Abstract
The methylerythritol phosphate (MEP) pathway is essential in most prokaryotes and some lower eukaryotes but absent from human cells, and is a validated target for antimicrobial drug development. The formation of MEP is catalyzed by 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR). MEP pathway genes have been identified in many category A and B biothreat agents, including Francisella tularensis, which causes the zoonosis tularemia. Fosmidomycin (Fos) inhibits purified Francisella DXR. This compound also inhibits the growth of F. tularensis NIH B38, F. novicida and F. tularensis subsp. holarctica LVS bacteria. Related compounds such as FR900098 and the lipophilic prodrug of FR900098 (compound 1) have been developed to improve the bioavailability of these DXR inhibitors. In performing disk-inhibition assays with these compounds, we observed breakthrough colonies of F. novicida in the presence of Fos, suggesting spontaneous development of Fos resistance (FosR). FosR bacteria had decreased sensitivity to both Fos and FR900098. The two most likely targets for the development of mutants would be the DXR enzyme itself or the glycerol-3-phosphate transporter (GlpT) that allows entry of Fos into the bacteria. Sensitivity of FosR F. novicida bacteria to compound 1 was not abated suggesting that spontaneous resistance is not due to mutation of DXR. We thus predicted that the glpT transporter may be mutated leading to this resistant phenotype. Supporting this, transposon insertion mutants at the glpT locus were also found to be resistant to Fos. DNA sequencing of four different spontaneous FosR colonies demonstrated a variety of deletions in the glpT coding region. The overall frequency of FosR mutations in F. novicida was determined to be 6.3 × 10−8. Thus we conclude that one mechanism of resistance of F. novicida to Fos is caused by mutations in GlpT. This is the first description of spontaneous mutations in Francisella leading to FosR.
Keywords: Francisella, fosmidomycin, resistance, glycerol-3-phosphate transporter
Introduction
Francisella tularensis is the etiologic agent of tularemia, a zoonotic disease that occurs in much of the northern hemisphere including North America. Its potential use as a biological weapon has given this zoonotic organism much attention (Foley and Nieto, 2010). Ulceroglandular forms of the disease can be contracted by humans following interaction with small rodents and lagomorphs, as well as through mechanical and vector transmission by biting arthropods (Akimana and Kwaik, 2011; Potz-Biedermann et al., 2011). In addition, pneumonic cases of tularemia are occasionally seen in humans following inhalation of aerosols containing the bacteria (Matyas et al., 2007; Ojeda et al., 2008). The outcome of these various forms of disease is determined by the availability of prompt treatment with a variety of antibiotics (Ikaheimo et al., 2000; Greenfield and Bronze, 2004). These small, Gram-negative coccobacilli are susceptible to treatment with antimicrobial agents, including streptomycin, gentamicin, doxycycline, quinolones, and chloramphenicol (Scheel et al., 1992; Ikaheimo et al., 2000; Johansson et al., 2002). F. tularensis subsp. holarctica, which is found in the northern parts of Europe and Asia, consists of Biovars I and II that are susceptible or resistant to erythromycin, respectively (Kudelina and Olsufiev, 1980; Gestin et al., 2010). Resistance to penicillin-class β-lactams has been routinely demonstrated from naturally occurring isolates of Francisella due to the expression of β-lactamase (Bina et al., 2006).
The methylerythritol phosphate (MEP) pathway is essential in most prokaryotes and some lower eukaryotes but absent from human cells, and is a validated target for antimicrobial drug development (Wiemer et al., 2010). The formation of MEP is catalyzed by 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR). MEP pathway genes have been identified in many category A and B biothreat agents, including F. tularensis, Bacillus anthracis, and Yersinia pestis. There are two well-known inhibitors of DXR, an enzyme responsible for the formation of MEP used for the down-stream synthesis of the isoprenoids (Jawaid et al., 2009). One is fosmidomycin (Fos, 3-[formyl(hydroxy) amino] propylphosphonic acid), currently in clinical trials for treatment of malaria (Wiesner et al., 2003), and the second is FR900098 (3-[acetyl(hydroxy)amino] propylphosphonic acid). We have previously demonstrated that Fos inhibits Francisella growth and can target purified Francisella DXR enzyme (Jawaid et al., 2009). Because the MEP pathway is found in most prokaryotes and lower eukaryotes, but not in humans, and is essential for survival of these organisms, it has been described as a validated target for the development of new antimicrobial therapies (Rodriguez-Concepcion, 2004; Singh et al., 2007; Davey et al., 2011).
The glycerol-3-phosphate transporter (GlpT) system has been well characterized in E. coli, B. subtilis, and Pseudomonas aeruginosa (Nilsson et al., 1994; Lemieux et al., 2005; Castaneda-Garcia et al., 2009). GlpT is a member of the Major Facilitator Superfamily (MFS) that functions as an antiporter moving glycerol-3-phosphate into the cell, and exporting intracellular phosphate. In E. coli, the structure of the transporter consists of 12 membrane-spanning sections in alpha-helix form, joined together on the cytoplasmic side by an extended loop structure (Lemieux et al., 2005). A similar arrangement is predicted for Francisella GlpT. We have previously shown that the Francisella genome contains glpT, and that Fos effectiveness is mediated by the action of this transporter (McKenney et al., in press). The polar nature of Fos requires it be transported across the bacterial cell membrane via a transporter such as GlpT. Organisms that have DXR but lack such transporters, such as Mycobacterium tuberculosis and Brucella abortus, cannot transport Fos, and thus are functionally resistant (Brown and Parish, 2008; Sangari et al., 2010). However, when E. coli GlpT is expressed in B. abortus, Fos is transported into the bacterial cell, interacts with the DXR-like enzyme, and leads to decreased bacterial growth (Sangari et al., 2010). A series of acyloxyalkyl ester prodrug derivatives of FR900098, including compound 1 (Figure 1), have been generated which are cleaved to FR900098 by the bacterial esterases. They are more lipophilic in nature, demonstrate improved in vitro activity against M. tuberculosis (Uh et al., 2011), and improved in vivo activity (Ortmann et al., 2003) against malaria. We have demonstrated that the lipophilic prodrug of FR900098, compound 1, is able to bypass the Fos transporter and exert antimicrobial effects regardless of mutations in glpT that lead to Fos resistance (FosR; McKenney et al., in press).
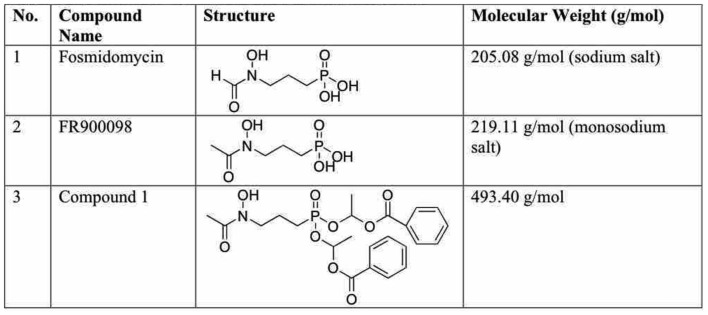
Figure 1.
Structure of inhibitors used in this study. (1) Fosmidomycin (Fos, 3-[formyl(hydroxy) amino] propylphosphonic acid). (2) FR900098 (3-[acetyl(hydroxy)amino] propylphosphonic acid). (3) Compound 1: acyloxyalkyl ester prodrug derivative of FR900098 (Ortmann et al., 2003).
Francisella species are not known to be highly multi-drug resistant, or to rapidly develop resistance. Francisella are susceptible to many common antibiotics, except penicillins (Ikaheimo et al., 2000; Urich and Petersen, 2008). Francisella has two TolC-like proteins, TolC and the highly related FltC (Gil et al., 2006); mutations in these genes increase the sensitivity of F. tularensis LVS to various antibiotics, suggesting at least some role for drug efflux in the baseline sensitivity of Francisella to antibiotics. The sensitivity to macrolides varies between Francisella strains (Ahmad et al., 2010). F. tularensis LVS has a point mutation in Domain V of the 23S rRNA, rendering it more resistant to erythromycin than F. novicida or F. tularensis Schu S4 (Biswas et al., 2008). In the North American Type A Francisella strains, erythromycin MICs range from 0.5 to 4 μg/ml, while F. tularensis LVS has an MIC > 256 μg/ml (Marinov et al., 2009). Spontaneous antibiotic resistance to spectinomycin has been reported in Francisella at a low frequency of incidence (Kormilitsyna and Marakusha, 1983).
Here we report the spontaneous development of a Fos-resistant phenotype in Francisella. Spontaneous FosR has not been previously reported for this organism, and it is of concern to have a biothreat agent spontaneously acquire resistance to new classes of antibiotics. Thus we sought to understand the mechanism of FosR in Francisella.
Materials and Methods
Strains and growth of bacteria
Francisella tularensis subsp. novicida Utah 112 (F. novicida; BEI Resources # NR13) was grown in trypticase soy broth with 0.1% cysteine HCl (TSB-C), on TSB-C agar plates, or Chocolate II Agar plates (CHAB – BD Biosciences) as noted for each assay. F. tularensis subsp. tularensis NIH B38 (ATCC 6223; BEI Resources # NR50, deposited as the type strain for F. tularensis tularensis) was grown on TSB-C agar plates at 37°C with 5% CO2 for 48 h, harvested by scraping into 20% glycerol, and stored at −80°C. Francisella on plates was grown at 37°C in an atmosphere of 5% CO2. Minimum inhibitory concentration (MIC) assays were performed in cation-adjusted Mueller–Hinton broth (MHB) according to the guidelines published by the Clinical and Laboratory Standards Institute; M11-A7.
Transposon insertion mutants in the glpT locus [FTN_0636; F. novicida tnfn1_pw060323p08q150 BEI Catalog #NR-5683 and F. novicida tnfn1_pw060418p01q161 BEI Catalog #NR-6558, both Type T20 (ISFn2/FRT)] (Gallagher et al., 2007) were obtained through the NIH Biodefense and Emerging Infectious Disease Research Repository, NIAID, NIH: F. novicida, “Two-Allele” Transposon Mutant Library. F. novicida U112 glpT-1 and glpT-2 (data not shown) transposon insertion mutants were grown as above, but in the presence of kanamycin (10 μg/ml) to select for the mutants. Sequences for the transposon mutants are shown in Figure A1 in Appendix (Gallagher et al., 2007).
Preparation of stock solutions of Fos, FR900098, and compound 1
All inhibitor stocks were made at high starting concentration between 10 and 20 mg/ml. Fos sodium salt (Invitrogen #F-23103) and FR900098 monosodium salt (Sigma–Aldrich # F8307) were obtained as dry powders and were dissolved in water. Compound 1 (kind gift from C. Dowd) was dissolved in 100% DMSO to the final concentrations indicated.
Disk-inhibition assay
Following the Kirby–Bauer disk diffusion assay protocols (Bauer et al., 1966), inhibitor-saturated disks were placed on a lawn of bacteria to display a zone of growth inhibition around the disk indicating susceptibility. A concentration of an overnight culture of bacteria equal to a 0.5 McFarland standard was spread on Chocolate II Agar plate (BD Biosciences) using a sterile cotton swab. Inhibitor disks were made by absorbing 20 μl of each inhibitor stock (10 mg/ml) to a sterile, thick Whatman filter paper 6-mm disk, so that each disk contained 200 μg. Three disks of the same inhibitor were added to each plate, except for NIHB38, for which only one disk was used per plate. For NIHB38 experiments, both 200 and 100 μg per disk were tested. The plates were wrapped in tin foil to protect the inhibitors from light and placed in the 37°C incubator for 48 h. The zone of inhibition was determined by measuring the diameter of the zone of no growth, including the 6-mm disk. Each zone was measured three times at different points of the zone (n = 9). A 6-mm zone of inhibition indicates no inhibition.
MIC assays for Fos and lipophilic analogs against wild-type F. novicida U112 and transposon insertion mutants of GlpT
In accordance with CLSI standards, 150 μl cation-adjusted MHB was inoculated with 1.5 × 105 CFU of F. novicida. For MIC assays against wild-type F. novicida, each well of column 1 was amended with 150 μl of Fos or compound 1 to achieve a final concentration of 300 and 200 μg/ml, respectively. Subsequent MIC assays had a starting concentration of 200 μg/ml regardless of the compound tested. Twofold serial dilutions of column 1 were performed to fill columns 2–11 of a 96-well plate. For compound 1, DMSO vehicle control wells were included in side-by-side assays. Column 12 received no drug and acts as the positive growth control for the assay. Absorbances (A600) were read for each well before incubation to record background. Plates were incubated at 37°C plus 5% CO2, A600 were determined at 24 and 48 h, and IC50 and MIC were determined.
Genomic DNA preparation, PCR amplification, and sequencing
Overnight cultures were centrifuged to form bacterial pellets and supernatants were removed. Genomic DNA was prepared using the Wizard Genomic DNA kit (Promega, Madison, WI, USA). Briefly, pellets were re-suspended in 400 μl of nuclei lysis solution and incubated at 80°C for 15 min to lyse bacteria. Total DNA concentrations were analyzed using the Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and adjusted to 40 ng/μl. Primers used for PCR confirmation that colonies were indeed F. novicida were designed to amplify 16S rRNAs (16SrRNAF: CTGTCGTCAGCTCGTGTTGT; 16SrRNA R: CGTAAGGGCCATGATGACTT; Forsman et al., 1994). Primers used to amplify the entire ORF of the glpT gene (glpTF: AACAGCGGTTTAGCTATTTTCAA; glpTR: TGCAATCTGAGCTGACTGAAG) were designed using Primer 3 through the National Center for Biotechnology Information (NCBI). Real-time PCR was performed on the Biorad MyIQ instrument with iQ SYBR Green Supermix in 50 μl reactions (Biorad, Hercules, CA, USA). Thermal cycler profile was set to 95°C for 3 min for initial denaturation and enzyme activation followed by 40 cycles of 95°C for 10 s and 55°C for 30 s. Sequencing primers were the same as the ORF amplification primers, but were synthesized and used by Functional Biosciences (Madison, WI, USA).
Spontaneous mutation frequency for FosR
A stock culture of F. novicida U112 was grown in TSB-C broth overnight at 37°C with continuous shaking at 165 rpm. Viable bacteria from overnight cultures were enumerated by serial dilution in sterile phosphate-buffered saline (PBS) and plated on TSB-C non-selective agar plates. TSB-C plates were incubated for >24 h, and colony forming units were counted to determine bacterial density of the culture. A diluted subculture was created to achieve a final concentration of 1 × 103 CFU/ml. Parallel cultures prepared as 1 ml aliquots from the subculture (1 × 103 CFU) were incubated overnight at 37°C with continuous shaking at 165 rpm (Rosche and Foster, 2000). The number of mutations that resulted in FosR was determined by plating 0.1 ml aliquots of the parallel cultures (n = 5) on TSB-C plates containing 122 μM (25 μg/ml) Fos. The total number of bacteria in the cultures was determined by serial dilution and plating on non-selective media (Rosche and Foster, 2000).
Results
We have previously reported the in vitro antimicrobial activity of Fos and FR900098 (Figure 1) against Francisella, and the inhibition of purified Francisella DXR enzyme by these compounds (Jawaid et al., 2009; McKenney et al., in press). We have also demonstrated that a lipophilic prodrug of FR900098 (compound 1) is able to bypass the GlpT transporter and exert antimicrobial effects regardless of mutations in glpT that lead to FosR, and that compound 1 is effective in an in vivo model of F. novicida infection (McKenney et al., in press).
Wild-type F. novicida
The Kirby–Bauer disk-inhibition assay was used to determine the susceptibility of wild-type F. novicida U112 to Fos and FR900098. The zone of inhibition around the disk containing 200 μg of Fos was 37 ± 1 mm (Figure 2A). The zone of inhibition around the disk containing 200 μg of FR900098 was 29 ± 2 mm (Figure 2C). For F. tularensis NIHB38, the type strain for Type A tularemia, both of these compounds lead to a large zone of inhibition. For a disk with 100 μg Fos, the zone of inhibition for F. tularensis NIHB38 was 72 ± 2 mm (data not shown), suggesting that F. tularensis Type A strains are sensitive to Fos. A zone of inhibition of 6 mm (the size of the disk) indicated no inhibition, which was observed for a water-only disk (data not shown).
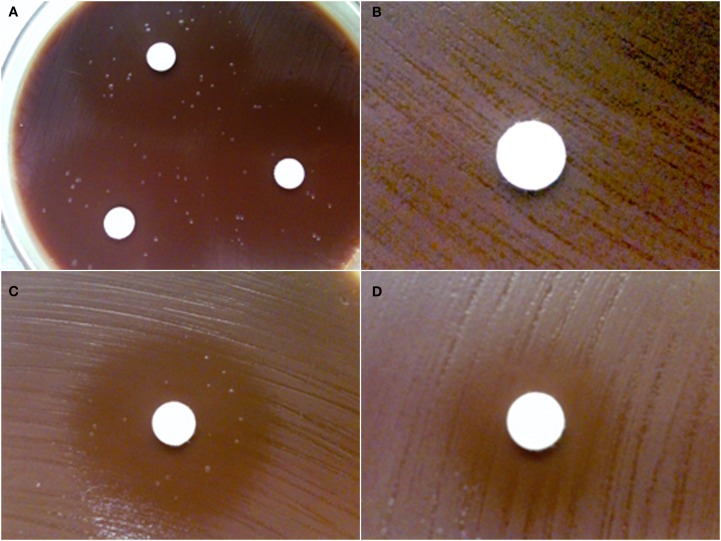
Figure 2.
Disk-inhibition assay. Each disk contains 200 μg of antibiotic. (A) Wild-type F. novicida has a 37 ± 1-mm zone of inhibition around the disk containing fosmidomycin with multiple breakthrough colonies shown. (B) GlpT-1 transposon mutant of F. novicida is completely resistant to fosmidomycin (zone of inhibition = 6 mm). (C) Wild-type F. novicida has a 29 ± 2-mm zone of inhibition around the disk containing FR900098. Breakthrough colonies can also be seen in the inhibition zone. (D) GlpT-1 transposon mutant of F. novicida has intermediate resistance to FR900098 (zone of inhibition = 15 mm), but does not demonstrate any breakthrough colonies. This experiment was performed three times, with representative examples shown.
Spontaneous FosR mutants
While performing the Kirby–Bauer disk-inhibition assay with F. novicida against Fos and FR900098, breakthrough colonies were observed growing inside the zone of inhibition (Figure 2A – Fos, Figure 2C – FR900098). However, the zone of inhibition is still clearly defined. The colonies within the zone of inhibition are slightly different in their appearance than the wild-type F. novicida that is growing outside the zone of inhibition. The colonies are slightly larger and more mucoid, but are the same color as the wild-type colonies. These colonies (from Figures 2A,C) were confirmed to be F. novicida by real-time PCR using F. novicida specific 16S rRNA primers (Forsman et al., 1994; data not shown). Numerous breakthrough colonies were also observed for F. tularensis NIHB38 with both of these compounds, indicating that this “type strain” for F. tularensis Type A strains may also develop spontaneous resistance to Fos.
Based on our results in the disk-inhibition assay as well as our previous studies (McKenney et al., in press), we hypothesized that the two most likely bacterial targets for the development of mutants would be mutations of the DXR enzyme itself or mutations of GlpT that allow entry of Fos into the bacteria.
MICs for wild-type and FosR mutants
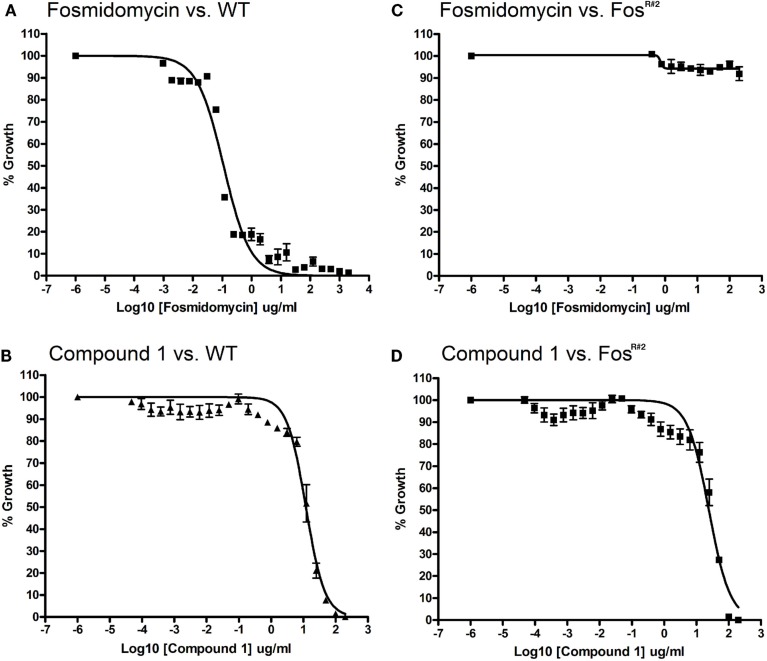
We found that FosR mutants are resistant to Fos but are sensitive to compound 1, the lipophilic prodrug of FR900098. The MIC for Fos against wild-type F. novicida is 121 μM (IC50 is 0.101 μg/ml with a 95% confidence interval (CI) = 0.079–0.130 mg/l; Figure 3A). The MIC for the lipophilic compound 1 against F. novicida is 100 μg/ml or 203 μM (IC50 is 11.99 μg/ml, 95% CI = 9.84–14.62 μg/ml; Figure 3B). A Fos-resistant colony (FosR#2) isolated from our disk-inhibition assays was grown overnight as described, and the MICs and IC50s of Fos and compound 1 were determined. Concentrations of inhibitor used in this assay ranged from 0.4 to 200 μg/ml. FosR#2-resistant bacteria grew well at all concentrations of Fos tested, with significant growth in all wells at or near the levels of no-drug control (Figure 3C). Conversely, the MIC of compound 1 for FosR#2 was 100 μg/ml or 203 μM (IC50 of compound 1 for FosR#2 is 24.38 μg/ml, 95% CI = 18.75–31.70 μg/ml; Figure 3D). The MIC was identical between wild-type and FosR#2 and the IC50s were very close. The IC50 curves can almost be superimposed.
Figure 3.
Minimum inhibitory concentration assay for F. novicida and FosR#2 mutant. Wild-type F. novicida susceptibility to fosmidomycin (A) and compound 1 (B). Wild-type MIC for fosmidomycin is 121 μM. [WT IC50 for fosmidomycin is 0.092 mg/l (95% CI = 0.079–0.107 mg/l)]. Wild-type MIC for compound 1 is 203 μm. Wild-type IC50 for compound 1 is 11.99 mg/l (95% CI = 9.84–14.62 mg/l, R2 = 0.972). FosR#2 mutant resistance to fosmidomycin (C) FosR#2 and susceptibility to compound 1 (D). F. novicida glpT mutant MIC or IC50 for fosmidomycin could not be defined due to lack of inhibition (R2 value for fitness = 0.38). FosR#2 mutant MIC for compound 1 is approximately 203 μM. The IC50 for compound 1 is 24.38 mg/l (95% CI = 18.75–31.70 mg/l, R2 = 0.944).
Is DXR mutated in FosR bacteria?
Fosmidomycin-resistant bacteria retained sensitivity to compound 1. Since compound 1 is a lipophilic prodrug it will get metabolized to FR900098 inside the bacteria, which will then target bacterial DXR. This suggests that spontaneous resistance is not due to mutation of DXR, since FosR bacteria are inhibited by compound 1. No transposon insertion mutants are available for DXR as this enzyme is thought to be essential to Francisella growth, so the role of DXR could not be directly tested in this study. We thus hypothesized that the mutations that lead to the Fos-resistant phenotype are not mutations in DXR. We then predicted that the GlpT transporter may be mutated, leading to this resistant phenotype. Supporting this, transposon insertion mutants at the glpT locus were also found to be resistant to Fos.
Disk inhibition with glpT transposon insertion mutants
The susceptibility of the glpT transposon insertion mutants to Fos and FR900098 was also tested in this manner. There was no inhibition of the glpT transposon mutant by Fos (Figure 2B), supporting the critical role of glpT in Fos transport across the bacterial membranes in order for it to reach the DXR enzyme target inside the bacteria. glpT mutants with FR900098 disks demonstrated intermediate inhibition (15 ± 1 mm zone; Figure 2D). This effect of FR900098 on the glpT mutant is consistent with our previous studies and published reports suggesting improved bacterial penetration of this compound (McKenney et al., in press). Interestingly, as can be seen in Figure 2D, there were no breakthrough colonies observed in the glpT transposon mutant experiments in multiple repeats, perhaps suggesting that the locus was the origination of the instability, leading to the breakthrough colonies observed with the wild-type F. novicida, and that other loci are perhaps not as susceptible to mutations leading to breakthrough colonies.
Antibiotic sensitivity of glpT Tn mutants
glpT transposon mutants are resistant to Fos and sensitive to compound 1. The glpT transposon insertion mutants were tested for their susceptibility to Fos and compound 1 in a liquid broth growth inhibition assay. As expected, the transporter-independent lipophilic prodrug compound 1 was effective at inhibiting growth of the glpT transposon insertion mutants (glpT-1 shown) as well as wild-type F. novicida, while Fos at twice the MIC was not (Figure 4). This result supports the hypothesis that mutation in the glpT transporter sequence can have a significant detrimental effect on transport of Fos across the bacterial membrane, preventing its interaction with the intended target enzyme, DXR. Intermediate inhibition of the glpT mutant by FR900098 was observed in this assay (gray bars) consistent with the disk-inhibition assay results (see Figure 2D), perhaps due to the slightly more lipophilic nature of FR900098 compared to Fos (see Figure 1 for structures), which may allow some bacterial penetration.
Figure 4.
Sensitivity of wild-type F. novicida and glpT transposon insertion mutants to fosmidomycin, FR900098, and compound 1. Sensitivity of F. novicida is shown to all three compounds, to fosmidomycin (white bar), FR900098 (gray bar), and the lipophilic prodrug of FR900098, or compound 1 (black bar). Fosmidomycin, FR900098, and compound 1 were tested at concentrations of 200 μg/ml. Resistance of glpT transposon insertion mutants to fosmidomycin (white bar) and to a lesser extent FR900098 (gray bar), but not compound 1 (black bar) is shown. No inhibition was seen using fosmidomycin and the transposon mutants. For this experiment, n = 3 and error bars represent SEM, and in some cases are too small to be seen.
Bioinformatic identification of GlpT
The glpT coding region (GlpT) was identified in the F. tularensis tularensis SchuS4 (FTT0725c) and F. novicida (FTN_0636) genome (accession numbers YP_169738.1 and YP_898283.1) via BLAST search using the E. coli K12 homologous sequence (accession number NP_416743.1) as the query. All other Francisella sequences also have a highly conserved glpT homolog (e.g., FTM_1358, FTW_1513, FTF0725c, FTA_1594, FTH_1462, FTL_1510, Fphi_0200). In addition, bioinformatics examination of Francisella genomes revealed no homologous gene for fsr, an efflux pump responsible for removal of the Fos from E. coli (Messiaen et al., 2011). Also, by our bioinformatic analysis, the published genomes for F. novicida, F. tularensis Schu S4, and F. tularensis LVS also do not contain the uhpT transporter (an alternate transporter that could substitute for glpT), although the related species F. philomiragia, which has an environmental habitat and is not a known pathogen for humans, has a gene that may be uhpT (Fphi_0883).
Sequencing of glpT in the FosR colonies
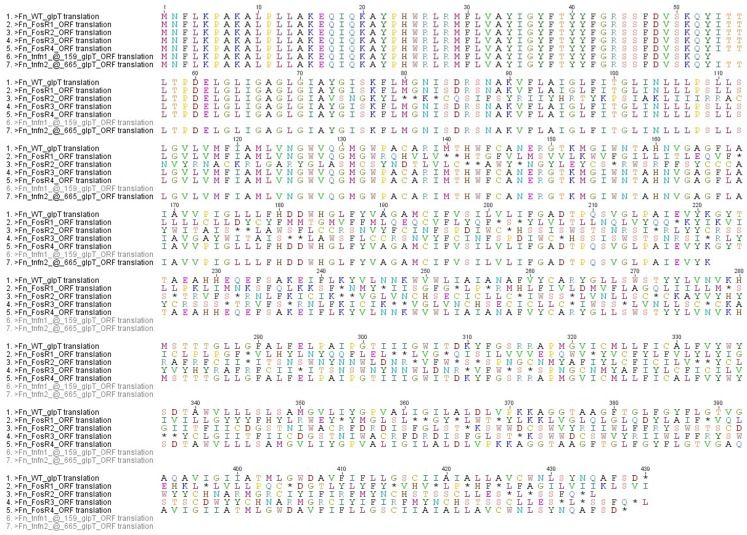
In order to examine the sequence of glpT in the FosR colonies, we isolated the glpT region by PCR and sequenced glpT from four different spontaneous FosR colonies. Colonies were picked and grown in liquid culture overnight in TSB-C to create stocks and small aliquots were removed for genomic DNA preparation and sequencing of glpT. Comparison of sequences of the four selected FosR mutants with wild-type sequence data reveals an array of nucleotide deletions and additions from various portions of the glpT gene (Figure 5; Figure A1 in Appendix). The 14 nucleotide deletion from mutant FnFosR#2 results in a deletion of amino acids in the translated protein as well as the formation of additional stop codons. Similarly, the addition of two nucleotides and one nucleotide in FnFosR#1 and FnFosR#3, respectively, results in missense mutation by amino acid substitutions and the appearance of premature stop codons resulting in truncated proteins. These abbreviated proteins are likely non-functional. FnFosR#4 has a single deletion of six nucleotides at position 1126 of the open reading frame, corresponding to the deletion of a Leu347 and Ser348 amino acid residues in the protein (Figure 5; Figure A2 in Appendix). When the amino acid sequence of GlpT from the FnFosR#4 is compared with E. coli, the deleted amino acids occur in a region that is analogous to a peripheral alpha-helix number H9 that spans the lipid membrane (Lemieux et al., 2005) but is not associated with the formation of the pore through which Fos is transported into the cell.
Figure 5.
Sequence of FosRF. novicidaglpT vs. WT. DNA Sequencing of the entire ORF for glpT from F. novicida reveals nucleotide deletions and additions that result in missense frame shifts and stop codon insertion mutations from isolated colonies grown in the presence of fosmidomycin. GlpT transposon mutants are shown for comparison (Gallagher et al., 2007). (A) FnFosR#1 – GC insertion at nt 401/402 results in frame shift after Try133 and stop codon insert at nt 418/420. FnFosR#2 – 14 bp deletions at nt 216 results in frame shift at Ala72 and stop codon insert at nt 238/240. FnFosR#3 – G insertions at nt 517 results in frame shift after Val171 and stop codon insert at 541/543. FnFosR#4 – 6 bp deletion at nt 1042 results in deletion of Ser346 and Leu347 from the alpha-helix H9. (B) Resulting amino acid alignment from the FosR mutants vs. WT F. novicida U112.
Rate and frequency of fosmidomycin resistance of F. novicida
We determined the rate and frequency of FosR of F. novicida. Using the Drake (1969) formula, the mutation frequency (f) and the mutation rate (μ) resulting in F. novicida FosR are approximately 6.3 × 10−8 and 0.173, respectively. The f is defined as the number of observed colonies in the presence of the inhibitor divided by the total number of cells in culture (Nt). The μ is determined by the number of observed colonies (m), the frequency of mutation (f), and the Nt (Table 1). Similarly, the rate determined by the Maximum Likelihood Method is 0.211 (95% CI = 0.159–0.268; Rosche and Foster, 2000).
Table 1.
Frequency of mutants resistant to fosmidomycin (122 μM) from the wild-type strain of F. novicida U112.
| Total bacteria (Nt) | Resistant colonies | Frequency (f) | Rate (μ) |
|---|---|---|---|
| 4.81 × 1010 | 270 | ||
| 5.18 × 1010 | 1500 | ||
| 3.66 × 1010 | 2310 | 6.3 × 10−8 | 0.173 |
| 3.96 × 1010 | 2520 | ||
| 4.17 × 1010 | 2820 |
First column represents five different aliquots of bacterial culture as described, and the second column represents the number of resistant colonies obtained when plated on 122 μM fosmidomycin. Bold signifies the value selected to determine Rate and Frequency.
Discussion
Francisella DXR is inhibited by Fos and FR900098. We have previously established that Fos inhibits purified DXR from F. tularensis LVS with half maximal activity of 247 nM (Jawaid et al., 2009), comparable to its effect against DXR from M. tuberculosis (310 nM; Dhiman et al., 2005). In addition, we previously demonstrated that FR900098 was found to have an IC50 of 230 nM against purified F. tularensis LVS DXR (McKenney et al., in press). In this study, we have demonstrated the sensitivity of F. tularensis tularensis (NIHB38) and F. novicida to Fos and FR900098 through the use of Kirby–Bauer disk-inhibition assays.
Francisella species are sensitive to Fos through their ability to transport the drug via the glycerol-3-phosphate transporter (GlpT) and expression of the target enzyme DXR. Spontaneous mutants (FosR) of Francisella (including Type A) were observed that contained mutations in glpT, suggesting a transporter-based mechanism of resistance. Fos has been shown to be effective against Plasmodium falciparum, the causative agent of malaria (Jomaa et al., 1999), and some Gram-negative bacteria in vivo (Neu and Kamimura, 1981). Spontaneous resistance to Fos has been observed with E. coli (Kanimoto and Greenwood, 1987). Known mechanisms of resistance to Fos include overexpression of the target enzymes DXR or IspD (Zhang et al., 2011), overexpression of efflux pumps (fsr) to remove the drug from the bacterial cell (Messiaen et al., 2011), and mutation or down-regulation of the GlpT transporter (Fujisaki et al., 1996; Sakamoto et al., 2003; Brown and Parish, 2008; Messiaen et al., 2011).
The proposed mechanism of resistance to Fos in Burkholderia cepacia involves both the lack of a functional GlpT transporter and an increased expression of an efflux pump (fsr) responsible for removal of the drug from the cell (Messiaen et al., 2011). Bioinformatics examination of Francisella genomes revealed no homologous fsr efflux pump. Upon bioinformatic analysis, the published genomes for F. novicida, F. tularensis Schu S4, and F. tularensis LVS also do not contain the uhpT transporter (an alternate transporter that could substitute for glpT), although the related environmental species F. philomiragia does appear to have uhpT. The reason for this difference is unknown.
Thus, the discovery and development of transporter-independent Fos derivatives could represent a viable strategy to combat zoonotic bacterial infections, or microbes that have been developed as bioweapons. Although Staphylococcus aureus and some other organisms lack a functional DXR homolog, bacteria that contain DXR include B. anthracis (Ravel et al., 2009) and Y. pestis (Pieper et al., 2008). FosR is perhaps surprising to see in Francisella given that there is little evidence that spontaneous resistance to other antimicrobials occurs readily in this organism (Kormilitsyna and Marakusha, 1983).
1-Deoxy-d-xylulose 5-phosphate reductoisomerase is an essential enzyme in organisms that use the MEP pathway, as mutations have been shown to be lethal (Gallagher et al., 2007; Brown and Parish, 2008). For this reason, antibiotics that target DXR have a great potential as broad-spectrum therapeutics. While mutations could occur in the DXR active site that would alter drug activity, complete knockouts of this enzyme are unlikely due to it being an essential enzyme. Interestingly, there were no breakthrough colonies observed in the disk assays with the glpT transposon mutants, suggesting that this locus may be responsible for the relatively high number of breakthrough colonies observed with the wild-type F. novicida. This finding suggests that mutations of DXR do not readily occur, even under selection with Fos, supporting the use of DXR as a validated target for future antimicrobial drug development. This also supports the study of lipophilic prodrugs such as compound 1 as potential broad-spectrum antimicrobial compounds (Obiol-Pardo et al., 2011; Uh et al., 2011; Ponaire et al., 2012).
It is shown here that mutations in the glpT gene can render F. novicida resistant to high levels of Fos (both FosR and glpT transposon insertion mutants). This is supported by the data showing that the transposon insertion mutants of glpT are also resistant to Fos. By using the lipophilic prodrug of FR900098 (compound 1), it is evident that both the spontaneous glpT transposon insertion mutants and FosR mutants are still susceptible to compound 1, consistent with its transporter-independent mechanism of entry into the bacteria. This result also suggests that the spontaneous resistance is not due to mutations in DXR, the intracellular bacterial target of Fos, FR900098, and compound 1. It has recently been shown that Fos also targets the enzyme following DXR in the MEP pathway, 2-C-methylerythritol-4-phoshpate cytidyltransferase (or IspD) in E. coli and P. falciparum (Zhang et al., 2011). This is a promising discovery, as there is a lower frequency of resistance to antibiotics that have multiple targets (Silver, 2011).
Developing novel therapeutics based on lipophilic derivatives of phosphonic acids such as Fos and FR900098 represents a new approach to antimicrobial development with the potential for broad-spectrum activity against various zoonotic agents (Ortmann et al., 2003; Jawaid et al., 2009; Wiemer et al., 2010; Davey et al., 2011). Compound 1 was recently shown to be effective against a wide range of bacteria (Obiol-Pardo et al., 2011; Uh et al., 2011; Ponaire et al., 2012). Such compounds may also be effective against many biothreat agents (Jawaid et al., 2009). The conservation of DXR and glpT in the genus Francisella suggests that transporter-dependence of any Fos-derived compounds should be considered early in the development of these compounds as potential antibiotics. Furthermore, the development of such compounds to be transporter-independent would be most desirable. Importantly, these compounds would retain their effectiveness against strains that may be engineered for other mechanisms of antimicrobial resistance due to the fundamental requirement for DXR in most bacterial biothreat agents.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Francisella novicida transposon insertion mutants were obtained through the NIH Biodefense and Emerging Infectious Disease Research Repository, NIAID, NIH. The authors gratefully acknowledge Dr. Cynthia Dowd, Department of Chemistry, George Washington University, for the kind gift of Compound 1 (NIH RC1AI086453 to C.S.D.). We also thank Dr. Serguei Popov, George Mason University, for helpful discussions about mutation frequency. Ryan S. Mackie was partially supported by the Naval Surface Warfare Center – Dahlgren Division Academic Fellowship program. Monique L. van Hoek was supported by the Defense Threat Reduction Agency HDTRA1-11-1-054.
Appendix
Figure A1.
Full DNA sequence data for FosR mutants vs. WT F. novicida. Detailed sequence results demonstrating the deletion and addition of nucleotides in a variety of locations in the full length open reading frame compared to the WT F. novicida glpT gene.
Figure A2.
Full amino acid sequence data for FosR mutants vs. WT F. novicida. Translation of the full length DNA sequences including the positions of amino acid substitution and appearance of early stop codon insertions.
References
- Ahmad S., Hunter L., Qin A., Mann B. J., Van Hoek M. L. (2010). Azithromycin effectiveness against intracellular infections of Francisella. BMC Microbiol. 10, 123. 10.1186/1471-2180-10-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimana C., Kwaik Y. A. (2011). Francisella-arthropod vector interaction and its role in patho-adaptation to infect mammals. Front. Microbiol. 2:34. 10.3389/fmicb.2011.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496 [PubMed] [Google Scholar]
- Bina X. R., Wang C., Miller M. A., Bina J. E. (2006). The Bla2 beta-lactamase from the live-vaccine strain of Francisella tularensis encodes a functional protein that is only active against penicillin-class beta-lactam antibiotics. Arch. Microbiol. 186, 219–228 10.1007/s00203-006-0133-5 [DOI] [PubMed] [Google Scholar]
- Biswas S., Raoult D., Rolain J. M. (2008). A bioinformatic approach to understanding antibiotic resistance in intracellular bacteria through whole genome analysis. Int. J. Antimicrob. Agents 32, 207–220 10.1016/S0924-8579(09)70004-4 [DOI] [PubMed] [Google Scholar]
- Brown A. C., Parish T. (2008). DXR is essential in Mycobacterium tuberculosis and fosmidomycin resistance is due to a lack of uptake. BMC Microbiol. 8, 78. 10.1186/1471-2180-8-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda-Garcia A., Rodriguez-Rojas A., Guelfo J. R., Blazquez J. (2009). The glycerol-3-phosphate permease GlpT is the only fosfomycin transporter in Pseudomonas aeruginosa. J. Bacteriol. 191, 6968–6974 10.1128/JB.00748-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M. S., Tyrrell J. M., Howe R. A., Walsh T. R., Moser B., Toleman M. A., Eberl M. (2011). A promising target for treatment of multidrug-resistant bacterial infections. Antimicrob. Agents Chemother. 55, 3635–3636 10.1128/AAC.00382-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman R. K., Schaeffer M. L., Bailey A. M., Testa C. A., Scherman H., Crick D. C. (2005). 1-Deoxy-D-xylulose 5-phosphate reductoisomerase (IspC) from Mycobacterium tuberculosis: towards understanding mycobacterial resistance to fosmidomycin. J. Bacteriol. 187, 8395–8402 10.1128/JB.187.24.8395-8402.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W. (1969). Comparative rates of spontaneous mutation. Nature 221, 1132. 10.1038/2211132a0 [DOI] [PubMed] [Google Scholar]
- Foley J. E., Nieto N. C. (2010). Tularemia. Vet. Microbiol. 140, 332–338 10.1016/j.vetmic.2009.07.017 [DOI] [PubMed] [Google Scholar]
- Forsman M., Sandstrom G., Sjostedt A. (1994). Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int. J. Syst. Bacteriol. 44, 38–46 10.1099/00207713-44-1-38 [DOI] [PubMed] [Google Scholar]
- Fujisaki S., Ohnuma S., Horiuchi T., Takahashi I., Tsukui S., Nishimura Y., Nishino T., Kitabatake M., Inokuchi H. (1996). Cloning of a gene from Escherichia coli that confers resistance to fosmidomycin as a consequence of amplification. Gene 175, 83–87 10.1016/0378-1119(96)00128-X [DOI] [PubMed] [Google Scholar]
- Gallagher L. A., Ramage E., Jacobs M. A., Kaul R., Brittnacher M., Manoil C. (2007). A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. U.S.A. 104, 1009–1014 10.1073/pnas.0700163104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestin B., Valade E., Thibault F., Schneider D., Maurin M. (2010). Phenotypic and genetic characterization of macrolide resistanc. in Francisella tularensis subsp. holarctica biovar I. J. Antimicrob. Chemother. 65, 2359–2367 10.1093/jac/dkq315 [DOI] [PubMed] [Google Scholar]
- Gil H., Platz G. J., Forestal C. A., Monfett M., Bakshi C. S., Sellati T. J., Furie M. B., Benach J. L., Thanassi D. G. (2006). Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc. Natl. Acad. Sci. U.S.A. 103, 12897–12902 10.1073/pnas.0602582103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield R. A., Bronze M. S. (2004). Current therapy and the development of therapeutic options for the treatment of diseases due to bacterial agents of potential biowarfare and bioterrorism. Curr. Opin. Investig. Drugs 5, 135–140 [PubMed] [Google Scholar]
- Ikaheimo I., Syrjala H., Karhukorpi J., Schildt R., Koskela M. (2000). In vitro antibiotic susceptibility of Francisella tularensis isolated from humans and animals. J. Antimicrob. Chemother. 46, 287–290 10.1093/jac/46.2.287 [DOI] [PubMed] [Google Scholar]
- Jawaid S., Seidle H., Zhou W., Abdirahman H., Abadeer M., Hix J. H., Van Hoek M. L., Couch R. D. (2009). Kinetic characterization and phosphoregulation of the Francisella tularensis 1-deoxy-D-xylulose 5-phosphate reductoisomerase (MEP synthase). PLoS ONE 4, e8288. 10.1371/journal.pone.0008288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A., Urich S. K., Chu M. C., Sjostedt A., Tarnvik A. (2002). In vitro susceptibility to quinolones of Francisella tularensis subspecies tularensis. Scand. J. Infect. Dis. 34, 327–330 10.1080/00365540110080773 [DOI] [PubMed] [Google Scholar]
- Jomaa H., Wiesner J., Sanderbrand S., Altincicek B., Weidemeyer C., Hintz M., Turbachova I., Eberl M., Zeidler J., Lichtenthaler H. K., Soldati D., Beck E. (1999). Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 285, 1573–1576 10.1126/science.285.5433.1573 [DOI] [PubMed] [Google Scholar]
- Kanimoto Y., Greenwood D. (1987). Activity of fosmidomycin in an in vitro model of the treatment of bacterial cystitis. Infection 15, 465–468 10.1007/BF01647239 [DOI] [PubMed] [Google Scholar]
- Kormilitsyna M. I., Marakusha B. I. (1983). Biological characteristics of spectinomycin-resistant strains of the tularemia pathogen. Antibiotiki 28, 434–436 [PubMed] [Google Scholar]
- Kudelina R. I., Olsufiev N. G. (1980). Sensitivity to macrolide antibiotics and lincomycin in Francisella tularensis holarctica. J. Hyg. Epidemiol. Microbiol. Immunol. 24, 84–91 [PubMed] [Google Scholar]
- Lemieux M. J., Huang Y., Wang Da N. (2005). Crystal structure and mechanism of GlpT, the glycerol-3-phosphate transporter from E. coli. J. Electron Microsc. (Tokyo) 54(Suppl. 1), i43–i46 10.1093/jmicro/dfh102 [DOI] [PubMed] [Google Scholar]
- Marinov K. T., Georgieva E. D., Ivanov I. N., Kantardjiev T. V. (2009). Characterization and genotyping of strains of Francisella tularensis isolated in Bulgaria. J. Med. Microbiol. 58, 82–85 10.1099/jmm.0.003426-0 [DOI] [PubMed] [Google Scholar]
- Matyas B. T., Nieder H. S., Telford S. R., III. (2007). Pneumonic tularemia on Martha’s Vineyard: clinical, epidemiologic, and ecological characteristics. Ann. N. Y. Acad. Sci. 1105, 351–377 10.1196/annals.1409.013 [DOI] [PubMed] [Google Scholar]
- Messiaen A. S., Verbrugghen T., Declerck C., Ortmann R., Schlitzer M., Nelis H., Van Calenbergh S., Coenye T. (2011). Resistance of the Burkholderia cepacia complex to fosmidomycin and fosmidomycin derivatives. Int. J. Antimicrob. Agents 38, 261–264 [DOI] [PubMed] [Google Scholar]
- Neu H. C., Kamimura T. (1981). In vitro and in vivo antibacterial activity of FR-31564, a phosphonic acid antimicrobial agent. Antimicrob. Agents Chemother. 19, 1013–1023 10.1128/AAC.19.6.1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R. P., Beijer L., Rutberg B. (1994). The glpT and glpQ genes of the glycerol regulon in Bacillus subtilis. Microbiology 140(Pt 4), 723–730 10.1099/00221287-140-4-723 [DOI] [PubMed] [Google Scholar]
- Obiol-Pardo C., Rubio-Martinez J., Imperial S. (2011). The methylerythritol phosphate (MEP) pathway for isoprenoid biosynthesis as a target for the development of new drugs against tuberculosis. Curr. Med. Chem. 18, 1325–1338 10.2174/092986711795029582 [DOI] [PubMed] [Google Scholar]
- Ojeda S. S., Wang Z. J., Mares C. A., Chang T. A., Li Q., Morris E. G., Jerabek P. A., Teale J. M. (2008). Rapid dissemination of Francisella tularensis and the effect of route of infection. BMC Microbiol. 8, 215. 10.1186/1471-2180-8-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann R., Wiesner J., Reichenberg A., Henschker D., Beck E., Jomaa H., Schlitzer M. (2003). Acyloxyalkyl ester prodrugs of FR900098 with improved in vivo anti-malarial activity. Bioorg. Med. Chem. Lett. 13, 2163–2166 10.1016/S0960-894X(03)00354-8 [DOI] [PubMed] [Google Scholar]
- Pieper R., Huang S. T., Clark D. J., Robinson J. M., Parmar P. P., Alami H., Bunai C. L., Perry R. D., Fleischmann R. D., Peterson S. N. (2008). Characterizing the dynamic nature of the Yersinia pestis periplasmic proteome in response to nutrient exhaustion and temperature change. Proteomics 8, 1442–1458 10.1002/pmic.200700923 [DOI] [PubMed] [Google Scholar]
- Ponaire S., Zinglé C., Tritsch D., Grosdemange-Billiard C., Rohmer M. (2012). Growth inhibition of Mycobacterium smegmatis by prodrugs of deoxyxylulose phosphate reducto-isomerase inhibitors, promising anti-mycobacterial agents. Eur. J. Med. Chem. 51, 277–285 10.1016/j.ejmech.2012.02.031 [DOI] [PubMed] [Google Scholar]
- Potz-Biedermann C., Schwendemann L., Schroppel K., Sonnichsen K., Schmidt D., Schaller M. (2011). Ulceroglandular tularemia. J. Dtsch. Dermatol. Ges. 9, 806–808 10.1111/j.1610-0387.2011.07670.x [DOI] [PubMed] [Google Scholar]
- Ravel J., Jiang L., Stanley S. T., Wilson M. R., Decker R. S., Read T. D., Worsham P., Keim P. S., Salzberg S. L., Fraser-Liggett C. M., Rasko D. A. (2009). The complete genome sequence of Bacillus anthracis Ames “Ancestor.”. J. Bacteriol. 191, 445–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M. (2004). The MEP pathway: a new target for the development of herbicides, antibiotics and antimalarial drugs. Curr. Pharm. Des. 10, 2391–2400 10.2174/1381612043384006 [DOI] [PubMed] [Google Scholar]
- Rosche W. A., Foster P. L. (2000). Determining mutation rates in bacterial populations. Methods 20, 4–17 10.1006/meth.1999.0901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y., Furukawa S., Ogihara H., Yamasaki M. (2003). Fosmidomycin resistance in adenylate cyclase deficient (cya) mutants of Escherichia coli. Biosci. Biotechnol. Biochem. 67, 2030–2033 10.1271/bbb.67.2030 [DOI] [PubMed] [Google Scholar]
- Sangari F. J., Perez-Gil J., Carretero-Paulet L., Garcia-Lobo J. M., Rodriguez-Concepcion M. (2010). A new family of enzymes catalyzing the first committed step of the methylerythritol 4-phosphate (MEP) pathway for isoprenoid biosynthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 14081–14086 10.1073/pnas.1001962107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel O., Reiersen R., Hoel T. (1992). Treatment of tularemia with ciprofloxacin. Eur. J. Clin. Microbiol. Infect. Dis. 11, 447–448 10.1007/BF01961860 [DOI] [PubMed] [Google Scholar]
- Silver L. L. (2011). Challenges of antibacterial discovery. Clin. Microbiol. Rev. 24, 71–109 10.1128/CMR.00030-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Cheve G., Avery M. A., Mccurdy C. R. (2007). Targeting the methyl erythritol phosphate (MEP) pathway for novel antimalarial, antibacterial and herbicidal drug discovery: inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) enzyme. Curr. Pharm. Des. 13, 1161–1177 10.2174/138161207780618939 [DOI] [PubMed] [Google Scholar]
- Uh E., Jackson E. R., San Jose G., Maddox M., Lee R. E., Boshoff H. I., Dowd C. S. (2011). Antibacterial and antitubercular activity of fosmidomycin, FR900098, and their lipophilic analogs. Bioorg. Med. Chem. Lett. 21, 6973–6976 10.1016/j.bmcl.2011.09.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich S. K., Petersen J. M. (2008). In vitro susceptibility of isolates of Francisella tularensis types A and B from North America. Antimicrob. Agents Chemother. 52, 2276–2278 10.1128/AAC.01584-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer A. J., Hsiao C. H., Wiemer D. F. (2010). Isoprenoid metabolism as a therapeutic target in Gram-negative pathogens. Curr. Top. Med. Chem. 10, 1858–1871 [DOI] [PubMed] [Google Scholar]
- Wiesner J., Borrmann S., Jomaa H. (2003). Fosmidomycin for the treatment of malaria. Parasitol. Res. 90(Suppl. 2), S71–S76 10.1007/s00436-002-0770-9 [DOI] [PubMed] [Google Scholar]
- Zhang B., Watts K. M., Hodge D., Kemp L. M., Hunstad D. A., Hicks L. M., Odom A. R. (2011). A second target of the antimalarial and antibacterial agent fosmidomycin revealed by cellular metabolic profiling. Biochemistry 50, 3570–3577 10.1021/bi101958h [DOI] [PMC free article] [PubMed] [Google Scholar]