination for unknown reasons (unpublished observations).
The dendritic nucleation model is believed to constitute the major mechanism controlling lamellipod actin dynamics.54),55) F-actin is nucleated by Arp2/3 complex off the side of preexisting filaments.56)–59) Those nucleated filaments grow outward as actin filaments in lamellipodia direct their fast growing barbed end toward the cell periphery.11) Growth of barbed ends is terminated by capping protein.60) The filament network moves toward the cell center and disassembles.
Single-molecule observation of EGFP-actin revealed F-actin turnover kinetics in lamellipodia of XTC fibroblasts in detail. The observation of single-molecule fluorescent actin revealed that (i) 34% of newly assembled filaments disassemble within 10 seconds, (ii) F-actin lifetime distributes over a wide range of 4–148 seconds and (iii) F-actin migrates parallel to each other except for subtle local deformation of the actin network (See Supplementary Movie 3 in ref. 3). The observation (iii) is expected from the dense, crosslinked filament network in EM studies.47),61) The observation (iii), however, does not agree with the conclusion drawn in a study led by Waterman-Storer and Danuser which claimed a large population of long-lived F-actin migrating at a slow speed in lamellipodia.26) Their method called qFSM (quantitative fluorescence speckle microscopy) analyzes behavior of fluorescence speckles, a cluster of fluorechromes, by computer-assisted algorisms. Recently, it was disputed that several manual or automatic analyses, designed to eliminate tracking errors, fail to detect such long-lived, slowly-migrating F-actin species in the actin speckle images.62) In general, errors in automatic particle tracking may dramatically increase at high particle densities and for molecules that appear and disappear quickly. Moreover, for particles localized in narrow cell structures such as focal adhesions, correlation-based tracking may detect displacement of structures but not of the particles if the label density is too high. Therefore, care also needs to be taken in the interpretation of other qFSM-based flow rate analyses for focal adhesion proteins,63) actin regulators64) and so on. I suggest that reanalysis using image samples labeled at various particle densities may help understand the mechanics of error generation by computer-assisted algorisms. I also suggest that reanalysis by single-molecule observation may be required for validating the conclusions drawn by qFSM approaches. For such a purpose, it is important to carefully develop conditions that eliminate phototoxicity problems in each cell system.
Apart from these possible tracking problems, the original actin speckle images provided by Ponti et al.26) and other studies65),66) show coexistence of quickly appearing and disappearing actin speckles in the body of lamellipodia, which is consistent with single-molecule observation of EGFP-actin.3) The average dissociation rate of F-actin is ≈0.03 s−1. Slightly faster is the dissociation of Arp2/3 complex from the actin network which occurs at 0.048 s−1 (Fig. 4). Thus the lag between uncapping at the pointed end and disassembly of the entire filament is very limited. On the other hand, the dissociation of capping protein (CP) occurs astoundingly fast at 0.58 s−1. Importantly, this fast CP dissociation is specific to in vivo as the EGFP-tagged capping protein probe tightly caps the barbed end with the dissociation rate of 0.005 s−1 in vitro, which is comparable to native capping protein.67) The recent study, which strongly argued that lamellipod actin turnover is slow,39) also detected fast dissociation of CP with T1/2 of 7.2 seconds using FRAP. Thus the previously prevailing view that CP blocks barbed end growth until the end of lifetime of its bound filament needs to be revised. None of the other barbed end interacting proteins, Eps8, VASP, gelsolin,4) wild-type mDia19) and AIP1,5) shows persistent association with the lamellipod actin network. Consistently, the concentration of free barbed ends is high at ~1 μM in lamellipodia of permeabilized XTC cells.4) Furthermore, the actin elongation rate determined from the speed of processive actin polymerization by the mDia1 FH2 domain4),68) and its biochemical property69) is very fast (66 s−1). Combined, growth of barbed ends in lamellipodia is not restricted as thought before. Mathematical simulation of the treadmilling model using the above parameters points to the requirement of >90 fold acceleration of pointed end disassembly (unpublished results). In vitro, pointed end disassembly can be accelerated by a maximum of 22–30 fold with ADF and less efficiently with cofilin.70),71) Thus the single-molecule kinetics data suggest a mechanism other than the treadmilling model in which cofilin/ADF disassembles F-actin solely from the pointed end.
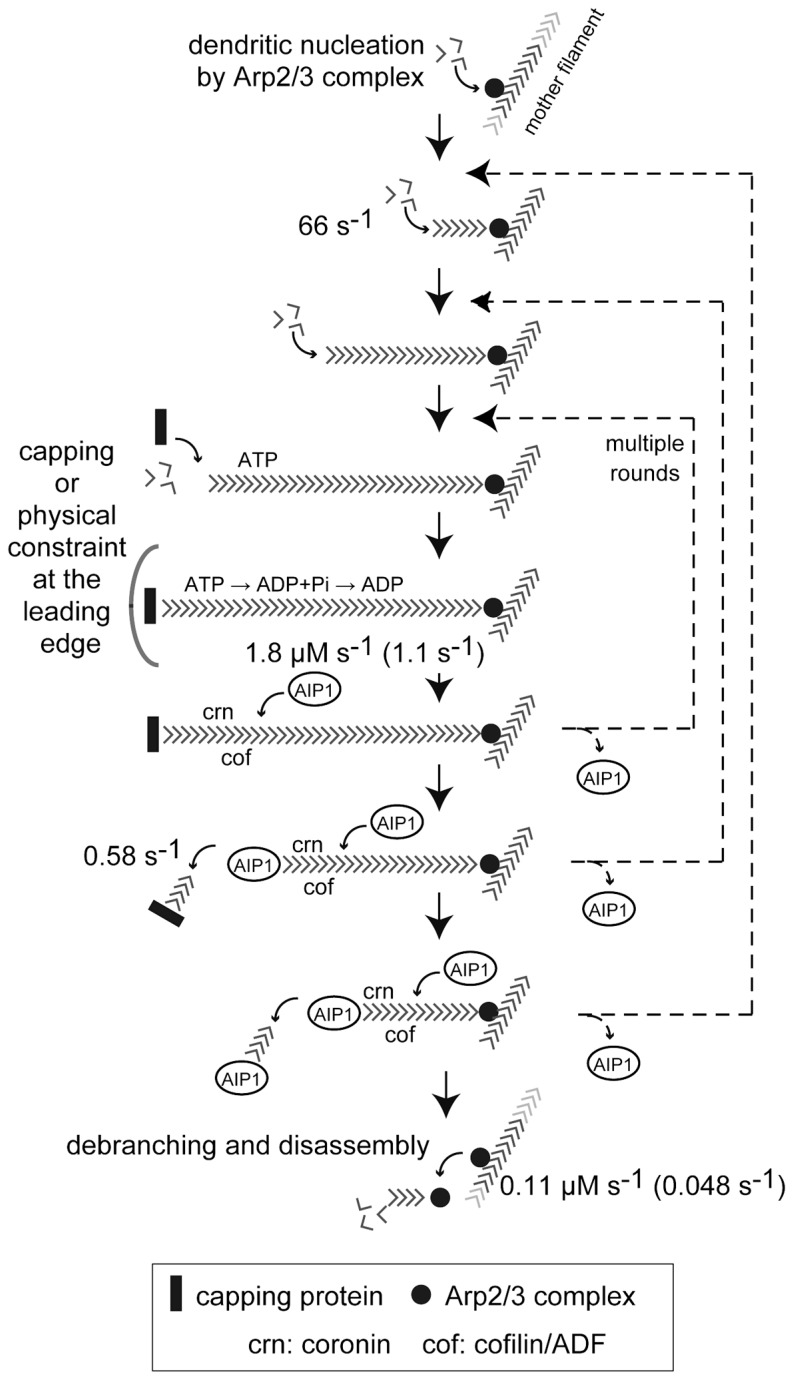
Fig. 4.
Summary of single-molecule kinetics in the dendritic actin network. The numbers in bold letters show kinetic parameters obtained in fluorescence single-molecule studies.3)–5) Overall, dynamics at the barbed end has turned out to be faster than previously thought. The dissociation of capping protein occurs at 0.58 s−1. Fast elongation of the free barbed end is derived from the speed of mDia1-catalyzed processive actin elongation in lamellipodia. In contrast, the pointed end side of F-actin, where Arp2/3 complex associates, disassembles much slower at the rate of 0.048 s−1. Certain mechanisms other than simple treadmilling must compensate the imbalance between fast growing barbed ends with slowly disassembling pointed ends. AIP1-mediated actin severing/disruption might be one of the mechanisms. It turned out to be ≈15 times more frequent than F-actin nucleation by Arp2/3 complex.
5. How might filament disassembly proceed in the cell?
1). Recent advance in actin depolymerization biochemistry
F-actin disassembly to G-actin recycling remains one of the most obscurely understood steps in the actin turnover cycle. The cofilin/ADF family72),73) is essential in cellular F-actin turnover.4),74),75) However, how actin disassembly proceeds (i.e. end depolymerization versus filament severing, the barbed end versus the pointed end) remains an open issue. Whether F-actin is frequently severed or not has attracted researchers’ interest because severing is a measure for the cell to generate newly-growing filaments. Currently it remains elusive whether cofilin/ADF functions exclusively by accelerating pointed end disassembly or its weak ‘non-catalyzing’ filament severing activity76) substantially contributes to actin disassembly processes. In addition, cofilin also promotes filament nucleation at a high concentration.77)
Cofilin/ADF by itself may not accomplish the fast actin disassembly observed in cells. Recent work, however, succeeded to purify a highly active tri-component actin disassembling system consisting of cofilin, AIP1 and coronin from calf thymus.78),79) The high actin disassembling activity by these three proteins appears to be promising towards the goal to elucidate the fast actin recycling mechanism.
AIP1 (actin interacting protein 1) was originally identified as a binding partner of both actin and cofilin (reviewed in ref. 80). AIP1 shows genetic interaction with cofilin in yeast,81),82) C. elegans83) and Drosophila,84) where they collaborate to disassemble F-actin.85),86) In mouse, two amino acid deletion in AIP1 causes neutrophil dysfunction and macro-thrombocytopenia.87) AIP1 caps the barbed end in a manner dependent on cofilin and inhibits elongation.88) AIP1 does not prevent reannealing of mechanically fragmented filaments.88) Hence AIP1 specifically recognizes and caps the barbed end generated by cofilin-catalyzed filament severing.
Recently, bipolar effects of coronin on cofilin-catalyzed actin disassembly have been demonstrated.89) Coronin prevents cofilin-induced severing of F-actin assembled from ATP-actin but promotes severing of F-actin assembled from ADP-actin. The authors interpreted that coronin protects ATP/ADP-Pi-F-actin from cofilin while helping cofilin sever the filament after ATP hydrolysis and γ-phosphate (Pi) release. Although this work provided important notions, their data (Fig. 2B in ref. 89) might not simply agree with the author’s interpretation. Hydrolysis of Mg-ATP occurs quickly upon poly-merization and Pi release proceeds over minutes.90) Pi release is prerequisite for filament disassembly because Pi stabilizes F-actin.91) Pi release is greatly accelerated by cofilin.92) At the start of the above filament disassembly experiments, a large fraction of F-actin should already be ADP-F-actin. Therefore coronin’s bipolar effects may not simply depend on the nucleotide bound to F-actin. Their microscopic observation (Fig. 2G and Movies S3 and S4 in ref. 89) shows weak F-actin bundling activity of coronin. I therefore speculate that the inhibitory effects of coronin on ATP-F-actin disassembly might be caused by other mechanisms such as coronin-induced filament bundling.93)
It is also known that coronin interferes with Arp2/3 complex-induced actin nucleation94),95) and promote debranching.96) These effects on Arp2/3 complex are week (typically, ≈2 fold changes) even with a saturating concentration of coronin (50–500 nM).94),96) Coronin may not exist free at these concentrations in cells because the dissociation constant of coronin with F-actin is ≈6 × 10−9 M.97) Thus these activities toward the Arp2/3 complex may not be the primary function of coronin under intracellular environment. Rather, the following actin disassembly activity might be the more important function of coronin because coronin, cofilin and AIP1 synergize effectively.
When combined together, cofilin, AIP1 and coronin achieve fast actin disassembly in a manner insensitive to polymerizable monomer.78) This system facilitates actin disassembly from both barbed and pointed ends by abruptly removing filaments of a mean size of 260 subunits.79) The authors of this study hypothesized that instantaneous subunit loss proceeds through cooperative strand separation. Further biochemical analysis of this tri-component actin disassembling system should provide a clue to solve how fast actin disassembly might be achieved in the cell.
2). Frequent severing triggers fast capping protein dissociation in living cells
In parallel with the above biochemical analyses, single-molecule analysis by my group has revealed frequent F-actin severing/disruption in lamellipodia.4),5) As mentioned, capping protein (CP) observed at the single-molecule level dissociates at 0.58 s−1, approximately 20 times faster than actin disassembly. Notably, F-actin stabilizing treatments such as jasplakinolide,98) overexpression of hLIMK1 which inactivates cofilin,99),100) and association with actin stress fibers all block the fast dissociation of CP. Jasplakinolide inhibits the interaction between cofilin and F-actin in live XTC cells.5) These data strongly indicate that filament severing by cofilin triggers dissociation of CP from the actin network.
The dissociation of CP from the barbed has been reported to be induced by phosphatidylinositol-4,5-bisphosphate (PIP2)67) or CARMIL.101),102) However, recent studies employing direct microscopic observation of the uncapping reaction reported that PIP2 induced-dissociation of CP is undetectable103) or occurs at a very slow rate (~10% uncapped by 50 μM PIP2 after 60 min).104) This markedly contrasts with the instantaneous uncapping by PIP2 observed in the previous reports,67) one of which also described the fast uncapping effect of CARMIL.101) The previous observations67),101) might be caused by filament shearing upon mixing and simultaneous blockade of free CP by PIP2. It would be necessary to reevaluate the effects of phospholipids and CARMIL using several other methods including direct microscopic observation.58),91),103) The binding site for phospholipids104) overlaps with the actin binding surface105) on the 3D structure of CP.106)
The finding of actin turnover-dependent fast CP dissociation led my research group to propose the frequent filament severing/end-to-end annealing hypothesis.4) In this model, CP dissociates accompanied by a barbed end actin oligomer. The other part of the filament is rescued by similarly frequent end-to-end annealing, escaping rapid collapse. The size of actin oligomers capable of diffusing off the actin network is unknown. An EM study47) observed the filament intercross every 14–16 nm, ≈5 actin subunits. The detergent-extracted actin comet tail induced by Listeria monocytogenes in macrophages largely consists of short filaments, typically ≈0.1 μm (Fig. 8 in ref. 107). The size of diffusible actin oligomers could thus be within the range between 5 and 30 subunits. To achieve fragmentation of F-actin to these sizes, a severing rate of 0.1–0.02 s−1 might be required. This problem was challenged in the following study.
Fig. 8.
Enhanced F-actin nucleation by mDia1 around the sites of vigorous actin disassembly in live XTC cells (adopted from ref. 9). Colocalization between the emergence site of the single-molecule mDia1 FH2 probe (circles), the catalytic core for its actin nucleation activity, and the distribution of AIP1 (mPlum-tagged; fluorescence image) is shown. AIP1 is a cofactor of cofilin and was used to monitor the distribution of cofilin-catalyzed actin disassembly. Scale bar, 5 μm. Adopted with permission from J. Cell Sci. 121, 3403–3412, Fig. 6 (2008).
3). AIP1-associated filament disruption in lamellipodia: 15 times more frequent than actin nucleation by Arp2/3 complex
A number of attempts have been made to quantitatively model actin-based cell edge protrusion. Regrettably, modeling studies108),109) omit filament severing despite the accumulating evidence.4),110) Such omission might have arisen from the lack of the knowledge regarding the in vivo frequency of filament severing. A recent study introducing an open source editable simulation database111) incorporated filament severing and annealing. This study built an extensive simulation with a large set of parameters. Yet the parameters used for several reactions including filament severing and uncapping are distinct from the data obtained in live cells (Fig. 4). The difficulty in predicting kinetic parameters only from in vitro reconstitution is evident.
To investigate filament severing in vivo, my research group turned to AIP1. AIP1 specifically recognizes the barbed end generated after cofilin-catalyzed filament severing/disruption. Several lines of evidence from single-molecule observation of AIP1 suggest the same property operating in vivo. First, LIMK1 overexpression decreased the F-actin association of AIP1. Second, the F-actin association of AIP1 is attenuated by jasplakinolide which blocks the cofilin-F-actin interaction simultaneously. Third, the dissociation of AIP1 slows upon jasplakinolide treatment, which is analogous to the reaction of CP to this drug.4) These notions support the use of AIP1 probes for monitoring cofilin/ADF-mediated filament severing/disruption.
Single-molecule observation of two AIP1 probes yielded 1.8 μM s−1 for the filament disruption rate in lamellipodia. There F-actin exists at ≈1000 μM.53) The frequency of AIP1-associated filament severing/disruption corresponds to 1 s−1 per ≈560 subunits long filament on average. Surprisingly, this frequency is ≈15 times more than that of actin nucleation by Arp2/3 complex, 0.11 μM s−1.4) Does AIP1 dissociate rapidly enough to allow barbed end growth?
In vitro, AIP1 dissociates from barbed end at τ < 10 s.79) AIP1 dissociates from barbed end at 0.1~0.2 s−1 in jasplakinolide-treated cells where severing-based release of CP is severely attenuated.4) If these observations reflect spontaneous dissociation of AIP1, 10~20% of dissociation of AIP1 may lead to the formation of free barbed ends. This mode of barbed end generation may account for the ubiquitous actin polymerization3),6),26) and the abundant free barbed ends4),15) throughout lamellipodia. At the leading edge, Arp2/3 complex adds new filament nuclei receiving signals downstream of Rac-WAVE/Scar and Cdc42-WASP.112) At the back of the leading edge, the AIP1-associated mechanism operates repeatedly along the retrograde flow. It is interesting to ask how the two opposite actions of AIP1, generation of the barbed end and breakdown of the polymer mass, might propagate in the actin array behind the leading edge.
4). Possible dynamic instability-like behavior of actin barbed end
To my regret, however, the above frequency of AIP1-associated filament disruption falls short of the rate predicted in the frequent filament severing-annealing theory by one order. Instead, this discrepancy opened up another possibility in the actin turnover regulation.5) The filament disassembly mechanism involving AIP1 might preferentially attack the filament near the barbed end. In vitro, both barbed and pointed ends are disassembled by cofilin, AIP1 and coronin faster than the other portion of the filament. In lamellipodia, the pointed-end side disassembles much more slowly than the barbed end as the dissociation of Arp2/3 complex occurs at 0.048 s−1 (Fig. 4).4) Thus the pointed end is more stabilized against the cellular actin disassembly activity than the barbed end. It is worthy mentioning that the dissociation rate of Arp2/3 complex is almost equal to the decay curve of lifetime distribution of fast disassembling F-actin species. Hence the whole actin network other than the barbed end filament disassembles with similar ‘slow’ kinetics (T1/2, ≈15 seconds) while the barbed end portion may undergo rapid filament disruption/severing cycles. This raises an intriguing possibility that the barbed end of cellular actin filaments may display growth and shrinkage behavior similar to dynamic instability (Fig. 5).
Fig. 5.
Possible dynamic instability-like behavior of F-actin plus end. Combined with recent progress in biochemical analysis of the actin disassembling machinery consisting of cofilin, AIP1 and coronin,79) the fast single-molecule kinetics of capping protein4) and AIP15) implies one-end growth and shrinkage behavior of F-actin, reminiscent of dynamic instability of the micro-tubules plus end.
Dynamic instability is characteristic of microtubules dynamics.113),114) At any point of time, a subset of microtubules are growing while others are rapidly shrinking. GTP-cap is thought to protect the plus end from disassembly. Once the plus end micro-tubules hydrolyze GTP, GDP-tubulin tends to be in a bent conformation, which triggers rapid disassembly from the plus end. While several molecules such as MCAK and XMAP215 enhance this growth and shrinkage dynamics,115) dynamic instability is an intrinsic property of microtubules.
In contrast, the actin filament is believed to undergo treadmilling. The dissociation of actin is the fastest at the barbed end of ADP-F-actin (7.2 s−1; ref. 46). However, in the presence of ATP-G-actin, this fast barbed end dissociation may not occur. ATP hydrolysis is fast but Pi release is slow after ATP-G-actin assembles at the barbed end.46),92) Pi abrogates the dissociation of ADP-F-actin at the barbed end.91) ATP, which is abundant in the cytoplasm, also prevents the barbed end depolymerization of ADP-F-actin.116) In the cell, profiln:actin complex exists at 22–40 μM.9),117),118) This complex can add to the barbed end as fast as free G-actin. Indeed, from the speed of processive movement of mDia1 FH2, the barbed end growth rate can be estimated to be 66 s−1 in lamellipodia.4) All together, depolymerization from the barbed end seems unlikely to contribute to filament disassembly. However, the identification of cofiln/AIP1/coronin system and its ultra-fast filament disruption from both ends of F-actin78),79) has now opened up the possibility that the barbed end may be the major site for filament disassembly in vivo. The fast barbed end and slow pointed end single-molecule kinetics of many filament end factors4),5) suggest the dynamic instability-like behavior of the barbed end (Fig. 5). Further extensive studies are required to prove this intriguing possibility.
6. How does actin reassemble? : actin nucleating factors
To form filaments de novo, actin undergoes two steps, nucleation and elongation.46) Filament nucleation occurs spontaneously in the physiological salt condition, but this step requires long time in typical in vitro experiments. The formation of actin dimers and trimers occurs frequently in solution. However, dissociation of subunits is also fast and the nucleation step barely proceeds forward. Once filament nuclei consisting of 4 actin subunits are formed, they act as a filament and actin elongation proceeds steadily. The rate of actin nucleation depends on the cubic of the free G-actin concentration. In the cell, actin nucleating factors are required for efficient filament nucleation.
1). Arp2/3 complex
The major actin nucleator at the leading edge is Arp2/3 complex. Arp2/3 complex nucleates the filaments off the side of pre-existing filaments.56),58),59) This branched actin network is abundantly found in lamellipodia.61) As revealed by single-molecule imaging, its activation zone is highly restricted in the narrow (<0.65 μm wide) area proximal to the leading edge.4) For activation, Arp2/3 complex requires three factors, the mother filament, actin monomer and activators such as WASP/WAVE family of proteins.112) Especially the WAVE complex119) is highly concentrated to the lamellipodium tip.120) Single-molecule observation revealed that distribution of the duration time of Arp2/3 association to the actin network fits with a single exponential curve. Thus a single rate-limiting step probably controls Arp2/3 complex dissociation. A cofilin/ADF protein actophorin121) and coronin96) accelerate debranching whereas cortactin inhibits debranching.122) Le Clainche et al.123) showed that ATP hydrolysis by Arp2/3 complex coincides with debranching, whereas Dayel and Mullins124) argued that Arp2/3 complex hydrolyzes ATP immediately upon nucleation. It remains to be solved how the timing of the dissociation of Arp2/3 complex is determined in the cell.
2). Formin family
On the other hand, the Formin family (Formins) is responsible for the formation of a different set of actin filaments such as actin stress fibers and cytokinetic contractile rings.125),126) Formins share conserved FH1 and FH2 domains in their C-terminal halves.127) With the FH2 domain, Formins nucleate actin filaments.128),129) Remarkably FH2 domains stay processively associated with the growing barbed end while the filament elongates. This property was initially recognized as ‘leaky cap’. At low concentrations, FH2 domains decrease the rate of actin subunit exchange at the barbed end to a certain extent while allowing subunit addition at higher concentrations.130),131) This leaky cap property led researchers to hypothesize the processive capping of the growing barbed end by Formins.131)
A proof of the processive actin capping by Formins was brought by the experiments demonstrating antagonism between Formins and CP.132) However, a possibility still remained for non-processive mechanisms such as a cluster of molecules associating with the side of the barbed end filament (See Fig. 1 in ref. 125). Recently, such a mechanism has been demonstrated for VASP.133) The VASP family134) has an actin nucleation activity which becomes apparent under non-physiological low salt conditions. VASP prevents capping protein from the barbed end. Certainly VASP and Formins share common properties. However, processive motion of VASP has not been observed (See for example supplementary video 6 in ref. 4; a cluster of VASP move forward in this movie, but this does not prove processive movement of individual VASP molecules).
A recent work provided compelling evidence for non-processive barbed end elongation by VASP.133) When VASP was clustered on the surface of beads, long range barbed end growth occurred in a manner insensitive to CP. Probably a cluster of VASP interacts one after the other with the barbed end and protects it from CP without processively moving along the growing barbed end. Formins can protect the barbed end at lower concentrations than VASP. This difference made it less likely that Formins use the same clustering mechanism as VASP. However in 2003, a direct proof was awaited for the processive mechanism of Formins.
My research brought a conclusion to this problem by direct visualization. Single-molecule observation of the FH1–FH2 domain of mDia1 directly demonstrated its directional movement in living cells.68) The FH1–FH2 unit structure moves over several tens of microns at 2 μm/s, which is comparable to the speed of kinesin and myosin in motility assays. The FH2 domain alone can move processively albeit at a slower speed of 0.13 μm/s. Moreover, processive actin assembly at the contact of recombinant mDia1 FH1–FH2 was reconstituted in vitro. Later, other groups employing similar in vitro single-filament elongation assays elucidated the acceleration mechanism of the barbed end growth by Formins. Profilin accelerates elongation of the FH1–FH2 bound barbed end remarkably fast, 5–15 times faster than the native filament69),135)–137) as it was predicted from the fast speed of mDia1 FH1–FH2.68) Formins can thus produce a long actin filament over the diameter of the cell in a short duration. Formins probably have evolved to enable rapid assembly of actin stress fibers and other structure-bridging F-actin.
Genetic studies have identified the roles of Formins in vivo. mDia1 is a mammalian homolog of Drosophila diaphanous, which is essential in cytokinesis. Among 3 mammalian isoforms, mDia2 is specifically essential in cytokinesis of certain cell types.138) mDia1−/− mice are viable, but exhibit abnormal proliferation and migration of T lymphocytes.139),140) mDia1−/− mice develop age-dependent myeloproliferative disorder.141) This phenotype is reminiscent of myelodysplastic syndrome (MDS) which is often associated with deletion in chromosome 5q. Human Dia1 gene is located in 5q31.3 and adds the list of candidates for a tumor suppressor such as EGR1, RPS14 and α-catenin in 5q− MDS.142) Mouse Formin-2 is essential in positioning of the meiotic spindle,143) an actin-dependent process. Over wide eukaryotic species including yeast, C. elegans and Drosophila, Formins play important roles in the cell polarity formation in early development and cytokinesis. Please refer to other review articles to follow the progress of the entire Formin field.125)–127),144),145)
3). Nucleators with tandem WH2 domains
This recently emerged group includes Spire,146) Cobl,147) VopF,148) VopL149) and Lmod150) which contain three or four tandem WH2 domains. The WH2 domain151),152) is also found in other types of actin regulators such as WASP, VASP, MIM,153)–155) IRSp53,156) thymosin β4 family and its related molecule ciboulot.157),158) Through WH2 domains, these molecules interact with either G-actin or F-actin and exert various effects such as monomer sequestration, nucleation, barbed end elongation and scaffolding. The WH2 domain consists of a short amino acid stretch (17–43 aa) with a conserved N-terminal helix and a four amino acid motif corresponding to the LKKT in thymosin β4.152) The N-terminal helix interacts with the cleft between actin subdomains 1 and 3 near the barbed end side of actin. The LKKT motif binds to a hydrophobic pocket on the actin surface.158)–160) The two to three WH2 domains in tandem stabilize an actin dimer or trimer along the long pitch helix of F-actin,161) illuminating the mode of filament nucleation. In N-WASP, the WH2 domain is followed by its closely related C motif. The C motif binds Arp2 (actin related protein 2) and stabilizes the association of the WH2-bound actin subunit with Arp2. Dominguez and his colleagues proposed that the C motif is a variant of the WH2 domain which has specifically evolved for Arp2. They further proposed that WASP/WAVE is conceptually viewed as actin filament nucleators that specifically use the actin related protein (Arp2) instead of actin to build filament nuclei.162)
On the other hand, the WCA region of WASP/WAVE dramatically increases the efficiency for Arp2/3 complex activation upon homo- or heterodimerization.163) The 2:1 complex formation between GST-WCA (VCA) and Arp2/3 complex implies an additional interface on the surface of Arp2/3 complex for the interaction with the second WCA region. Furthermore, the WH2 motif in N-WASP interacts with the barbed end filament, supporting attachment of F-actin to moving membranes.164) Thus diverse functions are brought by distinct combinations of WH2 domains. Together with the non-processive actin elongation by a cluster of VASP,133) a variety of activities by the cooperative actions of multiple WH2 domains within a single or combinations of WH2 proteins operate in the actin system, which are now being revealed.
7. A clue for activation mechanism of mDia1 and Formins: G-actin inhibitors
I describe a novel role of homeostasis between G- and F-actin which bridges the gap between actin disassembly and polymer reassembly.
The GTP-bound form of Rho binds N-terminal region of mDia1. mDia1 is an autoinhibited molecule, and the binding of Rho opens mDia1 to release the activity of the FH1–FH2 unit.32),165) On a mission to seek small molecule compounds that evoke processive actin assembly by mDia1, latrunculin B (LatB) was unexpectedly identified.9) LatB is a G-actin inhibitor and is widely used to probe actin-dependent cellular processes. LatB binds G-actin near its ATP binding site and inhibits nucleotide exchange and filament assembly.166) When administrated at a high concentration (>1 μM), LatB stops the movement of mDia1 because of a loss in the polymerization-competent G-actin.68) But at a lower dose, LatB dramatically increases the frequency of processively-moving mDia1. This induction is observed as early as 10 s after the perfusion of 100 nM LatB, accompanied by a reduction in the speed of mDia1.9)
The induction of processive mDia1 speckles is reproduced by analogous treatments such as swinholide A and unpolymerizable G13R and R62D actins. Swinholide A-bound G-actin forms an anti-parallel dimer.167) This dimer species is distinct from a readily polymerizing dimer obtained by cross-linking adjacent subunits in the F-actin helix.168) The anti-parallel dimer neither fits with the structure of actin in complex with the FH2 domain of Bni1p169) nor promotes actin nucleation.168) I therefore predict that swinholide A promotes actin nucleation by mDia1 not through dimer stabilization.
The FH1–FH2 or FH2 region of mDia1 alone responds to low-dose LatB and display increased processive movement. Rho is required for full-length mDia1 being in an open conformation, but is not a direct mediator of LatB-increased actin nucleation by mDia1. LatB thus increases the rate of actin nucleation by acting on the FH2 catalytic core domain of mDia1 in cells. In marked contrast, LatB has only negative effects on mDia1 FH2-catalyzed actin filament assembly in vitro.9)
8. Latrunculin paradox: inhibitor-induced increase in drug-free targets
The opposite effects of LatB on the mDia1-catalyzed actin nucleation between in cells and in vitro puzzled me. While profilin binds G-actin noncompetitively with latrunculin, the binding of latrunculin reduces the affinity between Tβ4 and G-actin.166) In one study, latrunculin-induced concentration changes of G-actin either free or bound to partners were extensively analyzed.170) The authors of this study concluded that latrunculin has little effect on the concentration of free G-actin. Even so, I reevaluated this issue by kinetic modeling. My research group came to notice the paradoxical increase in the free G-actin concentration induced by LatB.9) The simulation analysis predicted that LatB induces a several-fold increase in the concentration of free G-actin (Fig. 6). This increase in free G-actin is accompanied by the accumulation of several micromolar LatB in the cell compartment. The underlying key mechanism for this paradoxical drug effect is the accumulation of total G-actin. In addition to the primary effect of LatB to inactivate G-actin, LatB causes a 1.2–1.3 fold increase in the total G-actin concentration as a result of decreased actin assembly, which in turn raises the free G-actin concentration several fold. This latter effect is due to the saturation of the buffering capacity of G-actin sequestering proteins. This study highlight an unprecedented example of pharmacological effects in which an antagonist increases the concentration of drug-free targets in a complex biological system.9)
Fig. 6.
Latrunculin paradox.9) The left panel depicts the scheme of kinetic modeling used for estimating time-dependent concentration changes of each G-actin species either free or bound to LatB, Tβ4 and/or profilin. G, L, T, and P represent G-actin, LatB, thymosin-β4 (Tβ4) and profilin, respectively. The simulation results (middle and right graphs) revealed the paradoxical effect of latrunculin B. The primary action of latrunculin B is to bind G-actin and inhibit actin polymerization. In experiments, slowdown of actin elongation by mDia1 and a ~30% F-actin decrease were observed in cells treated with low-dose LatB. The change in the balance between F- and G-actin leads to the paradoxical, several fold increase in free G-actin accompanied by saturation of the G-actin sequestering activities in the simulation. Adopted with permission from J. Cell Sci. 121, 3403–3412, Fig. 4B (2008).
9. G-actin regulation of F-actin nucleation and reassembly
Figure 7 summarizes the model for dual regulation of mDia1-catalyzed actin assembly. Rho signaling determines the ratio of the opened and the closed forms of mDia1, whereas an increase in free G-actin effectively converts the opened mDia1 to the actin nucleating state, leading to fast long-range actin filament assembly. Since the efficiency of actin nucleation by Formins depends on the cubic of the free G-actin concentration in vitro, FH2 domains may serve as sensitive sensors for the concentration fluctuation of free G-actin in the cell. In accordance with this idea, a distantly-related member of Formins, FRL1, also exhibits frequent processive movement in response to low-dose LatB. In contrast, Arp2/3 complex does not respond to LatB.9) Activities of actin nucleators may thus be regulated differentially by the G-actin concentration in the cell.
Fig. 7.
Feedback actin polymer restoration mechanism involving mDia1. Rho signaling determines the ratio between the closed (left) and the opened (right) form of mDia1. An increase in free G-actin efficiently converts “opened” mDia1 to the fully-active actin nucleating state, leading to fast long-range actin filament assembly in cells. With this property, mDia1 responds to acute disruption of F-actin structures evoked by physical stress or by extracellular stimuli-induced active actin remodeling, and rapidly restores cellular actin polymers by their fast actin elongation ability.
Another important notion is that the emergence frequency of processive mDia1 correlates with the local concentration of AIP1. During the course of the initial work that proved the processive actin elongation by mDia1,68) it was noticed that the FH2 domain construct of mDia1 frequently appears in discrete foci found at the base of lamellipodia. From these spots, mDia1 FH2 mutants appear and move away as if such foci were a fountain of processive mDia1. This finding had not been explored for a while. Later, the similarity in the spatiotemporal dynamics between mDia1 FH2 and AIP1 was noticed. It was then confirmed that the local concentration of AIP1 correlates with the emergence frequency of both full-length and the FH2 mutant of mDia1 (Fig. 8).9) This coincidence is also observed between AIP1 and other processive Formins including FRL1 (unpublished results). The association of AIP1 with F-actin depends on the activity of cofilin. In vitro, the association of Formins to free barbed ends was described by direct microscopic observation, but this event appears to be much less frequent than filament nucleation on the image.69) Therefore appearance of processive mDia1 in the cell may mostly represent de novo filament nucleation. The above findings are best interpreted as showing that an increase in the local G-actin release upregulates the efficiency of filament nucleation by mDia1.
mDia1 demonstrates the greatest rates of accelerated actin elongation among Formins.69),135) mDia1ΔN3 elongates actin at the rate of 2 μm/s (≈720 subunits/s) in cells.68) Eukaryotic cells, whose diameter reaches several tens of microns, have probably evolved to obtain these fast actin elongating systems. For example, leukocytes in the blood flow need to adhere the vessel wall within a few seconds after stimulation by leukotrienes. Even with mDia1, it takes five seconds to form de novo actin filaments which bridge over the diameter of leukocytes. The property of mDia1 may also provide an efficient way to rapidly reassemble actin polymers when cells are subjected to physical stress. The alignment of actin stress fibers remarkably alters when cells are subjected to cyclic stretch.171),172) Rho is activated upon stretch of smooth muscle cells173) and plays a role in stretch-induced reorientation of stress fibers in endothelial cells.174) Mechanical stress-induced disassembly of actin stress fibers may possibly upregulate actin nucleation by mDia1 through an increase in G-actin. The role of the G-actin regulated actin polymer restoration mechanism is currently under investigation by the use of fluorescence single-molecule observation.
10. Perspective and future directions
In this article, I have focused on the accomplishment of live-cell fluorescence single-molecule imaging developed in my research, and described its implications in relation to findings from other studies. As the readers may notice, most of comparison is made against in vitro actin biochemistry rather than in vivo phenotype analysis. Direct comparison between the single-molecule data and the biochemical knowledge has led us to find the important links such as filament severing-mediated barbed end disruption (Fig. 5) and G-actin regulation of actin nucleation by Formins (Figs. 7 and 8). Single-molecule imaging has an enormous advantage in elucidating the coupling between the probe and its surrounding molecules under the complex intracellular environment. With its speeds and quantitative characteristics, single-molecule approach can be a powerful tool to elucidate elementary process in many dynamic biological systems.
On the other hand, there still remains a gap between molecular kinetics and determination of the cell behavior. To bridge over the gap, modeling and computational analysis are becoming more and more important. The example of the paradoxical latrunculin effect (chapter 7–9) highlights the importance of quantitative modeling. At the same time, this example warns us to be cautious before moving on to modeling of the body processes on the computer chip. We still do not fully understand biological systems as simple as the ‘latrunculin paradox’ case (Figs. 6 and 7).
Finally, I introduce another use of single-molecule imaging for bridging the gap over elementary and whole body processes. Pharmacological perturbation and real time in vivo molecular behavior analysis often synergize to reveal hidden molecular linkages. Recently, my research group has found the first example of inhibitor-induced conformational regulation of kinases.175) Imatinib, a very successful kinase inhibitor in the treatment of chronic myelogenous leukemia, induces rapid translocation of its target, Abelson kinase (c-abl) to the cell edge. This translocation is triggered by the drug-induced complex formation with unknown cofactor(s). Despite extensive studies employing biochemistry, genetics, cell-based assays and structural analysis, our findings had not been recognized for many years. Two characteristics of live-cell single-molecule imaging, (i) a whole set of molecules reconstituted in the assay and (ii) its capability of observing a specific molecular event, enabled us to discover this unanticipated effect of kinase inhibitors. Real time observation of the effect of small compounds on the target behavior may thus help elucidate the mechanism of action of the drugs. Conversely, small compounds provide tools to perturb molecular functions rapidly. This combination should deepen our understanding of molecular functions in vivo. Small compounds may be the key to bridge the gap of our knowledge between elementary and whole body processes in the post genomic era.
Acknowledgements
This work is supported in part by Grants-in-Aid form the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) and grants from the Uehara Memorial Foundation and the Human Frontier Science Program.
Abbreviations
- CCD
charge-coupled devise
- Formins
formin homology proteins
- PAF
photoactivation of fluorescence
- FRAP
fluorescence recovery after photobleaching
- CP
capping protein
- qFSM
quantitative fluorescence speckle microscopy
- EGFP
enhanced green fluorescent protein
- Pi
γ-phosphate
- FH1
formin homology 1
- FH2
formin homology 2
- Arp2
actin related protein 2
- LatB
latrunculin B.
Profile
Naoki Watanabe was born in 1964. After graduating from Kyoto University Faculty of Medicine, he worked as a clinical doctor for three years. He then started his research career in 1993 with studies on cell signaling under the supervision of Prof. Shuh Narumiya. He received Ph.D. from Kyoto University in 1998 for the studies on the cell signaling cascades downstream of the small GTPase Rho. He became a JSPS research fellow, and then he was promoted to assistant professor in 1998. From 1999 to 2002, he joined Prof. Timothy J. Mitchison’s laboratory at Harvard Medical School as a postdoctoral research fellow and developed fluorescence single-molecule speckle microscopy. He moved back to Kyoto University and was promoted to associate professor in 2002. He also worked as a researcher at Precursory Research for Embryonic Science and Technology (PRESTO) for three years from 2002. His research focuses on the complexity and dynamicity of actin cytoskeleton remodeling in living cells. The invention of live-cell fluorescence single-molecule imaging in 2002 and the discovery of the processive actin elongation mechanism by formin homology proteins in 2004 are the highlights of his study. He was awarded the first JSPS prize (FY2004).

References
- 1).Vale R.D. (2008) Microscopes for fluorimeters: the era of single molecule measurements. Cell 135, 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Monypenny J., Zicha D., Higashida C., Oceguera-Yanez F., Narumiya S., Watanabe N. (2009) Cdc42 and Rac family GTPases regulate mode and speed but not direction of primary fibroblast migration during platelet-derived growth factor-dependent chemotaxis. Mol. Cell. Biol. 29, 2730–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Watanabe N., Mitchison T.J. (2002) Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science 295, 1083–1086 [DOI] [PubMed] [Google Scholar]
- 4).Miyoshi T., Tsuji T., Higashida C., Hertzog M., Fujita A., Narumiya S., et al. (2006) Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J. Cell Biol. 175, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Tsuji T., Miyoshi T., Higashida C., Narumiya S., Watanabe N. (2009) An order of magnitude faster AIP1-associated actin disruption than nucleation by the Arp2/3 complex in lamellipodia. PLoS ONE 4, e4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Theriot J.A., Mitchison T.J. (1991) Actin microfilament dynamics in locomoting cells. Nature 352, 126–131 [DOI] [PubMed] [Google Scholar]
- 7).McGrath J.L., Tardy Y., Dewey C.F., Jr, Meister J.J., Hartwig J.H. (1998) Simultaneous measurements of actin filament turnover, filament fraction, and monomer diffusion in endothelial cells. Biophys. J. 75, 2070–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Mallavarapu A., Mitchison T. (1999) Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J. Cell Biol. 146, 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Higashida C., Suetsugu S., Tsuji T., Monypenny J., Narumiya S., Watanabe N. (2008) G-actin regulates rapid induction of actin nucleation by mDia1 to restore cellular actin polymers. J. Cell Sci. 121, 3403–3412 [DOI] [PubMed] [Google Scholar]
- 10).Mitchison T.J., Cramer L.P. (1996) Actin-based cell motility and cell locomotion. Cell 84, 371–379 [DOI] [PubMed] [Google Scholar]
- 11).Small J.V., Isenberg G., Celis J.E. (1978) Polarity of actin at the leading edge of cultured cells. Nature 272, 638–639 [DOI] [PubMed] [Google Scholar]
- 12).Ishikawa H., Bischoff R., Holtzer H. (1969) Formation of arrowhead complexes with heavy meromyosin in a variety of cell types. J. Cell Biol. 43, 312–328 [PMC free article] [PubMed] [Google Scholar]
- 13).Symons M.H., Mitchison T.J. (1991) Control of actin polymerization in live and permeabilized fibroblasts. J. Cell Biol. 114, 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Okabe S., Hirokawa N. (1989) Incorporation and turnover of biotin-labeled actin microinjected into fibroblastic cells: an immunoelectron microscopic study. J. Cell Biol. 109, 1581–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Bailly M., Macaluso F., Cammer M., Chan A., Segall J.E., Condeelis J.S. (1999) Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J. Cell Biol. 145, 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Peskin C.S., Odell G.M., Oster G.F. (1993) Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys. J. 65, 316–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Mogilner A., Oster G. (1996) Cell motility driven by actin polymerization. Biophys. J. 71, 3030–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Forscher P., Smith S.J. (1988) Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J. Cell Biol. 107, 1505–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Mogilner A, Oster G. (2003) Polymer motors: pushing out the front and pulling up the back. Curr. Biol. 13, R721–R733 [DOI] [PubMed] [Google Scholar]
- 20).Lammermann T., Sixt M. (2009) Mechanical modes of ‘amoeboid’ cell migration. Curr. Opin. Cell Biol. 21, 636–644 [DOI] [PubMed] [Google Scholar]
- 21).Martin P., Parkhurst S.M. (2004) Parallels between tissue repair and embryo morphogenesis. Development 131, 3021–3034 [DOI] [PubMed] [Google Scholar]
- 22).Paluch E., Piel M., Prost J., Bornens M., Sykes C. (2005) Cortical actomyosin breakage triggers shape oscillations in cells and cell fragments. Biophys. J. 89, 724–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Charras G.T., Yarrow J.C., Horton M.A., Mahadevan L., Mitchison T.J. (2005) Non-equilibration of hydrostatic pressure in blebbing cells. Nature 435, 365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Paluch E., Sykes C., Prost J., Bornens M. (2006) Dynamic modes of the cortical actomyosin gel during cell locomotion and division. Trends Cell Biol. 16, 5–10 [DOI] [PubMed] [Google Scholar]
- 25).Mitchison T.J., Charras G.T., Mahadevan L. (2008) Implications of a poroelastic cytoplasm for the dynamics of animal cell shape. Semin. Cell Dev. Biol. 19, 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Ponti A., Machacek M., Gupton S.L., Waterman-Storer C.M., Danuser G. (2004) Two distinct actin networks drive the protrusion of migrating cells. Science 305, 1782–1786 [DOI] [PubMed] [Google Scholar]
- 27).Martin P., Lewis J. (1992) Actin cables and epidermal movement in embryonic wound healing. Nature 360, 179–183 [DOI] [PubMed] [Google Scholar]
- 28).Ridley A.J., Hall A. (1992) The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389–399 [DOI] [PubMed] [Google Scholar]
- 29).Kishi K., Sasaki T., Kuroda S., Itoh T., Takai Y. (1993) Regulation of cytoplasmic division of Xenopus embryo by rho p21 and its inhibitory GDP/GTP exchange protein (rho GDI). J. Cell Biol. 120, 1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Mabuchi I., Hamaguchi Y., Fujimoto H., Morii N., Mishima M., Narumiya S. (1993) A rho-like protein is involved in the organisation of the contractile ring in dividing sand dollar eggs. Zygote 1, 325–331 [DOI] [PubMed] [Google Scholar]
- 31).Watanabe N., Madaule P., Reid T., Ishizaki T., Watanabe G., Kakizuka A., et al. (1997) p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 16, 3044–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S. (1999) Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1, 136–143 [DOI] [PubMed] [Google Scholar]
- 33).Leung T., Manser E., Tan L., Lim L. (1995) A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J. Biol. Chem. 270, 29051–29054 [DOI] [PubMed] [Google Scholar]
- 34).Matsui T., Amano M., Yamamoto T., Chihara K., Nakafuku M., Ito M., et al. (1996) Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. EMBO J. 15, 2208–2216 [PMC free article] [PubMed] [Google Scholar]
- 35).Kimura K., Ito M., Amano M., Chihara K., Fukata Y., Nakafuku M., et al. (1996) Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273, 245–248 [DOI] [PubMed] [Google Scholar]
- 36).Ishizaki T., Maekawa M., Fujisawa K., Okawa K., Iwamatsu A., Fujita A., et al. (1996) The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J. 15, 1885–1893 [PMC free article] [PubMed] [Google Scholar]
- 37).Uehata M., Ishizaki T., Satoh H., Ono T., Kawahara T., Morishita T., et al. (1997) Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature 389, 990–994 [DOI] [PubMed] [Google Scholar]
- 38).Wang Y.L. (1985) Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J. Cell Biol. 101, 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Lai F.P., Szczodrak M., Block J., Faix J., Breitsprecher D., Mannherz H.G., et al. (2008) Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27, 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Abercrombie M., Heaysman J.E., Pegrum S.M. (1970) The locomotion of fibroblasts in culture. 3. Movements of particles on the dorsal surface of the leading lamella. Exp. Cell Res. 62, 389–398 [DOI] [PubMed] [Google Scholar]
- 41).Ingram V.M. (1969) A side view of moving fibroblasts. Nature 222, 641–644 [DOI] [PubMed] [Google Scholar]
- 42).Mitchison T., Kirschner M. (1988) Cytoskeletal dynamics and nerve growth. Neuron 1, 761–772 [DOI] [PubMed] [Google Scholar]
- 43).Shimada T., Toriyama M., Uemura K., Kamiguchi H., Sugiura T., Watanabe N., et al. (2008) Shootin1 interacts with actin retrograde flow and L1-CAM to promote axon outgrowth. J. Cell Biol. 181, 817–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Bard L., Boscher C., Lambert M., Mege R.M., Choquet D., Thoumine O. (2008) A molecular clutch between the actin flow and N-cadherin adhesions drives growth cone migration. J. Neurosci. 28, 5879–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Kametani Y., Takeichi M. (2007) Basal-to-apical cadherin flow at cell junctions. Nat. Cell Biol. 9, 92–98 [DOI] [PubMed] [Google Scholar]
- 46).Pollard T.D., Blanchoin L., Mullins R.D. (2000) Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29, 545–576 [DOI] [PubMed] [Google Scholar]
- 47).Small J.V., Herzog M., Anderson K. (1995) Actin filament organization in the fish keratocyte lamellipodium. J. Cell Biol. 129, 1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Bryce N.S., Schevzov G., Ferguson V., Percival J.M., Lin J.J., Matsumura F., et al. (2003) Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol. Biol. Cell 14, 1002–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Bernstein B.W., Bamburg J.R. (1982) Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF). Cell Motil. 2, 1–8 [DOI] [PubMed] [Google Scholar]
- 50).Brieher W.M., Coughlin M., Mitchison T.J. (2004) Fascin-mediated propulsion of Listeria monocytogenes independent of frequent nucleation by the Arp2/3 complex. J. Cell Biol. 165, 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Nakagawa H., Terasaki A.G., Suzuki H., Ohashi K., Miyamoto S. (2006) Short-term retention of actin filament binding proteins on lamellipodial actin bundles. FEBS Lett. 580, 3223–3228 [DOI] [PubMed] [Google Scholar]
- 52).Tardy Y., McGrath J.L., Hartwig J.H., Dewey C.F. (1995) Interpreting photoactivated fluorescence microscopy measurements of steady-state actin dynamics. Biophys. J. 69, 1674–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Abraham V.C., Krishnamurthi V., Taylor D.L., Lanni F. (1999) The actin-based nanomachine at the leading edge of migrating cells. Biophys. J. 77, 1721–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Carlier M.F., Le Clainche C., Wiesner S., Pantaloni D. (2003) Actin-based motility: from molecules to movement. Bioessays 25, 336–345 [DOI] [PubMed] [Google Scholar]
- 55).Pollard T.D., Borisy G.G. (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 56).Mullins R.D., Heuser J.A., Pollard T.D. (1998) The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Nat. Acad. Sci. USA 95, 6181–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Welch M.D., Rosenblatt J., Skoble J., Portnoy D.A., Mitchison T.J. (1998) Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science 281, 105–108 [DOI] [PubMed] [Google Scholar]
- 58).Amann K.J., Pollard T.D. (2001) Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Nat. Acad. Sci. USA 98, 15009–15013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59).Fujiwara I., Suetsugu S., Uemura S., Takenawa T., Ishiwata S. (2002) Visualization and force measurement of branching by Arp2/3 complex and N-WASP in actin filament. Biochem. Biophys. Res. Commun. 293, 1550–1555 [DOI] [PubMed] [Google Scholar]
- 60).Wear M.A., Cooper J.A. (2004) Capping protein: new insights into mechanism and regulation. Trends Biochem. Sci. 29, 418–428 [DOI] [PubMed] [Google Scholar]
- 61).Svitkina T.M., Borisy G.G. (1999) Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145, 1009–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Vallotton P., Small J.V. (2009) Shifting views on the leading role of the lamellipodium in cell migration: speckle tracking revisited. J. Cell Sci. 122, 1955–1958 [DOI] [PubMed] [Google Scholar]
- 63).Hu K., Ji L., Applegate K.T., Danuser G., Waterman-Storer C.M. (2007) Differential transmission of actin motion within focal adhesions. Science 315, 111–115 [DOI] [PubMed] [Google Scholar]
- 64).Iwasa J.H., Mullins R.D. (2007) Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr. Biol. 17, 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Vallotton P., Gupton S.L., Waterman-Storer C.M., Danuser G. (2004) Simultaneous mapping of filamentous actin flow and turnover in migrating cells by quantitative fluorescent speckle microscopy. Proc. Nat. Acad. Sci. USA 101, 9660–9665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Ponti A., Matov A., Adams M., Gupton S., Waterman-Storer C.M., Danuser G. (2005) Periodic patterns of actin turnover in lamellipodia and lamellae of migrating epithelial cells analyzed by quantitative fluorescent speckle microscopy. Biophys. J. 89, 3456–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Schafer D.A., Jennings P.B., Cooper J.A. (1996) Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Higashida C., Miyoshi T., Fujita A., Oceguera-Yanez F., Monypenny J., Andou Y., et al. (2004) Actin polymerization-driven molecular movement of mDia1 in living cells. Science 303, 2007–2010 [DOI] [PubMed] [Google Scholar]
- 69).Kovar D.R., Harris E.S., Mahaffy R., Higgs H.N., Pollard T.D. (2006) Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435 [DOI] [PubMed] [Google Scholar]
- 70).Maciver S.K., Pope B.J., Whytock S., Weeds A.G. (1998) The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur. J. Biochem. 256, 388–397 [DOI] [PubMed] [Google Scholar]
- 71).Carlier M.F., Laurent V., Santolini J., Melki R., Didry D., Xia G.X., et al. (1997) Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72).Maciver S.K. (1998) How ADF/cofilin depolymerizes actin filaments. Curr. Opin. Cell Biol. 10, 140–144 [DOI] [PubMed] [Google Scholar]
- 73).Bamburg J.R. (1999) Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15, 185–230 [DOI] [PubMed] [Google Scholar]
- 74).Hotulainen P., Paunola E., Vartiainen M.K., Lappalainen P. (2005) Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell 16, 649–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75).Kiuchi T., Ohashi K., Kurita S., Mizuno K. (2007) Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J. Cell Biol. 177, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76).Moriyama K., Yahara I. (1999) Two activities of cofilin, severing and accelerating directional depolymerization of actin filaments, are affected differentially by mutations around the actin-binding helix. EMBO J. 18, 6752–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77).Andrianantoandro E., Pollard T.D. (2006) Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23 [DOI] [PubMed] [Google Scholar]
- 78).Brieher W.M., Kueh H.Y., Ballif B.A., Mitchison T.J. (2006) Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J. Cell Biol. 175, 315– 324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79).Kueh H.Y., Charras G.T., Mitchison T.J., Brieher W.M. (2008) Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J. Cell Biol. 182, 341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80).Ono S. (2003) Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: new blades for twisted filaments. Biochemistry 42, 13363–13370 [DOI] [PubMed] [Google Scholar]
- 81).Clark M.G., Amberg D.C. (2007) Biochemical and genetic analyses provide insight into the structural and mechanistic properties of actin filament disassembly by the Aip1p cofilin complex in Saccharomyces cerevisiae. Genetics 176, 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82).Iida K., Yahara I. (1999) Cooperation of two actin-binding proteins, cofilin and Aip1, in Saccharomyces cerevisiae. Genes Cells 4, 21–32 [DOI] [PubMed] [Google Scholar]
- 83).Ono S. (2001) The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 152, 1313–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84).Ren N., Charlton J., Adler P.N. (2007) The flare gene, which encodes the AIP1 protein of Drosophila, functions to regulate F-actin disassembly in pupal epidermal cells. Genetics 176, 2223– 2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85).Mohri K., Ono S. (2003) Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J. Cell Sci. 116, 4107–4118 [DOI] [PubMed] [Google Scholar]
- 86).Okada K., Ravi H., Smith E.M., Goode B.L. (2006) Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments. Mol. Biol. Cell 17, 2855–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87).Kile B.T., Panopoulos A.D., Stirzaker R.A., Hacking D.F., Tahtamouni L.H., Willson T.A., et al. (2007) Mutations in the cofilin partner Aip1/Wdr1 cause autoinflammatory disease and macrothrom-bocytopenia. Blood 110, 2371–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88).Okada K., Blanchoin L., Abe H., Chen H., Pollard T.D., Bamburg J.R. (2002) Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J. Biol. Chem. 277, 43011–43016 [DOI] [PubMed] [Google Scholar]
- 89).Gandhi M., Achard V., Blanchoin L., Goode B.L. (2009) Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol. Cell 34, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90).Carlier M.F., Pantaloni D. (1986) Direct evidence for ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin filaments. Biochemistry 25, 7789–7792 [DOI] [PubMed] [Google Scholar]
- 91).Fujiwara I., Vavylonis D., Pollard T.D. (2007) Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc. Nat. Acad. Sci. USA 104, 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92).Blanchoin L., Pollard T.D. (1999) Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 274, 15538–15546 [DOI] [PubMed] [Google Scholar]
- 93).Spoerl Z., Stumpf M., Noegel A.A., Hasse A. (2002) Oligomerization, F-actin interaction, and membrane association of the ubiquitous mammalian coronin 3 are mediated by its carboxyl terminus. J. Biol. Chem. 277, 48858–48867 [DOI] [PubMed] [Google Scholar]
- 94).Humphries C.L., Balcer H.I., D’Agostino J.L., Winsor B., Drubin D.G., Barnes G., et al. (2002) Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J. Cell Biol. 159, 993–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95).Cai L., Marshall T.W., Uetrecht A.C., Schafer D.A., Bear J.E. (2007) Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell 128, 915–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96).Cai L., Makhov A.M., Schafer D.A., Bear J.E. (2008) Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell 134, 828–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97).Goode B.L., Wong J.J., Butty A.C., Peter M., McCormack A.L., Yates J.R., et al. (1999) Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol. 144, 83–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98).Bubb M.R., Spector I., Beyer B.B., Fosen K.M. (2000) Effects of jasplakinolide on the kinetics of actin polymerization. An explanation for certain in vivo observations. J. Biol. Chem. 275, 5163–5170 [DOI] [PubMed] [Google Scholar]
- 99).Yang N., Higuchi O., Ohashi K., Nagata K., Wada A., Kangawa K., et al. (1998) Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393, 809–812 [DOI] [PubMed] [Google Scholar]
- 100).Arber S., Barbayannis F.A., Hanser H., Schneider C., Stanyon C.A., Bernard O., et al. (1998) Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393, 805–809 [DOI] [PubMed] [Google Scholar]
- 101).Yang C., Pring M., Wear M.A., Huang M., Cooper J.A., Svitkina T.M., et al. (2005) Mammalian CARMIL inhibits actin filament capping by capping protein. Dev. Cell 9, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102).Uruno T., Remmert K., Hammer J.A., 3rd (2006) CARMIL is a potent capping protein antagonist: identification of a conserved CARMIL domain that inhibits the activity of capping protein and uncaps capped actin filaments. J. Biol. Chem. 281, 10635–10650 [DOI] [PubMed] [Google Scholar]
- 103).Kuhn J.R., Pollard T.D. (2007) Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J. Biol. Chem. 282, 28014–28024 [DOI] [PubMed] [Google Scholar]
- 104).Kim K., McCully M.E., Bhattacharya N., Butler B., Sept D., Cooper J.A. (2007) Structure/function analysis of the interaction of phosphatidylinositol 4,5-bisphosphate with actin-capping protein: implications for how capping protein binds the actin filament. J. Biol. Chem. 282, 5871–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105).Narita A., Takeda S., Yamashita A., Maeda Y. (2006) Structural basis of actin filament capping at the barbed-end: a cryo-electron microscopy study. EMBO J. 25, 5626–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106).Yamashita A., Maeda K., Maeda Y. (2003) Crystal structure of CapZ: structural basis for actin filament barbed end capping. EMBO J. 22, 1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107).Tilney L.G., DeRosier D.J., Tilney M.S. (1992) How Listeria exploits host cell actin to form its own cytoskeleton. I. Formation of a tail and how that tail might be involved in movement. J. Cell Biol. 118, 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108).Dawes A.T., Bard Ermentrout G., Cytrynbaum E.N., Edelstein-Keshet L. (2006) Actin filament branching and protrusion velocity in a simple 1D model of a motile cell. J. Theor. Biol. 242, 265–279 [DOI] [PubMed] [Google Scholar]
- 109).Schaus T.E., Borisy G.G. (2008) Performance of a population of independent filaments in lamellipodial protrusion. Biophys. J. 95, 1393–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110).Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D.S., Condeelis J.S. (2004) Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304, 743–746 [DOI] [PubMed] [Google Scholar]
- 111).Ditlev J.A., Vacanti N.M., Novak I.L., Loew L.M. (2009) An open model of actin dendritic nucleation. Biophys. J. 96, 3529–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112).Takenawa T., Suetsugu S. (2007) The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8, 37–48 [DOI] [PubMed] [Google Scholar]
- 113).Mitchison T., Kirschner M. (1984) Dynamic instability of microtubule growth. Nature 312, 237– 242 [DOI] [PubMed] [Google Scholar]
- 114).Burbank K.S., Mitchison T.J. (2006) Microtubule dynamic instability. Curr. Biol. 16, R516–R517 [DOI] [PubMed] [Google Scholar]
- 115).Howard J., Hyman A.A. (2007) Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 19, 31–35 [DOI] [PubMed] [Google Scholar]
- 116).Teubner A., Wegner A. (1998) Kinetic evidence for a readily exchangeable nucleotide at the terminal subunit of the barbed ends of actin filaments. Biochemistry 37, 7532–7538 [DOI] [PubMed] [Google Scholar]
- 117).Southwick F.S., Young C.L. (1990) The actin released from profilin-actin complexes is insufficient to account for the increase in F-actin in chemoattractant-stimulated polymorphonuclear leukocytes. J. Cell Biol. 110, 1965–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118).Pring M., Weber A., Bubb M.R. (1992) Profilinactin complexes directly elongate actin filaments at the barbed end. Biochemistry 31, 1827–1836 [DOI] [PubMed] [Google Scholar]
- 119).Eden S., Rohatgi R., Podtelejnikov A.V., Mann M., Kirschner M.W. (2002) Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790–793 [DOI] [PubMed] [Google Scholar]
- 120).Nakagawa H., Miki H., Ito M., Ohashi K., Takenawa T., Miyamoto S. (2001) N-WASP, WAVE and Mena play different roles in the organization of actin cytoskeleton in lamellipodia. J. Cell Sci. 114, 1555–1565 [DOI] [PubMed] [Google Scholar]
- 121).Blanchoin L., Pollard T.D., Mullins R.D. (2000) Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10, 1273–1282 [DOI] [PubMed] [Google Scholar]
- 122).Weaver A.M., Karginov A.V., Kinley A.W., Weed S.A., Li Y., Parsons J.T., et al. (2001) Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11, 370– 374 [DOI] [PubMed] [Google Scholar]
- 123).Le Clainche C., Pantaloni D., Carlier M.F. (2003) ATP hydrolysis on actin-related protein 2/3 complex causes debranching of dendritic actin arrays. Proc. Nat. Acad. Sci. USA 100, 6337– 6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124).Dayel M.J., Mullins R.D. (2004) Activation of Arp2/3 complex: addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2, E91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125).Watanabe N., Higashida C. (2004) Formins: processive cappers of growing actin filaments. Exp. Cell Res. 301, 16–22 [DOI] [PubMed] [Google Scholar]
- 126).Evangelista M., Zigmond S., Boone C. (2003) Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116, 2603– 2611 [DOI] [PubMed] [Google Scholar]
- 127).Higgs H.N. (2005) Formin proteins: a domain-based approach. Trends Biochem. Sci. 30, 342–353 [DOI] [PubMed] [Google Scholar]
- 128).Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., et al. (2002) Role of formins in actin assembly: nucleation and barbed-end association. Science 297, 612–615 [DOI] [PubMed] [Google Scholar]
- 129).Sagot I., Rodal A.A., Moseley J., Goode B.L., Pellman D. (2002) An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4, 626–631 [DOI] [PubMed] [Google Scholar]
- 130).Li F., Higgs H.N. (2003) The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 131).Pring M., Evangelista M., Boone C., Yang C., Zigmond S.H. (2003) Mechanism of formin-induced nucleation of actin filaments. Biochemistry 42, 486–496 [DOI] [PubMed] [Google Scholar]
- 132).Zigmond S.H., Evangelista M., Boone C., Yang C., Dar A.C., Sicheri F., et al. (2003) Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13, 1820–1823 [DOI] [PubMed] [Google Scholar]
- 133).Breitsprecher D., Kiesewetter A.K., Linkner J., Urbanke C., Resch G.P., Small J.V., et al. (2008) Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 27, 2943–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134).Bear J.E., Gertler F.B. (2009) Ena/VASP: towards resolving a pointed controversy at the barbed end. J. Cell Sci. 122, 1947–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135).Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., Carlier M.F. (2004) Formin Is a Processive Motor that Requires Profilin to Accelerate Actin Assembly and Associated ATP Hydrolysis. Cell 119, 419–429 [DOI] [PubMed] [Google Scholar]
- 136).Vavylonis D., Kovar D.R., O’Shaughnessy B., Pollard T.D. (2006) Model of formin-associated actin filament elongation. Mol. Cell 21, 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137).Paul A.S., Pollard T.D. (2008) The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138).Watanabe S., Ando Y., Yasuda S., Hosoya H., Watanabe N., Ishizaki T., et al. (2008) mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol. Biol. Cell 19, 2328–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139).Eisenmann K.M., West R.A., Hildebrand D., Kitchen S.M., Peng J., Sigler R., et al. (2007) T cell responses in mammalian diaphanous-related formin mDia1 knock-out mice. J. Biol. Chem. 282, 25152–25158 [DOI] [PubMed] [Google Scholar]
- 140).Sakata D., Taniguchi H., Yasuda S., Adachi-Morishima A., Hamazaki Y., Nakayama R., et al. (2007) Impaired T lymphocyte trafficking in mice deficient in an actin-nucleating protein, mDia1. J. Exp. Med. 204, 2031–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141).Peng J., Kitchen S.M., West R.A., Sigler R., Eisenmann K.M., Alberts A.S. (2007) Myeloproliferative defects following targeting of the Drf1 gene encoding the mammalian diaphanous related formin mDia1. Cancer Res. 67, 7565–7571 [DOI] [PubMed] [Google Scholar]
- 142).Eisenmann K.M., Dykema K.J., Matheson S.F., Kent N.F., Deward A.D., West R.A., et al. (2009) 5q-myelodysplastic syndromes: chromosome 5q genes direct a tumor-suppression network sensing actin dynamics. Oncogene 28, 3429–3441 [DOI] [PubMed] [Google Scholar]
- 143).Leader B., Lim H., Carabatsos M.J., Harrington A., Ecsedy J., Pellman D., et al. (2002) Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat. Cell Biol. 4, 921–928 [DOI] [PubMed] [Google Scholar]
- 144).Goode B.L., Eck M.J. (2007) Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593–627 [DOI] [PubMed] [Google Scholar]
- 145).Blanchoin L, Staiger C.J. (2008) Plant formins: Diverse isoforms and unique molecular mechanism. Biochim. Biophys. Acta. 10.1016/j.bbamcr.2008.09.015 (in press). [DOI] [PubMed]
- 146).Quinlan M.E., Heuser J.E., Kerkhoff E., Mullins R.D. (2005) Drosophila Spire is an actin nucleation factor. Nature 433, 382–388 [DOI] [PubMed] [Google Scholar]
- 147).Ahuja R., Pinyol R., Reichenbach N., Custer L., Klingensmith J., Kessels M.M., et al. (2007) Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell 131, 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148).Tam V.C., Serruto D., Dziejman M., Brieher W., Mekalanos J.J. (2007) A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe 1, 95–107 [DOI] [PubMed] [Google Scholar]
- 149).Liverman A.D., Cheng H.C., Trosky J.E., Leung D.W., Yarbrough M.L., Burdette D.L., et al. (2007) Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc. Natl. Acad. Sci. USA 104, 17117–17122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150).Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D.B., Rebowski G., et al. (2008) Leiomodin is an actin filament nucleator in muscle cells. Science 320, 239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151).Carlier M.F., Hertzog M., Didry D., Renault L., Cantrelle F.X., van Heijenoort C, et al. (2007) Structure, function, and evolution of the beta-thymosin/WH2 (WASP-Homology2) actin-binding module. Ann. N.Y. Acad. Sci. 1112, 67–75 [DOI] [PubMed] [Google Scholar]
- 152).Dominguez R. (2007) The beta-thymosin/WH2 fold: multifunctionality and structure. Ann. N.Y. Acad. Sci. 1112, 86–94 [DOI] [PubMed] [Google Scholar]
- 153).Mattila P.K., Salminen M., Yamashiro T., Lappalainen P. (2003) Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J. Biol. Chem. 278, 8452–8459 [DOI] [PubMed] [Google Scholar]
- 154).Woodings J.A., Sharp S.J., Machesky L.M. (2003) MIM-B, a putative metastasis suppressor protein, binds to actin and to protein tyrosine phosphatase delta. Biochem. J. 371, 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155).Yamagishi A., Masuda M., Ohki T., Onishi H., Mochizuki N. (2004) A novel actin bundling/dilopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J. Biol. Chem. 279, 14929– 14936 [DOI] [PubMed] [Google Scholar]
- 156).Miki H., Yamaguchi H., Suetsugu S., Takenawa T. (2000) IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408, 732–735 [DOI] [PubMed] [Google Scholar]
- 157).Boquet I., Boujemaa R., Carlier M.F., Preat T. (2000) Ciboulot regulates actin assembly during Drosophila brain metamorphosis. Cell 102, 797– 808 [DOI] [PubMed] [Google Scholar]
- 158).Hertzog M., van Heijenoort C., Didry D., Gaudier M., Coutant J., Gigant B., et al. (2004) The β-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell 117, 611–623 [DOI] [PubMed] [Google Scholar]
- 159).Irobi E., Aguda A.H., Larsson M., Guerin C., Yin H.L., Burtnick L.D., et al. (2004) Structural basis of actin sequestration by thymosin-beta4: implications for WH2 proteins. EMBO J. 23, 3599–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160).Chereau D., Kerff F., Graceffa P., Grabarek Z., Langsetmo K., Dominguez R. (2005) Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Nat. Acad. Sci. USA 102, 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161).Rebowski G., Boczkowska M., Hayes D.B., Guo L., Irving T.C., Dominguez R. (2008) X-ray scattering study of actin polymerization nuclei assembled by tandem W domains. Proc. Nat. Acad. Sci. USA 105, 10785–10790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162).Boczkowska M., Rebowski G., Petoukhov M.V., Hayes D.B., Svergun D.I., Dominguez R. (2008) X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure 16, 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163).Padrick S.B., Cheng H.C., Ismail A.M., Panchal S.C., Doolittle L.K., Kim S., et al. (2008) Hierarchical regulation of WASP/WAVE proteins. Mol. Cell 32, 426–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164).Co C., Wong D.T., Gierke S., Chang V., Taunton J. (2007) Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell 128, 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165).Alberts A.S. (2001) Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 276, 2824–2830 [DOI] [PubMed] [Google Scholar]
- 166).Yarmola E.G., Somasundaram T., Boring T.A., Spector I., Bubb M.R. (2000) Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A. J. Biol. Chem. 275, 28120–28127 [DOI] [PubMed] [Google Scholar]
- 167).Bubb M.R., Spector I., Bershadsky A.D., Korn E.D. (1995) Swinholide A is a microfilament disrupting marine toxin that stabilizes actin dimers and severs actin filaments. J. Biol. Chem. 270, 3463–3466 [DOI] [PubMed] [Google Scholar]
- 168).Millonig R., Salvo H., Aebi U. (1988) Probing actin polymerization by intermolecular cross-linking. J. Cell Biol. 106, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169).Otomo T., Tomchick D.R., Otomo C., Panchal S.C., Machius M., Rosen M.K. (2005) Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433, 488–494 [DOI] [PubMed] [Google Scholar]
- 170).Pring M., Cassimeris L., Zigmond S.H. (2002) An unexplained sequestration of latrunculin A is required in neutrophils for inhibition of actin polymerization. Cell Motil. Cytoskeleton 52, 122–130 [DOI] [PubMed] [Google Scholar]
- 171).Takemasa T., Sugimoto K., Yamashita K. (1997) Amplitude-dependent stress fiber reorientation in early response to cyclic strain. Exp. Cell Res. 230, 407–410 [DOI] [PubMed] [Google Scholar]
- 172).Yoshigi M., Hoffman L.M., Jensen C.C., Yost H.J., Beckerle M.C. (2005) Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 171, 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173).Smith P.G., Roy C., Zhang Y.N., Chauduri S. (2003) Mechanical stress increases RhoA activation in airway smooth muscle cells. Am. J. Respir. Cell Mol. Biol. 28, 436–442 [DOI] [PubMed] [Google Scholar]
- 174).Kaunas R., Nguyen P., Usami S., Chien S. (2005) Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc. Nat. Acad. Sci. USA 102, 15895–15900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175).Fujita A., Shishido T., Yuan Y., Inamoto E., Narumiya S., Watanabe N. (2009) Imatinib mesylate (STI571)-induced cell edge translocation of kinase-active and kinase-defective Abelson kinase: requirements of myristoylation and src homology 3 domain. Mol. Pharmacol. 75, 75–84 [DOI] [PubMed] [Google Scholar]