Abstract
Lipoproteins fulfill diverse roles in antibiotic resistance, adhesion, protein secretion, signaling and sensing, and many also serve as the substrate binding protein (SBP) partner to ABC transporters for the acquisition of a diverse array of nutrients including peptides, sugars, and scarcely abundant metals. In the staphylococci, the iron-regulated SBPs are significantly upregulated during iron starvation and function to sequester and deliver iron into the bacterial cell, enabling staphylococci to circumvent iron restriction imposed by the host environment. Accordingly, this subset of lipoproteins has been implicated in staphylococcal pathogenesis and virulence. Lipoproteins also activate the host innate immune response, triggered through Toll-like receptor-2 (TLR2) and, notably, the iron-regulated subset of lipoproteins are particularly immunogenic. In this review, we discuss the iron-regulated staphylococcal lipoproteins with regard to their biogenesis, substrate specificity, and impact on the host innate immune response.
Keywords: Staphylococcus, lipoproteins, iron-regulated, iron acquisition, substrate binding protein, TLR2, Fur
Introduction
The staphylococci are a diverse group of Gram-positive, catalase-positive facultative anaerobes, consisting of approximately 36 species hosted largely by the human body (Kloos and Bannerman, 1994). Historically, the staphylococci have been broadly divided into two groups based on their coagulase activity with Staphylococcus aureus representing the most notable, coagulase-positive pathogen of the genus. S. aureus is often regarded as the leading cause of infections of the bloodstream, skin, soft tissue, and lower respiratory system (Moet et al., 2007). In contrast, coagulase-negative staphylococci (CoNS) were largely considered to be harmless commensals of the skin and mucous membranes until recent years, when the opportunistically pathogenic S. epidermidis has emerged as the most frequent cause of device-associated nosocomial infections (reviewed in Otto, 2012). The appearance of vancomycin-resistance in clinical isolates of both S. aureus and CoNS, in addition to the increasing prevalence of community-associated methicillin-resistant S. aureus (CA-MRSA), highlights the continued need to develop novel strategies to combat these global pathogens (Srinivasan et al., 2002; Vandenesch et al., 2003).
The success of the staphylococci has been attributed, in part, to their ability to acquire iron from the host. In contrast to its relative abundance in nature, iron represents a severely growth-limiting nutrient in vivo. Indeed, while the solubility for ferric iron is often cited as 10−18 M, an adjusted calculation suggests that the actual concentration is closer to 10−9–10−10 M at neutral pH in aerobic environments, based on Fe(OH)+2 being the primary species instead of Fe(OH)3 (Ratledge and Dover, 2000). The revised value is still orders of magnitude lower than required to support microbial growth and additionally, iron within the host is further sequestered in glycoproteins such as transferrin and lactoferrin, bound within storage proteins such as ferritin and hemosiderin, or complexed with heme in the form of hemoglobin and myoglobin (Ratledge, 2007). Sequestration of iron functions both in preventing the catalysis of reactions generating damaging free radicals and in providing nutritional immunity against bacterial infection (Schaible and Kaufmann, 2004).
To circumvent the aforementioned restrictions, the staphylococci have evolved a plethora of mechanisms to acquire iron from the host, including the elaboration of multiple siderophores, utilization of xenosiderophores, acquisition of iron from heme and hemoproteins, and the uptake of inorganic free iron (Beasley and Heinrichs, 2010; Haley and Skaar, 2012; Hammer and Skaar, 2011). Each of these systems employs an iron-regulated membrane protein, almost always of the ATP-binding cassette (ABC) transporter superfamily, for import of iron or complexed-iron across the membrane. In addition to the required ABC-type membrane permease and ATPase (Davidson et al., 2008), these iron-regulated ABC transporters employ a high-affinity membrane-anchored lipoprotein, i.e., a protein bearing an N-terminal, covalently linked lipid (Hutchings et al., 2009), for the specific recognition and binding of the iron substrate; these lipoproteins are analogous to the periplasmic substrate binding proteins (SBPs) of Gram-negative bacteria. Acylation of the hydrophilic protein promotes localization of the lipid into the phospholipid bilayer, anchoring the protein in close proximity to the membrane where it is positioned, once bound by substrate, to interact with its cognate ABC transporter to facilitate translocation of substrate into the cell. Whereas many other functions have been attributed to lipoproteins in bacteria, including sensing and signaling, protein secretion, antibiotic resistance, adhesion and the uptake of many nutrients in addition to iron (Sutcliffe and Russell, 1995), herein we focus attention solely on the iron-regulated lipoproteins (IRLPs) expressed by the staphylococci, given their important roles in staphylococcal pathogenesis and immune stimulation. Of the approximately 50 total lipoproteins encoded by S. aureus, the species in which most of the work has been performed, studies from several laboratories have identified, to date, a total of 9 IRLPs—SirA, HtsA, SstD, FhuD1, FhuD2, IsdE, FepA, SitC, and Opp1A. These proteins will be the subject of this review.
Control of iron-regulated gene expression
Fur
The ferric uptake regulator (Fur) is a homodimeric metalloprotein that functions as a transcriptional regulator of iron homeostasis in many bacteria. When complexed with iron, Fur regulates the transcription of genes by binding a 19-bp inverted repeat sequence (5′-GATAATGATAATCATTATC-3′), known as the Fur box, within the operator/promoter region (Escolar et al., 1999; Baichoo and Helmann, 2002). The Fur protein mainly functions as a repressor, mediating global suppression of iron-responsive genes in response to iron-replete conditions (Xiong et al., 2000; Hantke, 2001; Baichoo et al., 2002). Among the Fur-repressed genes are those involved in iron acquisition; Fur-boxes controlling expression of staphylococcal IRLPs are shown in Figure 1A. Binding of Fur to the Fur box sequences is dependent upon concomitant Fe2+ binding to Fur and, together, the competent DNA-binding complex blocks association of RNA polymerase with promoters, thereby inhibiting transcription. Under iron restriction, intracellular Fe2+ depletion results in dissociation of Fur from DNA, allowing transcription to proceed.
Figure 1.
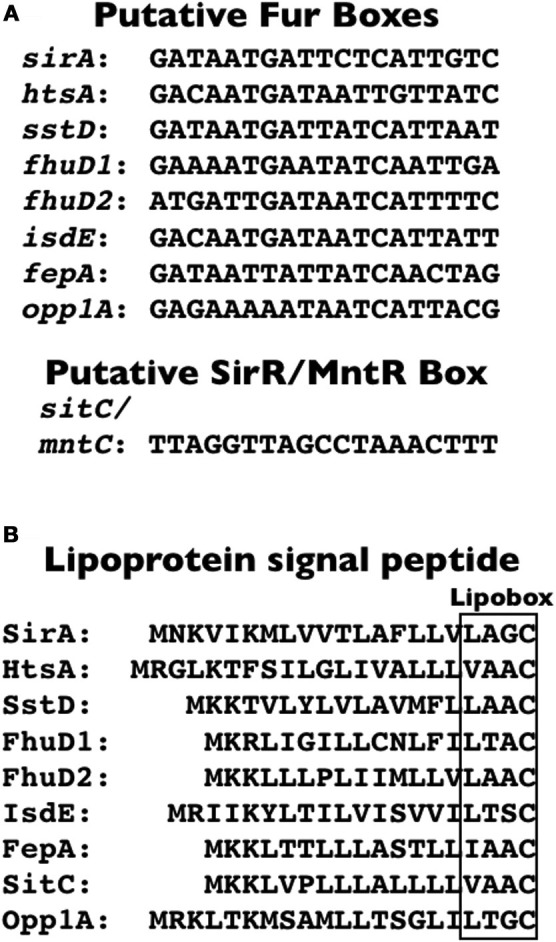
S. aureus Fur boxes, SirR/MntR box, and IRLP signal peptides. (A) Fur box sequences within the promoter/operator region of operons encoding the indicated S. aureus SBP-encoding gene and the SirR/MntR box located upstream of sitC/mntC. (B) Signal peptides for the indicated IRLPs, with the lipobox indicated by the box. Nucleotide and protein sequences are all derived from S. aureus COL.
SirR/MntR
The notable exception to Fur regulation of the IRLPs is the staphylococcal iron transporter, SitABC/MntABC. SitABC and SirR were initially discovered in S. epidermidis (Cockayne et al., 1998), and in Bacillus subtilis where the SitABC and SirR homologues were named MntABCD and MntR, respectively, based on work of Helmann and colleagues in confirming their role in manganese uptake and homeostasis (Que and Helmann, 2000; Glasfeld et al., 2003; Lee and Helmann, 2007). The homologous region in S. aureus was likewise identified and referred to as MntABC and MntR (Horsburgh et al., 2002). Expression of sitABC/mntABC is controlled by the diphtheria toxin repressor (DtxR)-like homologue, SirR/MntR (Hill et al., 1998; Horsburgh et al., 2002; Ando et al., 2003). Transcription of sitABC is repressed by SirR in the presence of either Mn2+ or Fe2+ in S. epidermidis (Hill et al., 1998), although there is some contention over whether both Mn2+ or Fe2+, or just Mn2+ controls expression in S. aureus (Horsburgh et al., 2002; Ando et al., 2003). Like with Fur, transcription is blocked in the presence of these divalent metal ions because the metal ions facilitate the binding of SirR/MntR to a conserved SirR (or MntR) box (Figure 1A), a region of dyad symmetry in the promoter/operator region of sitABC/mntABC (Hill et al., 1998; Ando et al., 2003). Repression of transcription is relieved when the metal ion concentration is depleted.
Lipoprotein biogenesis
A detailed and comprehensive analysis of lipoprotein biogenesis is outside the scope of this review, but we refer the reader to a recent review, and references therein, for an excellent summary of lipoprotein biogenesis in bacterial pathogens (Kovacs-Simon et al., 2011). Bacterial preprolipoproteins bear both an N-terminal signal peptide, characteristic of secreted proteins, and, within the C-region of the signal peptide, a conserved sequence [L/V/I]−3−[A/S/T/V/I]−2−[G/A/S]−1−C+1 which is referred to as the lipobox motif (see Figure 1B for lipobox motif of S. aureus IRLPs) (Kovacs-Simon et al., 2011). In the first biogenesis reaction, the lipoprotein diacylglyceryl transferase (Lgt) covalently links a diacylglycerol moiety to the thiol group on the side chain of the essential +1 cysteine of the lipobox. The subsequent reaction involves the cleavage of the signal peptide from the diacylated prolipoprotein by lipoprotein signal peptidase (Lsp; alternatively called signal peptidase II), an activity that appears contingent upon lipidation by Lgt in Gram-negative bacteria (Tokunaga et al., 1982), but is not a prerequisite in Gram-positive bacteria (Baumgartner et al., 2007). In a third reaction, highly conserved among Gram-negative bacteria and the Actinomycetes, the enzyme lipoprotein N-acyl transferase (Lnt) catalyzes the addition of a third fatty acid to the free amino group of the cysteine, resulting in a triacylated protein. With few exceptions, the lgt, lsp, and lnt genes are indispensible in Gram-negative bacteria, yet in contrast, these genes are dispensable in Gram-positive bacteria (reviewed in Kovacs-Simon et al., 2011).
Given the apparent lack of lnt homologues within Firmicute genomes, it was presumed that lipoproteins within this phylum were diacylated, a notion supported by the identification of diacylated lipoproteins among the staphylococci (Tawaratsumida et al., 2009). In contradistinction, SitC (MntC), the predominant lipoprotein of S. aureus and SBP for SitABC (MntABC), was found to be triacylated in multiple S. aureus strains and in S. epidermidis (Kurokawa et al., 2009; Asanuma et al., 2011). Furthermore, four other major staphylococcal lipoproteins were found to be triacylated (Asanuma et al., 2011), including the same lipoprotein earlier identified to be diacylated by Tawaratsumida et al. (2009). The discrepancies between these studies could be attributed to differences in culture conditions and preparation of the lipoproteins, given that Asanuma et al. noted a minor presence of diacylated SitC (MntC) lipopeptides in cultures grown at elevated temperatures (Asanuma et al., 2011). The notion of global N-acylation among the Firmicutes is further bolstered by a recent report of triacylated lipoproteins among the class Mollicute in Acholeplasma laidlawii, despite the lack of a recognizable Lnt homolog in this bacterium (Serebryakova et al., 2011). Consequently, the nature by, and degree to which, staphylococcal lipoproteins are N-acylated remains unknown, and it is possible that an as-yet unidentified enzyme, with no similarity to Gram-negative Lnt enzymes, exists within the Firmicutes that is responsible for this elusive activity.
The staphylococcal IRLPs: functions in iron acquisition
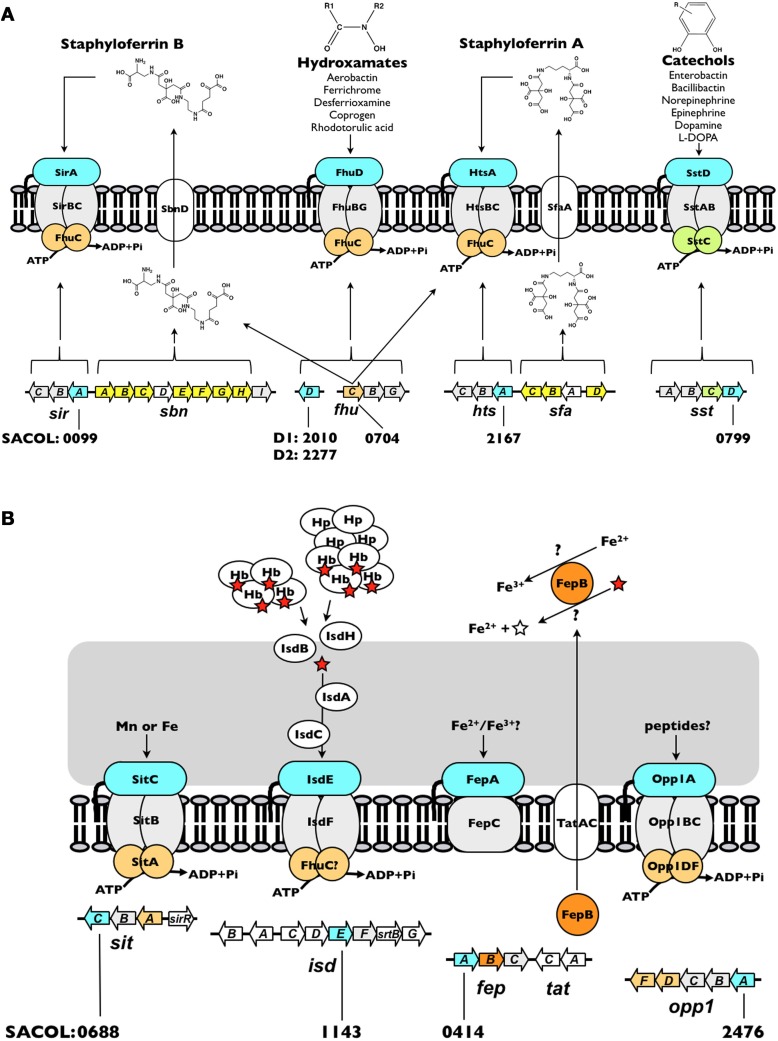
The thus-far identified nine IRLPs in the staphylococci are involved in, or at least implicated in, the uptake of iron through either siderophore (Figure 2A) or non-siderophore (Figure 2B) based systems. Together, these systems engender staphylococci with versatility in that they are able to recognize and utilize a broad range of iron substrates. Unsurprisingly, the common function of these IRLP is reflected in their largely conserved overall protein structure.
Figure 2.
Iron acquisition systems in S. aureus. (A) Siderophore-dependent iron uptake systems. (B) Siderophore-independent iron uptake systems. Each system is discussed in the text. The open reading frame accession number, based on S. aureus COL genome, for each of the IRLPs (blue) is indicated. The directionality indicated for each of the genes is as in the genome. The red star represents heme and the open star represents protoporphyrin IX. Hb, hemoglobin; Hp, haptoglobin.
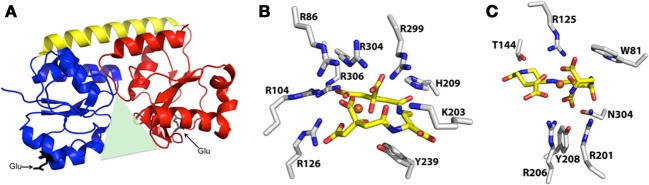
As a result of a recent revisitation of the classification scheme to segregate SBPs based upon structural characteristics (Berntsson et al., 2010), the majority of the staphylococcal IRLPs, with the exception of FepA and Opp1A (discussed below), fall into cluster A, which consists of class III SBPs associated with ABC transporters. These proteins have an approximately 20 residue long α-helical backbone that joins two independently folded domains, each of which consists of central β-strands surrounded by α-helices; substrate is bound into the groove formed between the two domains (Figure 3A). Relative to members of other SBP clusters, hinge motion, upon binding of the substrate, between the two domains of cluster A proteins is restricted by the helical spine. Docking of the cluster A SBPs with their cognate membrane ABC-type permeases is facilitated by salt-bridges formed between conserved glutamic acid residues on the lobes of the SBP (Figure 3A) and patches of positive charge on the exterior surface of the permease, formed by three conserved arginine/lysine residues (Borths et al., 2002).
Figure 3.
Representative structure of cluster A SBPs, and structures of protein-bound Fe-SA and Fe-SB. (A) Representative ribbon diagram to highlight some of the key characteristics of the cluster A SBPs, including the helical spine (yellow) connecting the alpha/beta N-terminal (blue) and C-terminal (red) lobes that surround the binding pocket (green triangle), and the two conserved glutamic acid residues (black) that interact with the membrane permease. (B) Structure of iron-bound staphyloferrin A with coordinating residues from the S. aureus HtsA SBP (PDB 3LI2). (C) Structure of iron-bound staphyloferrin B with coordinating residues from the S. aureus SirA SBP (PDB 3MWF).
HtsA and SirA—receptors for the ferrated staphyloferrin siderophores
Siderophores are secreted, low-molecular-weight molecules that have high-affinity for ferric iron. Among members of the staphylococci, two siderophores may be produced, staphyloferrin A (SA) and staphyloferrin B (SB) (Konetschny-Rapp et al., 1990; Meiwes et al., 1990; Drechsel et al., 1993; Haag et al., 1994). Enzymes for the synthesis of SA and SB are encoded from the sfaABCD and sbnABCDEFGHI loci, respectively (Dale et al., 2004a,b; Beasley et al., 2009; Cheung et al., 2009; Cotton et al., 2009) (Figure 2A). Adjacent to each of these biosynthetic loci, but transcribed separately, are operons encoding the requisite ABC-transporters for uptake of Fe-SA and Fe-SB complexes (Dale et al., 2004a,b; Beasley et al., 2009, 2011; Beasley and Heinrichs, 2010). The transport systems for the two siderophores have been shown to be non-interchangeable, where HtsABC specifically uptakes Fe-SA and SirABC specifically uptakes Fe-SB (Beasley et al., 2009). While each of the transporter-encoding operons codes for a SBP (HtsA and SirA) and a heterodimeric permease component (HtsBC and SirBC), both lack a gene encoding the ATPase component of the ABC transporter. Instead, at least in S. aureus, FhuC, expressed from the fhuCBG operon (Figure 2A) serves as the ATPase for import of both Fe-SA and Fe-SB (Speziali et al., 2006; Beasley et al., 2009). The notion of a common ATPase between multiple iron acquisition pathways is not unprecedented; the YusV ATPase in B. subtilis acts in the uptake of siderophores through both feuABC and yfiYZyfhA (Ollinger et al., 2006). Currently available genomic sequences reveal that while the hts-sfa locus is found in all staphylococcal genomes (incl. S. aureus, S. epidermidis, S. lugdunensis, S. saprophyticus, S. haemolyticus, S. pseudintermedius, S. warneri, S. capitis, S. caprae, S. hominis, S. carnosus and S. xylosus), the sir-sbn locus is only found in the genomes of S. aureus and S. pseudintermedius. Curiously, however, SB was identified in the supernatants of several members of CoNS, and was first identified in coagulase-variable S. hyicus (Konetschny-Rapp et al., 1990; Meiwes et al., 1990; Drechsel et al., 1993); the reason for the discrepancy between these findings and the available genome information remains unknown. Genomes of S. lugdunensis, although possessing htsABC, carry a deletion of sfaA and sfaD, suggesting that this species may utilize Fe-SA as an iron source, but likely does not synthesize either staphyloferrin molecule.
Given that both SA and SB are highly hydrophilic, α-hydroxycarboxylate type siderophores (Beasley and Heinrichs, 2010), highly specific binding to their cognate receptors would be necessary to maintain the aforementioned uptake specificity. Without the specific recognition and binding of Fe-SA and Fe-SB to HtsA and SirA, respectively, one might anticipate that transporters for these structurally similar siderophores would be interchangeable. Recent structural characterizations of both HtsA and SirA have shed significant light on the reason for this specificity. HtsA and SirA both adopt the bilobed, α-helical backboned structure typical of the cluster A SBPs (see above), and their binding pockets are shallow and basic to accommodate the negatively charged siderophores, yet few residues within the substrate binding pocket are conserved between the two lipoproteins (Grigg et al., 2010a,b). HtsA coordinates Fe-SA through H-bonding between six arginine residues, directed into the binding pocket, and oxygen atoms in SA (Figure 3B) (Grigg et al., 2010b). Conserved residue R125 also anchors Fe-SB in SirA (Grigg et al., 2010a), however the remaining coordinating residues are unique (Figure 3C). The number of coordinating arginine residues is a reflection of the net negative charge on the associated siderophore, where up to six are required to neutralize the –5 net charge on SA, and three are required for the –3 net charge of SB. Furthermore, while siderophore binding to HtsA and SirA induces negligible hinge movement along the α-helix, significant and specific conformational changes occur within the C-terminal domain of both (Grigg et al., 2010a,b). The unique coordinating residues, in addition to the distinct conformational changes, likely maintain the specificity and affinity of these receptors for their cognate siderophores, and it is unsurprising that their ligand affinities fall within the nanomolar range (Grigg et al., 2010a,b)
Inactivation of any of the individual biosynthetic or transport loci for SA or SB yields an unimpressive, if any, phenotype when examining for defects in iron-restricted growth (Beasley et al., 2009, 2011). However, the combined inactivation of either sbn together with sfa or sir together with hts yields mutants with severe growth defects in serum (Beasley et al., 2009, 2011). Notably, the inactivation of multiple transport systems in S. aureus results in decreased bacterial fitness during infection, relative to inactivation of the corresponding biosynthetic loci (Beasley et al., 2011), likely due to the continued production of siderophores even in the absence of their cognate receptors, which would enhance iron-starvation in vivo (Beasley et al., 2011). Impairment of receptor function, therefore, represents a potential avenue for therapeutic intervention (see below).
SstD—receptor for ferrated catechols/catecholamines
Catechol siderophores, typified by the well-studied enterobactin (or enterochelin), bacillibactin, vibriobactin and salmochelin, are representative of a major class of bacterial siderophores (for a review, see Miethke and Marahiel, 2007). Additionally, the ability of catecholamine stress hormones to stimulate growth of bacteria, including members of CoNS, in the presence of serum (i.e., transferrin) has been studied over the past decade (Freestone et al., 2000, 2008; Neal et al., 2001). Indeed, recent work illuminated how hormones such as epinephrine and norepinephrine are capable of reducing transferrin-bound ferric iron to ferrous iron and thereby liberating iron from this central component of innate immunity (Sandrini et al., 2010). While catechol siderophores have not been identified in culture supernatants of staphylococci, staphylococci are capable of using both catechols and catecholamines as an iron source via the SstABCD ABC transporter (Beasley et al., 2011). The locus (Figure 2A), initially identified through screens for iron-regulated staphylococcal antigens (Morrissey et al., 2000), is highly conserved among S. aureus strains and present in the majority of CoNS. The function of this transporter remained uncharacterized until recently, when sst mutations were characterized in staphyloferrin-deficient strains (Beasley et al., 2011). The sst locus is required for growth promotion by catechol siderophores and catecholamine-liberated transferrin iron, and contributes to in vivo colonization of the murine liver and kidneys. With the exception of the colonization of murine hearts, these in vivo and in vitro effects were otherwise masked in strains synthesizing the staphyloferrins (Beasley et al., 2011).
SstD specifically binds both ferrated catechol siderophores and catecholamine stress hormones with micromolar and submicromolar affinities (Beasley et al., 2011). The reduced affinity of SstD for its ligands, relative to SirA and HtsA, is likely a trade-off in favor of an enhanced range of potential substrates. The closest homologs to SstD include the YclQ protein from B. subtilis and the CeuE protein from Campylobacter jejuni. Structural information exists for these proteins in complex with their ligands; YclQ binds the “stealth” siderophore petrobactin (Zawadzka et al., 2009), while CeuE binds iron complexed by the enterobactin analog, mecam (Muller et al., 2006). Some conservation in the binding mechanism of these proteins is indicated by shared binding pocket residues between SstD, CeuE, and YclQ, which in CeuE interact with the mecam substrate. Crystallographic information for liganded SstD unfortunately remains lacking, but this information would afford insight into the mechanism by which this receptor binds a diverse range of catechol ligands.
FhuD1 and FhuD2—receptors for ferric-hydroxamate siderophores
While the staphylococci have not been demonstrated to synthesize hydroxamate-type siderophores, they are known to be able to utilize them as iron sources. These include aerobactin (produced by some enterobacteriaceae), ferrichrome (produced by the basidiomycete fungus Ustilago sphaerogena), coprogen (produced by Neurospora crassa), and desferrioxamine B, produced by Streptomyces pilosus; the mesylate salt of desferrioxamine B is used clinically under the name Desferal™ (Brock and Ng, 1983; Sebulsky et al., 2000; Sebulsky and Heinrichs, 2001). Uptake of multiple hydroxamate-type siderophores was similarly demonstrated for B. subtilis (Schneider and Hantke, 1993), and a homologous system identified in the S. aureus (Sebulsky et al., 2000). Ferric-hydroxamate uptake is achieved through the concerted effort of the FhuC ATPase, FhuBG heterodimeric permease, and the independently transcribed substrate-binding lipoproteins, FhuD1 and FhuD2 (Figure 2A) (Sebulsky et al., 2000; Sebulsky and Heinrichs, 2001). All species of staphylococci, with the possible exception of S. hominis and S. xylosus, possess the Fhu uptake system.
While many hydroxamate ligands are common between FhuD1 and FhuD2, the two lipoproteins do possess some unique substrate specificities with FhuD2 exhibiting both a wider range of ligands and greater substrate affinity than FhuD1 (Sebulsky et al., 2003, 2004). While FhuD1 appears to be a less effective Fe3+-siderophore SBP than FhuD2, it is certainly possible that the substrates and/or conditions under which FhuD1 are optimal are not defined. While the structures of FhuD1 and FhuD2 are not yet available, structures have been determined for E. coli FhuD bound to several hydroxamate ligands (Clarke et al., 2000, 2002). In contrast to the binding pocket of proteins such as SirA and HtsA, which harbor key charged residues that interact with the siderophore molecule, the structures of E. coli FhuD show that the binding of ligands is largely dependent upon hydrophobic contacts; accommodation of several different hydroxamate substrates occurs only through subtle rearrangements of the FhuD protein side-chains.
IsdE—a heme receptor
isdE homologues have been identified in several Gram-positive pathogens including S. aureus, S. lugdunensis, Bacillus anthracis, Clostridium tetani, and Listeria monocytogenes (Skaar and Schneewind, 2004; Heilbronner et al., 2011). The iron-regulated surface determinant pathway (Isd) represents the predominant system for iron acquisition from heme and hemoproteins in S. aureus (Mazmanian et al., 2002, 2003). The proposed architectural arrangement of the nine Isd proteins, facilitated by anchoring to the cell wall through sortase A activity (for IsdA, IsdB, and IsdH), and isd locus-encoded sortase B activity (for IsdC), allows for heme, extracted from hemoglobin or haptoglobin-hemoglobin, at the cell surface to be shuttled proximal to the membrane where it is bound by the SBP IsdE, and transported into the cytoplasm through the ABC permease IsdF (Figure 2B) (Mazmanian et al., 2003; Skaar and Schneewind, 2004; Torres et al., 2006; Muryoi et al., 2008; Zhu et al., 2008; Grigg et al., 2010c). Once in the cytoplasm, heme may be degraded by IsdG and IsdI, releasing iron (Skaar et al., 2004a,b; Lee et al., 2008).
As a member of the cluster A SBPs (see above), IsdE possesses the characteristics depicted in Figure 3A but, unlike SirA and HtsA, bears a deep hydrophobic groove between the two lobes comprising a heme binding pocket (Grigg et al., 2007). A single intermediate or high-spin ferric or, preferentially, low-spin ferrous heme molecule is coordinated by the axial ligands H229 and M78 (Grigg et al., 2007; Pluym et al., 2007). As with several of the other iron ABC transporters in the staphylococci, the isdEF genes are not genetically linked with a gene encoding the obligatory ATPase for the transporter. It is possible that the promiscuous FhuC drives transport through IsdEF.
In vitro experiments using purified proteins demonstrated that IsdE could receive heme directly from IsdC (depicted in Figure 2B), as part of a larger study that provided support to the notion that the Isd proteins act together as a heme shuttle system within the bacterial cell wall and membrane (Muryoi et al., 2008). Moreover, growth impairment has been demonstrated for Isd mutants cultured on hemoglobin as a sole iron source (Torres et al., 2006; Pishchany et al., 2009, 2010); subsequent to hemoglobin binding at the cell surface and heme extraction, heme is likely never free from a protein and is shuttled through the wall and membrane via direct Isd-Isd protein contacts. On the contrary, it should be noted, that an isdE mutation had less of an impact when cultured in the presence of heme as a sole iron source (Grigg et al., 2007). The lack of a marked phenotype for the isdE mutant on heme raises the likely possibility that when presented with free heme, a growth-supporting amount of heme makes its way through the cell wall, bypassing the Isd proteins, and is taken into the cell via non-specific mechanisms or by secondary heme transporters, a notion currently under investigation.
FepA—an iron receptor
The translocation of staphylococcal preprolipoproteins was initially thought to occur exclusively through the dominant secretory pathway, Sec (Driessen and Nouwen, 2008; Natale et al., 2008). However, the twin-arginine translocation pathway (Tat) is another means of lipoprotein translocation across the membrane in a number of Gram-positive, high GC bacteria (McDonough et al., 2005; Widdick et al., 2006, 2011; Thompson et al., 2010), and was recently identified in some (incl. S. aureus, S. haemolyticus and S. carnosus), but not all, staphylococcal genomes. Pre-folded proteins bearing a twin arginine motif in their signal peptide are secreted by Tat (Meissner et al., 2007; Biswas et al., 2009), however the role of Tat in the staphylococci appears limited both in the apparent lack of complete Tat homologues in many CoNS, such as S. epidermidis and S. saprophyticus, and in identified substrates (Yamada et al., 2007; Biswas et al., 2009). Currently, the sole identified substrate of Tat is the iron-dependent DyP-family peroxidase, FepB (Biswas et al., 2009).
Dye-decolorizing (DyP)-type peroxidases form a unique heme peroxidase family (for a review, see Sugano, 2009), and members of the family have been reported to have several different substrates (Letoffe et al., 2009; Ahmad et al., 2011; Liers et al., 2011). Insofar as iron uptake is concerned, two Dyp paralogs in E. coli, namely YfeX and EfeB (cytoplasmic and periplasmic enzymes, respectively), were reported to have deferrochelatase activity on heme (Letoffe et al., 2009). Deferrochelation would provide bacteria with an iron source in the absence of dedicated heme uptake systems, however this is currently a subject of some debate. A recent study has argued that the E. coli YfeX protein is not a heme deferrochelatase but rather is a peroxidase that oxidizes porphyrinogens to porphyrins (Dailey et al., 2011). Should DyP-type peroxidases bear deferrochelatase activity, it is possible that, like YfeX and EfeB, staphylococcal FepB could also serve to release ferrous iron from heme.
In addition to FepB, the fepABC operon, located adjacent to tatAC, encodes an iron permease (FepC) and a putative iron-binding lipoprotein (FepA) (Figure 2B). The exact function of S. aureus fepABC in iron acquisition is still under investigation, and the substrate specificity of FepA has not yet been determined. Free ferrous iron released through the potential deferrocheletase activity of FepB could serve as a substrate for FepAC, similar to E. coli ferrous iron-specific efeUOB, which has been shown to be induced by growth in aerobic, low pH and low iron conditions (Grosse et al., 2006; Cao et al., 2007). Alternatively, fepABC may function in elemental iron uptake in a manner analogous to the Ftr1p/Fet3p ferric iron permease in Saccharomyces cerevisiae, where Fe2+ is oxidized to Fe3+ by the multicopper oxidase, Fet3p, and is subsequently transported into the cell by Ftr1p (Kwok et al., 2006). A similar mechanism was first proposed in B. subtilis for ywbLMN, a fepABC homologue, and, accordingly, the ywbLMN operon was shown essential to the normal growth of B. subtilis in iron-restricted media (Ollinger et al., 2006; van der Ploeg et al., 2011). Regardless, the presence of fep operon homologs in many different bacteria, including both Gram-positive and Gram-negative species, suggests a conserved and important functionality. Indeed, S. aureus fep-tat mutants are impaired in inorganic iron uptake and virulence in a murine renal abscess model (Biswas et al., 2009).
Although no structure of a staphylococcal FepA protein has yet been solved, homology searches and structural modeling indicate that it is rather unique among the IRLPs. The closest homologs to FepA are members of the imelysin-like superfamily (Xu et al., 2011). The canonical imelysin fold is all-helical, comprising two similar four-helix bundle domains with a predicted functional site at the domain interface (Xu et al., 2011). Specifically, a conserved GxHxxE motif, also present in staphylococcal FepA proteins, is located at the open end of the binding cleft; this motif has been implicated in the coordination of divalent metal cations (Rajasekaran et al., 2010a,b; Xu et al., 2011). With well over 100 members, the imelysin-like proteins are widely distributed in bacteria and are virtually always located next to a gene encoding a DyP-type peroxidase. Although biochemical evidence is still lacking, this reinforces the assumption that the FepABC-like systems are involved in iron uptake, where the peroxidase potentially converts ferrous iron to ferric iron, which is the ligand for the FepA-like binding proteins.
Opp1A
Despite being annotated as an oligopeptide permease, opp1ABCDF has no defined role in nitrogen metabolism (Hiron et al., 2007). However, Opp1A, the SBP of this complex, was shown to be significantly upregulated during iron-starvation, both by our laboratory (unpublished data) and others (Hempel et al., 2011), consistent with the identification of a putative Fur box upstream of Opp1A (Figure 1A). The role of oligopeptide permeases in metal ion and heme acquisition is not unprecedented; S. aureus Opp2BCDF and the orphan SBP Opp5A were recently renamed NikBCDE and NikA due to their role in nickel acquisition (Hiron et al., 2010), whereas in E. coli a di-peptide permease is involved in the uptake of both nickel and heme (DppA) (Letoffe et al., 2006, 2008; Shepherd et al., 2007). A potential role for Opp1A in iron acquisition is currently under investigation.
The opp1ABCDF operon encodes a prototypical oligopeptide permease: two homologous permease proteins (OppB and OppC) and two ATP-binding proteins (OppD and OppF), in addition to the peptide/SBP (Figure 2B). Like FepA, Opp1A is not a member of the cluster A SBPs, but instead is a member of cluster C which includes oligopeptide, di-peptide and nickel binding proteins (Berntsson et al., 2010). Cluster C members are characterized by a larger size (approx. 50–70 kDa) than the cluster A proteins (which are typically in the 30–35 kDa range), a large binding cavity, an extra domain that augments the binding cavity - in some cases to accommodate larger peptide ligands, and ligand binding via a “Venus flytrap” mechanism (Berntsson et al., 2009, 2010). The latter point indicates that these proteins capture their substrates through significant inter-domain hinge movement, unlike the relative lack of inter-domain movement afforded by the helical spine of the cluster A SBPs (see above).
According to Hiron et al. (Hiron et al., 2007), S. aureus possesses two additional oligopeptide permeases (opp3 and opp4) and one di/tri-peptide permease (dtpT), in addition to opp1 and nikA/nikBCDE. While opp3 and dtpT are involved in nitrogen metabolism (Hiron et al., 2007; Borezee-Durant et al., 2009), the roles of opp1 and opp4 are unknown and none of the lipoproteins have been investigated to determine if, like E. coli NikA, they are capable of binding multiple substrates. Although the role of opp1 is unknown, it has been implicated, along with opp2 and dtpT, in staphylococcal infectivity and survival through signature tagged-mutagenesis studies (Mei et al., 1997; Coulter et al., 1998). The exact mechanism by which these permeases contribute to virulence, however, is not known.
SitC/MntC—receptors for iron or manganese
SitC, the lipoprotein component of SitABC (Figure 2B), was first described as an immunogenic iron-repressible cytoplasmic membrane protein (IRMP) in S. epidermidis (Cockayne et al., 1998). The 32-kDa S. epidermidis SitC protein was found both to be expressed by S. epidermidis and to react strongly with antibodies in pooled human peritoneal dialysate (HPD) (Williams et al., 1988; Smith et al., 1991; Wilcox et al., 1991; Modun et al., 1992). HPD, the byproduct of continuous ambulatory peritoneal dialysis (CAPD) for renal failure, represents a severely iron-restricted environment (Wallaeys et al., 1986; Williams et al., 1988). Expression of SitC by S. epidermidis and S. aureus isolates known to cause peritonitis in CAPD patients highlighted a potential role for SitC in iron acquisition, and consequently in vivo survival of the staphylococci.
Sequence analysis of SitC revealed that it bears homology both to proteins involved in bacterial adhesion as well as to metal binding proteins (Cockayne et al., 1998). Two of the closest structural homologs to SitC are cluster A SBPs: the zinc-binding TroA protein from Treponema pallidum (PDB 1TOA) and the manganese-binding MntC protein from Synechocystis sp. (PDB 1XVL). Given this, and that SitC is lipid tethered to the cell membrane with distribution throughout the cell wall and minimal surface exposure (Smith et al., 1991; Wilcox et al., 1991; Cockayne et al., 1998), it is unlikely to play a role in bacterial adhesion, although involvement in adhesion has not been conclusively disproven. Greater surface exposure would be expected to promote adhesion, which is seen with the adhesins to which SitC was initially likened, but not for SitC itself (Jenkinson, 1992; Sutcliffe et al., 1993; Cockayne et al., 1998). The relatively porous nature of the cell wall, however, would render SitC accessible to metal cations, such as Fe2+, Mn2+ or Zn2+, as well as to antibodies. Additionally, the extracellular release of lipoproteins during iron starvation, such as SitC, would further contribute to immunogenicity (Cockayne et al., 1998). Despite a proposed role in divalent metal uptake, the fact that expression of sitC is repressed in the presence of excess Mn2+ or Fe2+ (Hill et al., 1998), and that SitC is highly expressed in vivo, the substrate of SitC in S. epidermidis remains unknown.
In contrast to S. epidermidis, the sitABC homologue in S. aureus has an identified function in the transport of Mn2+, and was consequently named mntABC (Horsburgh et al., 2002) to reflect this activity, following the nomenclature previously used in B. subtilis (Que and Helmann, 2000). While the presence of Mn2+ facilitates repression of mntABC through the DtxR-family regulator, MntR (SirR) (see regulation section, above), the role of iron in mntABC expression is still unclear (Horsburgh et al., 2002; Ando et al., 2003). Furthermore, the structure and characterization of the substrate binding properties of MntC (SitC) have yet to be elucidated. Regardless, with MntC (SitC) being identified as a predominant lipoprotein in S. aureus and S. epidermidis, and the observation that lipoprotein-deficient mutants are both inhibited in iron-restricted growth and in Toll-like receptor 2 (TLR2) activation in the host, it is not surprising that MntC (SitC) contributes an important role in staphylococcal pathogenesis and survival (Stoll et al., 2005; Bubeck Wardenburg et al., 2006; Schmaler et al., 2009, 2010; Muller et al., 2010).
Roles for IRLPs in immune recognition
The recognition of bacterial invaders by the innate immune system occurs through the identification of pathogen-associated molecular patterns (PAMPs) by Toll-like receptors (TLRs). Of the >10 human TLRs currently identified, TLR2, together with the adapter molecule MyD88, represents the main responsive element to Gram-positive cell wall components and, consequently, staphylococcal infection (Takeuchi et al., 1999, 2000a,b). Until recently, the predominant staphylococcal PAMP for induction of TLR2 and a potent inducer of cytokine release was considered to be lipoteichoic acid (LTA) (Schwandner et al., 1999; Morath et al., 2001, 2005). However, it has been shown that the LTA fractions in these experiments were likely contaminated by lipoproteins, the implied true stimuli for TLR2-mediated inflammation (Hashimoto et al., 2006a,b, 2007). The generation of staphylococcal lgt mutants (see biogenesis section, above) has revealed that lipid modification of prelipoproteins is essential to maximal activation of and subsequent NF-κB-dependent cytokine release via TLR2-MyD88, both in vitro and in vivo (Stoll et al., 2005; Bubeck Wardenburg et al., 2006; Kurokawa et al., 2009; Schmaler et al., 2009). Conversely, an LTA-depleted mutant was still capable of inducing TLR2 (Kurokawa et al., 2009). Furthermore, highly purified SitC (MntC) co-localized with and induced TLR2 (Kurokawa et al., 2009; Muller et al., 2010), showing direct evidence of IRLP recognition by the host immune system.
In vivo studies have revealed a paradox in the expression of lipoproteins during infection. Of particular note, sublethal challenge of mice with an lgt mutant resulted in increased mortality relative to the wild-type infection, suggesting that lipoproteins are required to initiate the host innate immune response (Bubeck Wardenburg et al., 2006; Schmaler et al., 2009, 2010). Conversely, the expression of lipoproteins increases bacterial burden during persistent tissue infections, shown to be due to their role in enhancing staphylococcal survival through iron acquisition (Schmaler et al., 2009, 2010). While SitC (MntC) represents the predominant iron-regulated staphylococcal lipoprotein, as previously discussed, S. aureus lacking SitC (MntC) can still stimulate TLR2 (Kurokawa et al., 2009). Given that staphylococcal lipoproteins involved in the acquisition of iron are highly upregulated during iron starvation and expressed in vivo and in vitro (Morrissey et al., 2000; Sebulsky and Heinrichs, 2001; Dale et al., 2004a,b; Skaar et al., 2004a,b; Allard et al., 2006; Hempel et al., 2011), they together likely serve as important PAMPs for TLR2 recognition. Together these results suggest that the elaboration of multiple and sometimes redundant IRLPs may ensure staphylococcal survival in an otherwise inflamed host, providing an intriguing option for the development of potential therapeutics.
Clinical applications
Given their cell surface exposure, propensity to be expressed in vivo and corresponding immunogenicity, the IRLPs represent a tangible target for staphylococcal vaccine development. Indeed, both Sanofi-Pasteur/Syntiron and Novartis are including IRLPs in pre-clinical multivalent vaccine preparations. A discussion of S. aureus vaccine attempts is beyond the scope of this review, so we refer the reader to several excellent, recent reviews on the subject (Otto, 2010; Daum and Spellberg, 2012; DeDent et al., 2012; Patti, 2011; Proctor, 2011). The focus of published passive and active immunization efforts targeting iron acquisition systems thus far have centered on the Isd cell wall anchored proteins IsdA, IsdB, and IsdH, with promising results in animal models of S. aureus infection (Clarke et al., 2006; Kuklin et al., 2006; Ebert et al., 2010; Kim et al., 2010; Ster et al., 2010; Arlian and Tinker, 2011; Daum and Spellberg, 2012; DeDent et al., 2012; Harro et al., 2012). Of the Isd proteins, Merck and Intercell took their single-antigen IsdB vaccine, V710 (Kuklin et al., 2006), through phase I clinical trials, demonstrating that the vaccine was immunogenic within 2 weeks of administration and had a good safety profile in humans (Harro et al., 2010, 2012). Phase II/III clinical trials were terminated following a statistical review of the data suggesting that V710 was unlikely to demonstrate a significant benefit to patients (Patti, 2011). Many experts in the field are now suggesting that as opposed to a single-antigen vaccine, multivalent strategies, at least in humans, offer the greatest chance for success. In support of this recommendation, IsdA and IsdB, when combined with two additional S. aureus surface antigens, offered greater protective immunity, at least in rodent models, than when the animals were immunized with the individual antigens alone (Stranger-Jones et al., 2006).
In addition to the challenges in mounting an antibody response against the staphylococci (Foster, 2005), the functional redundancy (in terms of supplying a vital iron source to the bacteria) and strain variation in iron-regulated proteins suggests multiple systems should be targeted through a combinatorial vaccine. Indeed, the reduced fitness of sir hts sst mutants relative to single or double mutants recommends targeting multiple iron acquisition lipoproteins to inhibit growth (Beasley et al., 2011). Moreover, effective antibody-based approaches may require antibodies that inhibit protein function, in addition to, or in lieu of, being opsonic. Certainly, protective anti-IsdA and anti-IsdB antibodies appear to function by inhibiting heme acquisition (Kim et al., 2010). Thus, puzzling out the substrate specificities, expression patterns, presence/absence in different strains or species, and relevance, alone or in combination, to in vivo growth/infectivity is paramount toward effective use of the IRLPs as therapeutic targets. Aside of vaccine strategies, other avenues do exist for taking advantage of iron acquisition systems for therapeutic intervention. It is possible that knowledge on the substrate specificities, along with detailed structural information, will lead to the use of the uptake systems as portals for toxic “trojan horse” compounds that kill or limit growth of the staphylococci.
Concluding remarks
IRLPs play an essential role in the acquisition of iron, and consequently the in vivo survival of the staphylococci. Paradoxically, these lipoproteins are also strongly immunogenic, inducing an inflammatory response through recognition by TLR2-MyD88. A balance, therefore, exists between the surface display of multiple IRLPs and evading immune recognition by the host. Both of these characteristics make IRLPs a good choice of targets for vaccine development, and the formulation of a combinatorial vaccine targeting multiple iron uptake systems has been suggested to maximize efficacy. Further research is required to identify the source of the activity responsible for the N-acylation of staphylococcal lipoproteins. More work should also focus on obtaining high-resolution structural information on each of the proteins discussed here. As iron represents an essential element to survival, the IRLPs remain at the interface of pathogenesis and potential therapeutic control of the often pathogenic staphylococci.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Jessica R. Sheldon is the recipient of a Canadian Institutes for Health Research Frederick Banting and Charles Best Doctoral Research Award (CIHR-DRA). Work in the authors' laboratory is supported by operating grants (to David E. Heinrichs) from the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada.
References
- Ahmad M., Roberts J. N., Hardiman E. M., Singh R., Eltis L. D., Bugg T. D. (2011). Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 50, 5096–5107 10.1021/bi101892z [DOI] [PubMed] [Google Scholar]
- Allard M., Moisan H., Brouillette E., Gervais A. L., Jacques M., Lacasse P., Diarra M. S., Malouin F. (2006). Transcriptional modulation of some Staphylococcus aureus iron-regulated genes during growth in vitro and in a tissue cage model in vivo. Microbes Infect. 8, 1679–1690 [DOI] [PubMed] [Google Scholar]
- Ando M., Manabe Y. C., Converse P. J., Miyazaki E., Harrison R., Murphy J. R., Bishai W. R. (2003). Characterization of the role of the divalent metal ion-dependent transcriptional repressor MntR in the virulence of Staphylococcus aureus. Infect. Immun. 71, 2584–2590 10.1128/IAI.71.5.2584-2590.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlian B. M., Tinker J. K. (2011). Mucosal immunization with a Staphylococcus aureus IsdA-cholera toxin A2/B chimera induces antigen-specific Th2-type responses in mice. Clin. Vaccine Immunol. 18, 1543–1551 10.1128/CVI.05146-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asanuma M., Kurokawa K., Ichikawa R., Ryu K. H., Chae J. H., Dohmae N., Lee B. L., Nakayama H. (2011). Structural evidence of alpha-aminoacylated lipoproteins of Staphylococcus aureus. FEBS J. 278, 716–728 10.1111/j.1742-4658.2010.07990.x [DOI] [PubMed] [Google Scholar]
- Baichoo N., Helmann J. D. (2002). Recognition of DNA by fur: a reinterpretation of the fur box consensus sequence. J. Bacteriol. 184, 5826–5832 10.1128/JB.184.21.5826-5832.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baichoo N., Wang T., Ye R., Helmann J. D. (2002). Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45, 1613–1629 10.1046/j.1365-2958.2002.03113.x [DOI] [PubMed] [Google Scholar]
- Baumgartner M., Karst U., Gerstel B., Loessner M., Wehland J., Jansch L. (2007). Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes. J. Bacteriol. 189, 313–324 10.1128/JB.00976-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley F. C., Heinrichs D. E. (2010). Siderophore-mediated iron acquisition in the staphylococci. J. Inorg. Biochem. 104, 282–288 10.1016/j.jinorgbio.2009.09.011 [DOI] [PubMed] [Google Scholar]
- Beasley F. C., Marolda C. L., Cheung J., Buac S., Heinrichs D. E. (2011). Staphylococcus aureus Transporters Hts, Sir, and Sst capture iron liberated from human transferrin by staphyloferrin A, staphyloferrin B, and catecholamine stress hormones, respectively, and contribute to virulence. Infect. Immun. 79, 2345–2355 10.1128/IAI.00117-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley F. C., Vines E. D., Grigg J. C., Zheng Q., Liu S., Lajoie G. A., Murphy M. E., Heinrichs D. E. (2009). Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol. Microbiol. 72, 947–963 10.1111/j.1365-2958.2009.06698.x [DOI] [PubMed] [Google Scholar]
- Berntsson R. P., Doeven M. K., Fusetti F., Duurkens R. H., Sengupta D., Marrink S. J., Thunnissen A. M., Poolman B., Slotboom D. J. (2009). The structural basis for peptide selection by the transport receptor OppA. EMBO J. 28, 1332–1340 10.1038/emboj.2009.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berntsson R. P., Smits S. H., Schmitt L., Slotboom D. J., Poolman B. (2010). A structural classification of substrate-binding proteins. FEBS Lett. 584, 2606–2617 10.1016/j.febslet.2010.04.043 [DOI] [PubMed] [Google Scholar]
- Biswas L., Biswas R., Nerz C., Ohlsen K., Schlag M., Schafer T., Lamkemeyer T., Ziebandt A. K., Hantke K., Rosenstein R., Gotz F. (2009). Role of the twin-arginine translocation pathway in Staphylococcus. J. Bacteriol. 191, 5921–5929 10.1128/JB.00642-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borezee-Durant E., Hiron A., Piard J. C., Juillard V. (2009). Dual role of the oligopeptide permease Opp3 during growth of Staphylococcus aureus in milk. Appl. Environ. Microbiol. 75, 3355–3357 10.1128/AEM.02819-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borths E. L., Locher K. P., Lee A. T., Rees D. C. (2002). The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter. Proc. Natl. Acad. Sci. U.S.A. 99, 16642–16647 10.1073/pnas.262659699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock J. H., Ng J. (1983). The effect of desferrioxamine on the growth of Staphylococcus aureus, Yersinia enterocolitica, and Streptococcus faecalis in human serum. Uptake of desferrioxamine-bound iron. FEMS Microbiol. Lett. 20, 439–442 [Google Scholar]
- Bubeck Wardenburg J., Williams W. A., Missiakas D. (2006). Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc. Natl. Acad. Sci. U.S.A. 103, 13831–13836 10.1073/pnas.0603072103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Woodhall M. R., Alvarez J., Cartron M. L., Andrews S. C. (2007). EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol. Microbiol. 65, 857–875 10.1111/j.1365-2958.2007.05802.x [DOI] [PubMed] [Google Scholar]
- Cheung J., Beasley F. C., Liu S., Lajoie G. A., Heinrichs D. E. (2009). Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol. Microbiol. 74, 594–608 10.1111/j.1365-2958.2009.06880.x [DOI] [PubMed] [Google Scholar]
- Clarke S. R., Brummell K. J., Horsburgh M. J., McDowell P. W., Mohamad S. A., Stapleton M. R., Acevedo J., Read R. C., Day N. P., Peacock S. J., Mond J. J., Kokai-Kun J. F., Foster S. J. (2006). Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J. Infect. Dis. 193, 1098–1108 10.1086/501471 [DOI] [PubMed] [Google Scholar]
- Clarke T. E., Braun V., Winkelmann G., Tari L. W., Vogel H. J. (2002). X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin. J. Biol. Chem. 277, 13966–13972 10.1074/jbc.M109385200 [DOI] [PubMed] [Google Scholar]
- Clarke T. E., Ku S. Y., Dougan D. R., Vogel H. J., Tari L. W. (2000). The structure of the ferric siderophore binding protein FhuD complexed with gallichrome. Nat. Struct. Biol. 7, 287–291 10.1038/74048 [DOI] [PubMed] [Google Scholar]
- Cockayne A., Hill P. J., Powell N. B., Bishop K., Sims C., Williams P. (1998). Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect. Immun. 66, 3767–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton J. L., Tao J., Balibar C. J. (2009). Identification and characterization of the Staphylococcus aureus gene cluster coding for staphyloferrin A. Biochemistry 48, 1025–1035 10.1021/bi801844c [DOI] [PubMed] [Google Scholar]
- Coulter S. N., Schwan W. R., Ng E. Y., Langhorne M. H., Ritchie H. D., Westbrock-Wadman S., Hufnagle W. O., Folger K. R., Bayer A. S., Stover C. K. (1998). Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol. Microbiol. 30, 393–404 10.1046/j.1365-2958.1998.01075.x [DOI] [PubMed] [Google Scholar]
- Dailey H. A., Septer A. N., Daugherty L., Thames D., Gerdes S., Stabb E. V., Dunn A. K., Dailey T. A., Phillips J. D. (2011). The Escherichia coli protein YfeX functions as a porphyrinogen oxidase, not a heme dechelatase. mBio 2, e00248–e00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale S. E., Doherty-Kirby A., Lajoie G., Heinrichs D. E. (2004a). Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect. Immun. 72, 29–37 10.1128/IAI.72.1.29-37.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale S. E., Sebulsky M. T., Heinrichs D. E. (2004b). Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J. Bacteriol. 186, 8356–8362 10.1128/JB.186.24.8356-8362.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum R. S., Spellberg B. (2012). Progress toward a Staphylococcus aureus vaccine. Clin. Infect. Dis. 54, 560–567 10.1093/cid/cir828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A. L., Dassa E., Orelle C., Chen J. (2008). Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364 10.1128/MMBR.00031-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDent A., Kim H. K., Missiakas D., Schneewind O. (2012). Exploring Staphylococcus aureus pathways to disease for vaccine development. Semin. Immunopathol. 34, 317–333 10.1007/s00281-011-0299-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel H., Freund S., Nicholson G., Haag H., Jung O., Zahner H., Jung G. (1993). Purification and chemical characterization of staphyloferrin B, a hydrophilic siderophore from staphylococci. Biometals 6, 185–192 [DOI] [PubMed] [Google Scholar]
- Driessen A. J., Nouwen N. (2008). Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667 10.1146/annurev.biochem.77.061606.160747 [DOI] [PubMed] [Google Scholar]
- Ebert T., Smith S., Pancari G., Clark D., Hampton R., Secore S., Towne V., Fan H., Wang X. M., Wu X., Ernst R., Harvey B. R., Finnefrock A. C., Wang F., Tan C., Durr E., Cope L., Anderson A., An Z., McNeely T. (2010). A fully human monoclonal antibody to Staphylococcus aureus iron regulated surface determinant B (IsdB) with functional activity in vitro and in vivo. Hum. Antibodies 19, 113–128 10.3233/HAB-2010-0235 [DOI] [PubMed] [Google Scholar]
- Escolar L., Perez-Martin J., De Lorenzo V. (1999). Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181, 6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J. (2005). Immune evasion by staphylococci. Nat. Rev. Microbiol. 3, 948–958 10.1038/nrmicro1289 [DOI] [PubMed] [Google Scholar]
- Freestone P. P., Lyte M., Neal C. P., Maggs A. F., Haigh R. D., Williams P. H. (2000). The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J. Bacteriol. 182, 6091–6098 10.1128/JB.182.21.6091-6098.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freestone P. P., Sandrini S. M., Haigh R. D., Lyte M. (2008). Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol. 16, 55–64 10.1016/j.tim.2007.11.005 [DOI] [PubMed] [Google Scholar]
- Glasfeld A., Guedon E., Helmann J. D., Brennan R. G. (2003). Structure of the manganese-bound manganese transport regulator of Bacillus subtilis. Nat. Struct. Biol. 10, 652–657 10.1038/nsb951 [DOI] [PubMed] [Google Scholar]
- Grigg J. C., Cheung J., Heinrichs D. E., Murphy M. E. (2010a). Specificity of Staphyloferrin B recognition by the SirA receptor from Staphylococcus aureus. J. Biol. Chem. 285, 34579–34588 10.1074/jbc.M110.172924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg J. C., Cooper J. D., Cheung J., Heinrichs D. E., Murphy M. E. (2010b). The Staphylococcus aureus siderophore receptor HtsA undergoes localized conformational changes to enclose staphyloferrin A in an arginine-rich binding pocket. J. Biol. Chem. 285, 11162–11171 10.1074/jbc.M109.097865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg J. C., Ukpabi G., Gaudin C. F., Murphy M. E. (2010c). Structural biology of heme binding in the Staphylococcus aureus Isd system. J. Inorg. Biochem. 104, 341–348 10.1016/j.jinorgbio.2009.09.012 [DOI] [PubMed] [Google Scholar]
- Grigg J. C., Vermeiren C. L., Heinrichs D. E., Murphy M. E. (2007). Heme coordination by Staphylococcus aureus IsdE. J. Biol. Chem. 282, 28815–28822 10.1074/jbc.M704602200 [DOI] [PubMed] [Google Scholar]
- Grosse C., Scherer J., Koch D., Otto M., Taudte N., Grass G. (2006). A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol. Microbiol. 62, 120–131 10.1111/j.1365-2958.2006.05326.x [DOI] [PubMed] [Google Scholar]
- Haag H., Fiedler H. P., Meiwes J., Drechsel H., Jung G., Zahner H. (1994). Isolation and biological characterization of staphyloferrin B, a compound with siderophore activity from staphylococci. FEMS Microbiol. Lett. 115, 125–130 [DOI] [PubMed] [Google Scholar]
- Haley K. P., Skaar E. P. (2012). A battle for iron: host sequestration and Staphylococcus aureus acquisition. Microbes. Infect. 14, 217–227 10.1016/j.micinf.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer N. D., Skaar E. P. (2011). Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 65, 129–147 10.1146/annurev-micro-090110-102851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. (2001). Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4, 172–177 10.1016/S1369-5274(00)00184-3 [DOI] [PubMed] [Google Scholar]
- Harro C., Betts R., Orenstein W., Kwak E. J., Greenberg H. E., Onorato M. T., Hartzel J., Lipka J., Dinubile M. J., Kartsonis N. (2010). Safety and immunogenicity of a novel Staphylococcus aureus vaccine: results from the first study of the vaccine dose range in humans. Clin. Vaccine Immunol. 17, 1868–1874 10.1128/CVI.00356-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harro C. D., Betts R. F., Hartzel J. S., Onorato M. T., Lipka J., Smugar S. S., Kartsonis N. A. (2012). The immunogenicity and safety of different formulations of a novel Staphylococcus aureus vaccine (V710): results of two Phase I studies. Vaccine 30, 1729–1736 10.1016/j.vaccine.2011.12.045 [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Furuyashiki M., Kaseya R., Fukada Y., Akimaru M., Aoyama K., Okuno T., Tamura T., Kirikae T., Kirikae F., Eiraku N., Morioka H., Fujimoto Y., Fukase K., Takashige K., Moriya Y., Kusumoto S., Suda Y. (2007). Evidence of immunostimulating lipoprotein existing in the natural lipoteichoic acid fraction. Infect. Immun. 75, 1926–1932 10.1128/IAI.02083-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Tawaratsumida K., Kariya H., Aoyama K., Tamura T., Suda Y. (2006a). Lipoprotein is a predominant Toll-like receptor 2 ligand in Staphylococcus aureus cell wall components. Int. Immunol. 18, 355–362 10.1093/intimm/dxh374 [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Tawaratsumida K., Kariya H., Kiyohara A., Suda Y., Krikae F., Kirikae T., Gotz F. (2006b). Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J. immunol. 177, 3162–3169 [DOI] [PubMed] [Google Scholar]
- Heilbronner S., Holden M. T., Van Tonder A., Geoghegan J. A., Foster T. J., Parkhill J., Bentley S. D. (2011). Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol. Lett. 322, 60–67 10.1111/j.1574-6968.2011.02339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel K., Herbst F. A., Moche M., Hecker M., Becher D. (2011). Quantitative proteomic view on secreted, cell surface-associated, and cytoplasmic proteins of the methicillin-resistant human pathogen Staphylococcus aureus under iron-limited conditions. J. Proteome Res. 10, 1657–1666 10.1021/pr1009838 [DOI] [PubMed] [Google Scholar]
- Hill P. J., Cockayne A., Landers P., Morrissey J. A., Sims C. M., Williams P. (1998). SirR, a novel iron-dependent repressor in Staphylococcus epidermidis. Infect. Immun. 66, 4123–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiron A., Borezee-Durant E., Piard J. C., Juillard V. (2007). Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 189, 5119–5129 10.1128/JB.00274-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiron A., Posteraro B., Carriere M., Remy L., Delporte C., La Sorda M., Sanguinetti M., Juillard V., Borezee-Durant E. (2010). A nickel ABC-transporter of Staphylococcus aureus is involved in urinary tract infection. Mol. Microbiol. 77, 1246–1260 10.1111/j.1365-2958.2010.07287.x [DOI] [PubMed] [Google Scholar]
- Horsburgh M. J., Wharton S. J., Cox A. G., Ingham E., Peacock S., Foster S. J. (2002). MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44, 1269–1286 10.1046/j.1365-2958.2002.02944.x [DOI] [PubMed] [Google Scholar]
- Hutchings M. I., Palmer T., Harrington D. J., Sutcliffe I. C. (2009). Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold ‘em, knowing when to fold ‘em. Trends Microbiol. 17, 13–21 10.1016/j.tim.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Jenkinson H. F. (1992). Adherence, coaggregation, and hydrophobicity of Streptococcus gordonii associated with expression of cell surface lipoproteins. Infect. Immun. 60, 1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K., DeDent A., Cheng A. G., McAdow M., Bagnoli F., Missiakas D. M., Schneewind O. (2010). IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine 28, 6382–6392 10.1016/j.vaccine.2010.02.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos W. E., Bannerman T. L. (1994). Update on clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev. 7, 117–140 10.1128/CMR.7.1.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konetschny-Rapp S., Jung G., Meiwes J., Zahner H. (1990). Staphyloferrin A: a structurally new siderophore from staphylococci. Eur. J. Biochem. 191, 65–74 10.1111/j.1432-1033.1990.tb19094.x [DOI] [PubMed] [Google Scholar]
- Kovacs-Simon A., Titball R. W., Michell S. L. (2011). Lipoproteins of bacterial pathogens. Infect. Immun. 79, 548–561 10.1128/IAI.00682-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuklin N. A., Clark D. J., Secore S., Cook J., Cope L. D., McNeely T., Noble L., Brown M. J., Zorman J. K., Wang X. M., Pancari G., Fan H., Isett K., Burgess B., Bryan J., Brownlow M., George H., Meinz M., Liddell M. E., Kelly R., Schultz L., Montgomery D., Onishi J., Losada M., Martin M., Ebert T., Tan C. Y., Schofield T. L., Nagy E., Meineke A., Joyce J. G., Kurtz M. B., Caulfield M. J., Jansen K. U., McClements W., Anderson A. S. (2006). A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect. Immun. 74, 2215–2223 10.1128/IAI.74.4.2215-2223.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K., Lee H., Roh K. B., Asanuma M., Kim Y. S., Nakayama H., Shiratsuchi A., Choi Y., Takeuchi O., Kang H. J., Dohmae N., Nakanishi Y., Akira S., Sekimizu K., Lee B. L. (2009). The triacylated ATP binding cluster transporter substrate-binding lipoprotein of Staphylococcus aureus functions as a native ligand for toll-like receptor 2. J. Biol. Chem. 284, 8406–8411 10.1074/jbc.M809618200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok E. Y., Severance S., Kosman D. J. (2006). Evidence for iron channeling in the Fet3p-Ftr1p high-affinity iron uptake complex in the yeast plasma membrane. Biochemistry 45, 6317–6327 10.1021/bi052173c [DOI] [PubMed] [Google Scholar]
- Lee J. W., Helmann J. D. (2007). Functional specialization within the Fur family of metalloregulators. Biometals 20, 485–499 10.1007/s10534-006-9070-7 [DOI] [PubMed] [Google Scholar]
- Lee W. C., Reniere M. L., Skaar E. P., Murphy M. E. (2008). Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J. Biol. Chem. 283, 30957–30963 10.1074/jbc.M709486200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letoffe S., Delepelaire P., Wandersman C. (2006). The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 12891–12896 10.1073/pnas.0605440103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letoffe S., Delepelaire P., Wandersman C. (2008). Functional differences between heme permeases: Serratia marcescens HemTUV permease exhibits a narrower substrate specificity (restricted to heme) than the Escherichia coli DppABCDF peptide-heme permease. J. Bacteriol. 190, 1866–1870 10.1128/JB.01636-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letoffe S., Heuck G., Delepelaire P., Lange N., Wandersman C. (2009). Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proc. Natl. Acad. Sci. U.S.A. 106, 11719–11724 10.1073/pnas.0903842106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liers C., Ullrich R., Hofrichter M., Minibayeva F. V., Beckett R. P. (2011). A heme peroxidase of the ascomyceteous lichen Leptogium saturninum oxidizes high-redox potential substrates. Fungal. Genet. Biol. 48, 1139–1145 10.1016/j.fgb.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K., Skaar E. P., Gaspar A. H., Humayun M., Gornicki P., Jelenska J., Joachmiak A., Missiakas D. M., Schneewind O. (2003). Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299, 906–909 10.1126/science.1081147 [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K., Ton-That H., Su K., Schneewind O. (2002). An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 2293–2298 10.1073/pnas.032523999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough J. A., Hacker K. E., Flores A. R., Pavelka M. S. Jr., Braunstein M. (2005). The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J. Bacteriol. 187, 7667–7679 10.1128/JB.187.22.7667-7679.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J.-M., Nourbakhsh F., Ford C. W., Holden D. W. (1997). Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26, 399–407 10.1046/j.1365-2958.1997.5911966.x [DOI] [PubMed] [Google Scholar]
- Meissner D., Vollstedt A., Van Dijl J. M., Freudl R. (2007). Comparative analysis of twin-arginine (Tat)-dependent protein secretion of a heterologous model protein (GFP) in three different Gram-positive bacteria. Appl. Microbiol. Biotechnol. 76, 633–642 10.1007/s00253-007-0934-8 [DOI] [PubMed] [Google Scholar]
- Meiwes J., Fiedler H.-P., Haag H., Zähner H., Konetschny-Rapp S., Jung G. (1990). Isolation and characterization of staphyloferrin A, a compound with siderophore activity from Staphylococcus hyicus DSM 20459. FEMS Microbiol. Lett. 67, 201–206 [DOI] [PubMed] [Google Scholar]
- Modun B., Williams P., Pike W. J., Cockayne A., Arbuthnott J. P., Finch R., Denyer S. P. (1992). Cell envelope proteins of Staphylococcus epidermidis grown in vivo in a peritoneal chamber implant. Infect. Immun. 60, 2551–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke M., Marahiel M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451 10.1128/MMBR.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moet G. J., Jones R. N., Biedenbach D. J., Stilwell M. G., Fritsche T. R. (2007). Contemporary causes of skin and soft tissue infections in North America, Latin America, and Europe: report from the SENTRY Antimicrobial Surveillance Program (1998–2004). Diagn. Microbiol. Infect. Dis. 57, 7–13 10.1016/j.diagmicrobio.2006.05.009 [DOI] [PubMed] [Google Scholar]
- Morath S., Geyer A., Hartung T. (2001). Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J. Exp. Med. 193, 393–397 10.1084/jem.193.3.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morath S., Von Aulock S., Hartung T. (2005). Structure/function relationships of lipoteichoic acids. J. Endotoxin. Res. 11, 348–356 10.1179/096805105X67328 [DOI] [PubMed] [Google Scholar]
- Morrissey J. A., Cockayne A., Hill P. J., Williams P. (2000). Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect. Immun. 68, 6281–6288 10.1128/IAI.68.11.6281-6288.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A., Wilkinson A. J., Wilson K. S., Duhme-Klair A. K. (2006). An [{Fe(mecam)}2]6-bridge in the crystal structure of a ferric enterobactin binding protein. Angew. Chem. Int. Ed. Engl. 45, 5132–5136 10.1002/anie.200601198 [DOI] [PubMed] [Google Scholar]
- Muller P., Muller-Anstett M., Wagener J., Gao Q., Kaesler S., Schaller M., Biedermann T., Gotz F. (2010). The Staphylococcus aureus lipoprotein SitC colocalizes with Toll-like receptor 2 (TLR2) in murine keratinocytes and elicits intracellular TLR2 accumulation. Infect. Immun. 78, 4243–4250 10.1128/IAI.00538-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muryoi N., Tiedemann M. T., Pluym M., Cheung J., Heinrichs D. E., Stillman M. J. (2008). Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J. Biol. Chem. 283, 28125–28136 10.1074/jbc.M802171200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale P., Bruser T., Driessen A. J. (2008). Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane–distinct translocases and mechanisms. Biochim. Biophys. Acta 1778, 1735–1756 10.1016/j.bbamem.2007.07.015 [DOI] [PubMed] [Google Scholar]
- Neal C. P., Freestone P. P., Maggs A. F., Haigh R. D., Williams P. H., Lyte M. (2001). Catecholamine inotropes as growth factors for Staphylococcus epidermidis and other coagulase-negative staphylococci. FEMS Microbiol. Lett. 194, 163–169 [DOI] [PubMed] [Google Scholar]
- Ollinger J., Song K. B., Antelmann H., Hecker M., Helmann J. D. (2006). Role of the Fur regulon in iron transport in Bacillus subtilis. J. Bacteriol. 188, 3664–3673 10.1128/JB.188.10.3664-3673.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2010). Novel targeted immunotherapy approaches for staphylococcal infection. Expert. Opin. Biol. Ther. 10, 1049–1059 10.1517/14712598.2010.495115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2012). Molecular basis of Staphylococcus epidermidis infections. Semin. Immunopathol. 34, 201–214 10.1007/s00281-011-0296-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti J. M. (2011). Will we ever see the approval of a Staphylococcus aureus vaccine? Expert Rev. Anti Infect. Ther. 9, 845–846 10.1586/eri.11.99 [DOI] [PubMed] [Google Scholar]
- Pishchany G., Dickey S. E., Skaar E. P. (2009). Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infect. Immun. 77, 2624–2634 10.1128/IAI.01531-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishchany G., McCoy A. L., Torres V. J., Krause J. C., Crowe J. E. Jr., Fabry M. E., Skaar E. P. (2010). Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe 8, 544–550 10.1016/j.chom.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluym M., Vermeiren C. L., Mack J., Heinrichs D. E., Stillman M. J. (2007). Heme binding properties of Staphylococcus aureus IsdE. Biochemistry 46, 12777–12787 10.1021/bi7009585 [DOI] [PubMed] [Google Scholar]
- Proctor R. (2011). Is there a future for a Staphylococcus aureus vaccine? Vaccine 21 [Epub ahead of print]. 10.1016/j.vaccine.2011.11.006 [DOI] [PubMed] [Google Scholar]
- Que Q., Helmann J. D. (2000). Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35, 1454–1468 10.1046/j.1365-2958.2000.01811.x [DOI] [PubMed] [Google Scholar]
- Rajasekaran M. B., Mitchell S. A., Gibson T. M., Hussain R., Siligardi G., Andrews S. C., Watson K. A. (2010a). Isolation and characterisation of EfeM, a periplasmic component of the putative EfeUOBM iron transporter of Pseudomonas syringae pv. syringae. Biochem. Biophys. Res. Commun. 398, 366–371 10.1016/j.bbrc.2010.06.072 [DOI] [PubMed] [Google Scholar]
- Rajasekaran M. B., Nilapwar S., Andrews S. C., Watson K. A. (2010b). EfeO-cupredoxins: major new members of the cupredoxin superfamily with roles in bacterial iron transport. Biometals 23, 1–17 10.1007/s10534-009-9262-z [DOI] [PubMed] [Google Scholar]
- Ratledge C. (2007). Iron metabolism and infection. Food Nutr. Bull. 28, S515–S523 [DOI] [PubMed] [Google Scholar]
- Ratledge C., Dover L. G. (2000). Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54, 881–941 10.1146/annurev.micro.54.1.881 [DOI] [PubMed] [Google Scholar]
- Sandrini S. M., Shergill R., Woodward J., Muralikuttan R., Haigh R. D., Lyte M., Freestone P. P. (2010). Elucidation of the mechanism by which catecholamine stress hormones liberate iron from the innate immune defense proteins transferrin and lactoferrin. J. Bacteriol. 192, 587–594 10.1128/JB.01028-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible U. E., Kaufmann S. H. (2004). Iron and microbial infection. Nat. Rev. Microbiol. 2, 946–953 10.1038/nrmicro1046 [DOI] [PubMed] [Google Scholar]
- Schmaler M., Jann N. J., Ferracin F., Landolt L. Z., Biswas L., Gotz F., Landmann R. (2009). Lipoproteins in Staphylococcus aureus mediate inflammation by TLR2 and iron-dependent growth in vivo. J. Immunol. 182, 7110–7118 10.4049/jimmunol.0804292 [DOI] [PubMed] [Google Scholar]
- Schmaler M., Jann N. J., Gotz F., Landmann R. (2010). Staphylococcal lipoproteins and their role in bacterial survival in mice. Int. J. Med. Microbiol. 300, 155–160 10.1016/j.ijmm.2009.08.018 [DOI] [PubMed] [Google Scholar]
- Schneider R., Hantke K. (1993). Iron-hydroxamate uptake systems in Bacillus subtilis: identification of a lipoprotein as part of a binding protein-dependent transport system. Mol. Microbiol. 8, 111–121 10.1111/j.1365-2958.1993.tb01208.x [DOI] [PubMed] [Google Scholar]
- Schwandner R., Dziarski R., Wesche H., Rothe M., Kirschning C. J. (1999). Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J. Biol. Chem. 274, 17406–17409 10.1074/jbc.274.25.17406 [DOI] [PubMed] [Google Scholar]
- Sebulsky M. T., Heinrichs D. E. (2001). Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 183, 4994–5000 10.1128/JB.183.17.4994-5000.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebulsky M. T., Hohnstein D., Hunter M. D., Heinrichs D. E. (2000). Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 182, 4394–4400 10.1128/JB.182.16.4394-4400.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebulsky M. T., Shilton B. H., Speziali C. D., Heinrichs D. E. (2003). The role of FhuD2 in iron(III)-hydroxamate transport in Staphylococcus aureus. Demonstration that FhuD2 binds iron(III)-hydroxamates but with minimal conformational change and implication of mutations on transport. J. Biol. Chem. 278, 49890–49900 10.1074/jbc.M305073200 [DOI] [PubMed] [Google Scholar]
- Sebulsky M. T., Speziali C. D., Shilton B. H., Edgell D. R., Heinrichs D. E. (2004). FhuD1, a ferric hydroxamate-binding lipoprotein in Staphylococcus aureus: a case of gene duplication and lateral transfer. J. Biol. Chem. 279, 53152–53159 10.1074/jbc.M409793200 [DOI] [PubMed] [Google Scholar]
- Serebryakova M. V., Demina I. A., Galyamina M. A., Kondratov I. G., Ladygina V. G., Govorun V. M. (2011). The acylation state of surface lipoproteins of mollicute Acholeplasma laidlawii. J. Biol. Chem. 286, 22769–22776 10.1074/jbc.M111.231316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd M., Heath M. D., Poole R. K. (2007). NikA binds heme: a new role for an Escherichia coli periplasmic nickel-binding protein. Biochemistry 46, 5030–5037 10.1021/bi700183u [DOI] [PubMed] [Google Scholar]
- Skaar E. P., Gaspar A. H., Schneewind O. (2004a). IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279, 436–443 10.1074/jbc.M307952200 [DOI] [PubMed] [Google Scholar]
- Skaar E. P., Humayun M., Bae T., Debord K. L., Schneewind O. (2004b). Iron-source preference of Staphylococcus aureus infections. Science 305, 1626–1628 10.1126/science.1099930 [DOI] [PubMed] [Google Scholar]
- Skaar E. P., Schneewind O. (2004). Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes. Infect. 6, 390–397 [DOI] [PubMed] [Google Scholar]
- Smith D. G., Wilcox M. H., Williams P., Finch R. G., Denyer S. P. (1991). Characterization of cell envelope proteins of Staphylococcus epidermidis cultured in human peritoneal dialysate. Infect. Immun. 59, 617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speziali C. D., Dale S. E., Henderson J. A., Vines E. D., Heinrichs D. E. (2006). Requirement of Staphylococcus aureus ATP-binding cassette-ATPase FhuC for iron-restricted growth and evidence that it functions with more than one iron transporter. J. Bacteriol. 188, 2048–2055 10.1128/JB.188.6.2048-2055.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A., Dick J. D., Perl T. M. (2002). Vancomycin resistance in staphylococci. Clin. Microbiol. Rev. 15, 430–438 10.1128/CMR.15.3.430-438.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ster C., Beaudoin F., Diarra M. S., Jacques M., Malouin F., Lacasse P. (2010). Evaluation of some Staphylococcus aureus iron-regulated proteins as vaccine targets. Vet. Immunol. Immunopathol. 136, 311–318 10.1016/j.vetimm.2010.03.010 [DOI] [PubMed] [Google Scholar]
- Stoll H., Dengjel J., Nerz C., Gotz F. (2005). Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect. Immun. 73, 2411–2423 10.1128/IAI.73.4.2411-2423.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger-Jones Y. K., Bae T., Schneewind O. (2006). Vaccine assembly from surface proteins of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 103, 16942–16947 10.1073/pnas.0606863103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano Y. (2009). DyP-type peroxidases comprise a novel heme peroxidase family. Cel. Mol. Life Sci. 66, 1387–1403 10.1007/s00018-008-8651-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe I. C., Russell R. R. B. (1995). Lipoproteins of Gram-positive bacteria. J. Bacteriol. 177, 1123–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe I. C., Tao L., Ferretti J. J., Russell R. R. B. (1993). MsmE, a lipoprotein involved in sugar transport in Streptococcus mutans. J. Bacteriol. 175, 1853–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Hoshino K., Akira S. (2000a). Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J. Immunol. 165, 5392–5396 [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., Akira S. (1999). Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11, 443–451 10.1016/S1074-7613(00)80119-3 [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Takeda K., Hoshino K., Adachi O., Ogawa T., Akira S. (2000b). Cellular responses to bacterial cell wall components are mediated through MyD88-dependent signaling cascades. Int. Immunol. 12, 113–117 10.1093/intimm/12.1.113 [DOI] [PubMed] [Google Scholar]
- Tawaratsumida K., Furuyashiki M., Katsumoto M., Fujimoto Y., Fukase K., Suda Y., Hashimoto M. (2009). Characterization of N-terminal structure of TLR2-activating lipoprotein in Staphylococcus aureus. J. Biol. Chem. 284, 9147–9152 10.1074/jbc.M900429200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J., Widdick D. A., Hicks M. G., Chandra G., Sutcliffe I. C., Palmer T., Hutchings M. I. (2010). Investigating lipoprotein biogenesis and function in the model Gram-positive bacterium Streptomyces coelicolor. Mol. Microbiol. 77, 943–957 10.1111/j.1365-2958.2010.07261.x [DOI] [PubMed] [Google Scholar]
- Tokunaga M., Tokunaga H., Wu H. C. (1982). Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc. Natl. Acad. Sci. U.S.A. 79, 2255–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres V. J., Pishchany G., Humayun M., Schneewind O., Skaar E. P. (2006). Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 188, 8421–8429 10.1128/JB.01335-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg R., Mader U., Homuth G., Schaffer M., Denham E. L., Monteferrante C. G., Miethke M., Marahiel M. A., Harwood C. R., Winter T., Hecker M., Antelmann H., Van Dijl J. M. (2011). Environmental salinity determines the specificity and need for Tat-dependent secretion of the YwbN protein in Bacillus subtilis. PloS One 6:e18140 10.1371/journal.pone.0018140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenesch F., Naimi T., Enright M. C., Lina G., Nimmo G. R., Heffernan H., Liassine N., Bes M., Greenland T., Reverdy M. E., Etienne J. (2003). Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9, 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallaeys B., Cornelis R., Mees L., Lameire N. (1986). Trace elements in serum, packed cells, and dialysate of CAPD patients. Kidney Int. 30, 599–604 [DOI] [PubMed] [Google Scholar]
- Widdick D. A., Dilks K., Chandra G., Bottrill A., Naldrett M., Pohlschroder M., Palmer T. (2006). The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 103, 17927–17932 10.1073/pnas.0607025103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdick D. A., Hicks M. G., Thompson B. J., Tschumi A., Chandra G., Sutcliffe I. C., Brulle J. K., Sander P., Palmer T., Hutchings M. I. (2011). Dissecting the complete lipoprotein biogenesis pathway in Streptomyces scabies. Mol. Microbiol. 80, 1395–1412 10.1111/j.1365-2958.2011.07656.x [DOI] [PubMed] [Google Scholar]
- Wilcox M. H., Williams P., Smith D. G., Modun B., Finch R. G., Denyer S. P. (1991). Variation in the expression of cell envelope proteins of coagulase-negative staphylococci cultured under iron-restricted conditions in human peritoneal dialysate. J. Gen. Microbiol. 137, 2561–2570 [DOI] [PubMed] [Google Scholar]
- Williams P., Denyer S. P., Finch R. G. (1988). Protein antigens of Staphylococcus epidermidis grown under iron-restricted conditions in human peritoneal dialysate. FEMS Microbiol. Lett. 50, 29–33 1783903 [Google Scholar]
- Xiong A., Singh V. K., Cabrera G., Jayaswal R. K. (2000). Molecular characterization of the ferric-uptake regulator, Fur, from Staphylococcus aureus. Microbiology 146, 659–668 [DOI] [PubMed] [Google Scholar]
- Xu Q., Rawlings N. D., Farr C. L., Chiu H. J., Grant J. C., Jaroszewski L., Klock H. E., Knuth M. W., Miller M. D., Weekes D., Elsliger M. A., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2011). Structural and sequence analysis of imelysin-like proteins implicated in bacterial iron uptake. PloS One 6:e21875 10.1371/journal.pone.0021875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Sanzen I., Ohkura T., Okamoto A., Torii K., Hasegawa T., Ohta M. (2007). Analysis of twin-arginine translocation pathway homologue in Staphylococcus aureus. Curr. Microbiol. 55, 14–19 10.1007/s00284-006-0461-3 [DOI] [PubMed] [Google Scholar]
- Zawadzka A. M., Kim Y., Maltseva N., Nichiporuk R., Fan Y., Joachimiak A., Raymond K. N. (2009). Characterization of a Bacillus subtilis transporter for petrobactin, an anthrax stealth siderophore. Proc. Natl. Acad. Sci. U.S.A. 106, 21854–21859 10.1073/pnas.0904793106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Xie G., Liu M., Olson J. S., Fabian M., Dooley D. M., Lei B. (2008). Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants system of Staphylococcus aureus. J. Biol. Chem. 283, 18450–18460 10.1074/jbc.M801466200 [DOI] [PMC free article] [PubMed] [Google Scholar]