Abstract
The large size of spectrin, the flexible protein promoting reversible deformation of red cells, has been an obstacle to elucidating the molecular mechanism of its function. By studying cloned fragments of the repeating unit domain, we have found a correspondence between positions of selected spectrin repeats in a tetramer with their stabilities of folding. Six fragments consisting of two spectrin repeats were selected for study primarily on the basis of the predicted secondary structures of their linker regions. Fragments with a putatively helical linker were more stable to urea- and heat-induced unfolding than those with a putatively nonhelical linker. Two of the less stably folded fragments, human erythroid α-spectrin repeats 13 and 14 (HEα13,14) and human erythroid β-spectrin repeats 8 and 9 (HEβ8,9), are located opposite each other on antiparallel spectrin dimers. At least partial unfolding of these repeats under physiological conditions indicates that they may serve as a hinge. Also less stably folded, the fragment of human erythroid α-spectrin repeats 4 and 5 (HEα4,5) lies opposite the site of interaction between the partial repeats at the C- and N-terminal ends of β- and α-spectrin, respectively, on the opposing dimer. More stably folded fragments, human erythroid α-spectrin repeats 1 and 2 (HEα1,2) and human erythroid α-spectrin repeats 2 and 3 (HEα2,3), lie nearly opposite each other on antiparallel spectrin dimers of a tetramer. These clusterings along the spectrin tetramer of repeats with similar stabilities of folding may have relevance for spectrin function, particularly for its well known flexibility.
The striking, elongated shape of members of the spectrin superfamily of cytoskeletal proteins reflects their formation of a chain from multiple repeating units. These repeats of ≈106 amino acids were first modeled by Speicher and Marchesi (1) as triple helical units connected by nonhelical linkers. The limited homology among the amino acid sequences of these repeats (the 21 and 16 repeating units, respectively, of the α and β monomers of spectrin are on average only 20-30% homologous) is likely to be relevant to functions of spectrin's erythroid (2, 3) and nonerythroid (4, 5) forms, because major steps in their evolution have been deduced in part from their sequences (6, 7). A relationship between the functions of repeats and their sequence differences is also supported by the specific affinities of different repeats in spectrin and its relatives, α-actinin and dystrophin, for a variety of domains (8).
Recent results of biophysical studies of cloned spectrin repeats have raised the prospect of relating some of their sequence variability to their differential stabilities of folding by focusing on their linker regions. Specifically, the finding by x-ray crystallography that the linker regions associated with two repeats of chicken brain α-spectrin (9) and two (10) and four (11) repeats of α-actinin are α-helical and the definitive demonstration that two connected repeats are more stably folded than a single repeat (12) led to the design of the study reported here. The selection of two-repeat fragments likely to vary in their stabilities of folding was based primarily on secondary structure predictions for the linker regions between repeats of human red cell α-spectrin (13), human red cell β-spectrin (14), and chicken brain α-spectrin (15).
Three pairs of repeats connected by linkers predicted to be α-helical by the discrimination of secondary structure class (dsc) program (16) were found to unfold at higher urea concentrations than three pairs of repeats connected by linkers predicted to adopt a random coil. This difference was corroborated by measuring thermal unfolding in the absence or presence of a physiological NaCl concentration, which occurred at 30-40°C for the less stably folded fragments but at 50-60°C for the more stably folded fragments. The 30-40°C unfolding temperatures of certain repeats support earlier fluorescence polarization (17), [1H]NMR (18) and EPR (19) measurements indicating that intact spectrin is unfolded to some degree under physiological conditions.
Interestingly, stabilities of folding of the two-repeat fragments were found to be correlated with their positions in a spectrin tetramer. Pairs of more stably folded repeats in one dimer of a tetramer were determined to be located nearly opposite similarly more stably folded pairs in the coaligned dimer, whereas the less stably folded pairs in a dimer of a tetramer were determined to be located either opposite similarly less stably folded pairs or opposite the site of interaction of the partial repeats at the C- and N-terminal ends of β- and α-spectrin, respectively, in the coaligned dimer. The at-least-partially unfolded state of the juxtaposed, less stably folded pair of repeats is compatible with hinge-like motion (20). Thus, variable stability of folding may be coordinated along the lengths of two associated, antiparallel erythroid spectrin dimers, perhaps for the purposes of modulating the flexibility of and/or promoting long-distance effects along these chains of repeats (21).
Materials and Methods
Secondary Structure Prediction. Amino acid sequences of repeating units of human erythroid α-spectrin (HEα) (13), human erythroid β-spectrin (HEβ) (14), and chicken brain α-spectrin (15) were phased by ascertaining the 14th residue to be the nearly invariant tryptophan, as in the x-ray crystal structures of the 16th and 17th repeating units of chicken brain α-spectrin (9). Repeats without a nearly invariant tryptophan, e.g., repeats 9 and 15 of HEβ, were phased by alignment with the dozen or so other residues highly conserved in most repeats and with the repeating heptad pattern of residues stabilizing each repeat (22). The aligned sequences were analyzed with the secondary structure prediction program DSC (available at http://workbench.sdsc.edu; ref. 16) or, subsequently, with the program psipred (ref. 23, available at www.expasy.org; ref. 24).
Cloning and Expression. Cloning was performed (9, 25) by ligating PCR products into pET3d [chicken brain α-spectrin repeats 15 and 16 (CBα15,16, residues 1662-1876), HEα2,3 (residues 160-371)], pET3c [HEα1,2 (residues 55-265), HEα13,14 (residues 1396-1606)], pET30b [“new” HEβ8,9 (residues 1063-1275)], pET23d [“old” HEβ8,9 (residues 1063-1275)], or pGEX 4T-1 [HEα4,5 (residues 1300-1936)] to transform BL21,DE3 Escherichia coli with the cDNA-containing plasmid. The old and new HEβ8,9 differ in their N-terminal amino acids, glutamic acid in the former but leucine in the latter. The two forms gave the same CD222 and tryptophan fluorescence intensities and stabilities of folding, but the yield of old HEβ8,9 was significantly greater than that of new HEβ8,9. The cDNAs for all proteins had the correct sequences except for (i) the previously mentioned N terminus of the high-yield HEβ8,9, (ii) those constructs ligated to pET3d vector in which the N-terminal residue was changed to valine to accommodate the restriction enzyme site for NCOI, and (iii) HEα4,5 with glycine and serine at N-terminal and n + 1 positions to create a glutathione transferase fusion protein. The 14th residue of HEα13,14, at which a polymorphism occurs (13), was tryptophan, instead of arginine.
Isolation and Purification. For constructs ligated to pET vectors, isolation and purification of the expressed protein was performed as described (9, 25). To purify the pGEX 4T-1 construct HEα4,5, cells in 20 mM Tris·HCl, pH 8.0/1 mM EDTA/0.15 mM PMSF/1 mM DTT were sonicated to release the expressed glutathione transferase fusion protein and centrifuged to remove cell debris, and the supernatant was adjusted to 0.14 M NaCl before affinity chromatography on a glutathione-Sepharose 4B column. HEα4,5 was isolated by thrombin cleavage of the fusion protein and further purified by Q-Sepharose and S-100 chromatography. Protein concentrations were determined from their extinction coefficients at 280 nm, which were calculated from their amino acid sequences. All cloned proteins were at least 99% pure as judged by SDS/PAGE, and their masses by electrospray mass spectrometry were within 0.08-0.6% of their calculated masses.
Analytical Ultracentrifugation. The apparent molecular weight of each protein in solution was obtained by analytical ultracentrifugation, as described (12). Proteins from 100 μg/ml to 1 mg/ml in 0.1 M NaCl/10 mM sodium phosphate, pH 7.5/1 mM EDTA were centrifuged to equilibrium at 21,100 rpm in a Beckman XL-A analytical ultracentrifuge at 20°C. Data were fit with sigmaplot 2000 to the equation for a single, ideally behaving species, 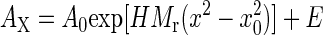 , where AX is the A280 at radius x, A0 is the A280 at the reference radius x0, H is a constant, and E is the baseline offset to obtain Mr, the apparent molecular weight.
, where AX is the A280 at radius x, A0 is the A280 at the reference radius x0, H is a constant, and E is the baseline offset to obtain Mr, the apparent molecular weight.
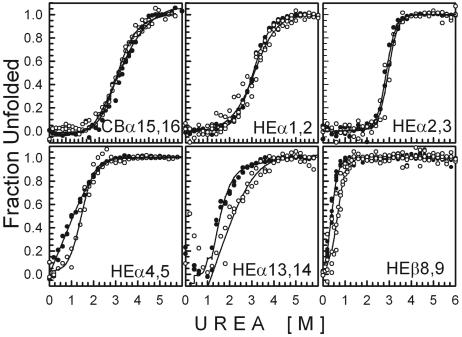
Urea-Induced Unfolding. Urea-induced unfolding was performed as described (12, 25). CD222 and tryptophan fluorescence intensities were collected from at least two sets of samples, corrected for background, and converted into fraction unfolded values presented in Fig. 1.
Fig. 1.
Fraction unfolded of each two-repeat spectrin fragment plotted against increasing concentrations of urea plus 10 mM sodium phosphate, pH 8. Filled circles indicate CD at 222 nm; open circles indicate tryptophan fluorescence.
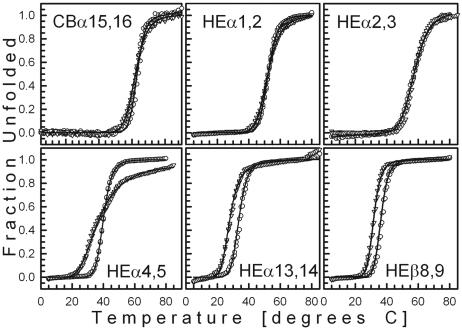
Temperature-Induced Unfolding. Temperature-induced unfolding was performed as described (12). CD222 measurements were performed on at least two sets of samples, corrected for background, and converted into fraction unfolded values presented in Fig. 2.
Fig. 2.
Fraction unfolded of each two-repeat spectrin fragment monitored by CD at 222 nm plotted against increasing temperature. Inverted triangles indicate 10 mM sodium phosphate, pH 8; open circles indicate 0.14 M NaCl plus 10 mM sodium phosphate, pH 7.5.
As described (12), the free energy of temperature-induced unfolding, ΔGUN, was determined for all of the fragments in 0.14 M NaCl/10 mM sodium phosphate, pH 7.4, the unfolding of which appeared to be a two-state, reversible process. (Because the unfolding of HEα4,5 in 10 mM sodium phosphate, pH 8, involved at least three states, the two-state model could not be applied uniformly to all of the fragments in this buffer.) As described (12), the free energy of unfolding, ΔGUN, can be expressed in terms of the CD signal, y, at any temperature, T, in K by
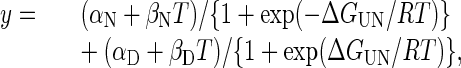 |
[1] |
where α is the y intercept of the native (N) or denatured (D) state, β is the slope of the native (N) or denatured (D) state and R is the gas constant, 1.987 cal·mol-1·K-1. This equation can be simplified to a form suitable for nonlinear regression fitting of the data with sigmaplot 2000
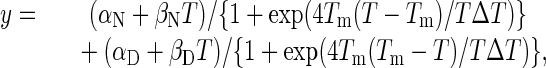 |
[2] |
where Tm is the melting temperature at the midpoint of thermal unfolding and ΔT is the width of the transition. After Tm and ΔT were thus obtained, ΔGUN was calculated from the equation
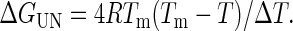 |
[3] |
Results
Secondary Structure Prediction by dsc. The beginning and end of each repeating unit in the cDNA sequences of chicken brain α-spectrin (15), HEα (13), and HEβ (14), or “phasing” of repeats (26), were identified as described in Materials and Methods. Listed in Table 1 are the DSC-predicted conformations of the 20 α-spectrin and 15 β-spectrin linkers connecting two repeating units, which are important, but not the sole, determinants of the stability of folding of each repeat. In 36 of the 55 linkers, all five of the linker amino acids were predicted by DSC to be α-helical, as had been found in the x-ray crystal structures of three linkers in chicken brain α-spectrin (9) and the three linkers in α-actinin (10, 11). (The linkers connecting the first repeat of HEα with the single α-helix at its N-terminal end, found by NMR to be nonhelical (27), and the 16th repeat of HEβ with the two α-helices at its C-terminal end were not included in this survey because their secondary structures may be suited more to linking the partial repeats of α- and β-spectrin than to facilitating spectrin flexibility.) In contrast with the amino acids in the putatively helical linkers, none of the linker region amino acids of fragments such as HEα4,5 and HEβ8,9 and only two of the five linker region amino acids of fragment HEα13,14 were predicted to adopt a helical conformation. Guided by the predictions in Table 1, evolutionary considerations (6, 7), and their positions within the spectrin tetramer, we chose to measure the stabilities of folding of three fragments of two repeats joined by a putatively helical linker: chicken brain α-spectrin repeats 15 and 16 (CBα15,16), HEα repeats 1 and 2 (HEα1,2), and HEα repeats 2 and 3 (HEα2,3); and three fragments of two repeats joined by a putatively nonhelical linker: HEα repeats 4 and 5 (HEα4,5), HEα repeats 13 and 14 (HEα13,14), and HEβ repeats 8 and 9 (HEβ8,9). It should be noted that, when the same spectrin repeat sequences were analyzed by psipred (23) as had been analyzed by DSC, all, not merely 65%, of the linkers were predicted to be α-helical. From the results to be described, however, reliance on the DSC method led to the desired selection of two-repeat fragments with either high or low stabilities of folding, although the DSC predictions themselves did not agree with those of the psipred method. Moreover, we have recently determined by x-ray crystallography that the linker of HEβ8,9, predicted to be nonhelical by DSC, is, in fact, α-helical (53), as predicted by psipred.
Table 1.
Secondary structure prediction of spectrin linker conformation by the dsc method (16)
| Linkers between repeats | HEα* | HEβ* | CBα* |
|---|---|---|---|
| 1 and 2 | 5.0 | 5.0 | 5.0 |
| 2 and 3 | 5.0 | 5.0 | 3.0 |
| 3 and 4 | 0.0 | 5.0 | 5.0 |
| 4 and 5 | 0.0 | 3.0 | 5.0 |
| 5 and 6 | 5.0 | 5.0 | 5.0 |
| 6 and 7 | 5.0 | 3.0 | 5.0 |
| 7 and 8 | 5.0 | 5.0 | 5.0 |
| 8 and 9 | 3.0 | 0.0 | 5.0 |
| 9 and 10 | 3.0 | 5.0 | 3.0 |
| 10 and 11 | 1.0 | 2.0 | 2.0 |
| 11 and 12 | 5.0 | 2.0 | 5.0 |
| 12 and 13 | 5.0 | 5.0 | 1.0 |
| 13 and 14 | 2.0 | 5.0 | 5.0 |
| 14 and 15 | 5.0 | 5.0 | 5.0 |
| 15 and 16 | 5.0 | 3.0 | 5.0 |
| 16 and 17 | 5.0 | NA | 5.0 |
| 17 and 18 | 3.0 | NA | 5.0 |
| 18 and 19 | 5.0 | NA | 5.0 |
| 19 and 20 | 5.0 | NA | 5.0 |
| 20 and 21 | 1.0 | NA | 3.0 |
NA, not applicable.
Numbers of helical residues among the five residues of the linker region
CBα15,16, HEα1,2, HEα2,3, HEα4,5, HEα13,14, and HEβ8,9 are folded and exist as monomers in solution. The cloned spectrin fragments are well folded, as indicated by their mean residue ellipticities in degrees·cm2·dmol-1 at 222 nm and 10°C listed in Table 2. The ellipticities range from -18,483 ± 568 for HEα4,5 to -29,216 ± 644 for HEβ8,9 or from 51% to 81% of the maximum for homopolymers of amino acids (28). Similar values were obtained previously for other one- and two-repeat fragments (12, 25). The presence of physiological concentrations of NaCl did not affect these values, nor did prior unfolding of the proteins in 6 M guanidine hydrochloride followed by refolding on removal of the denaturant by dialysis. Also indicating their folded condition, the wavelengths of maximum tryptophan fluorescence in Table 2 ranged from 328 nm for HEα2,3 to 340 nm for HEα4,5, HEα13,14, and HEβ8,9. The longer wavelengths of tryptophan emission of the last three proteins with putatively nonhelical linkers signify greater exposure of their tryptophans to the bulk phase than those of the fragments with putatively helical linkers, but not to the extent of urea denatured proteins with tryptophan emission maxima at 354 nm (25). Finally, molecular weights of the cloned spectrin fragments obtained by analytical ultracentrifugation in Table 2 are within 15% of their calculated molecular weights, thus indicating that these proteins are monomers in solution.
Table 2. Ellipticities at 222 nm and wavelengths of maximum tryptophan fluorescence of two-repeat fragments of spectrin.
| Fragment | θ* | Fluorescence†, nm | Mr‡, g/mol |
|---|---|---|---|
| CBα15,16 | -28,032 ± 287 | 330 | 26,035 ± 779 |
| HEα1,2 | -24,554 ± 3,539 | 335 | 27,435 ± 237 |
| HEα2,3 | -26,739 ± 4,528 | 328 | 21,856 ± 260 |
| HEα4,5 | -18,483 ± 568 | 340 | 25,362 ± 208 |
| HEβ8,9 | -29,216 ± 644 | 340 | 21,719 ± 80 |
| HEα13,14 | -18,618 ± 2,181 | 340 | 23,551 ± 514 |
Molar ellipticity ± standard error at 222 nm and 10°C in degree·cm2·dmol-1
Wavelength of maximum tryptophan emission
Molecular weight ± standard error by analytical ultracentrifugation; protein concentrations ranged from 0.1 to 1 mg/ml
Urea-Induced Unfolding of Cloned Spectrin Fragments. The stabilities of folding of the cloned spectrin fragments were measured by their CD222 (Fig. 1, filled circles) and tryptophan fluorescence (Fig. 1, open circles) on exposure to increasing urea concentrations, expressed as fraction unfolded values in Fig. 1. As indicated by the solid curves, related sets of data were fit simultaneously to the equation describing urea-induced unfolding as a two-state process (12, 25), according to the method of Jackson et al. (29). All curves exhibit the cooperative feature characteristic of protein unfolding. However, a striking difference between Fig. 1 Upper and Lower is that higher urea concentrations are necessary to unfold repeats connected by a putatively helical linker (Fig. 1 Upper) than to unfold repeats connected by a putatively nonhelical linker (Fig. 1 Lower). The urea concentration at a fraction unfolded value of 0.5 is ≈3-4.5 M for fragments predicted to possess a helical linker, whereas the urea concentration at a fraction unfolded value of 0.5 is <2 M for fragments predicted to possess a nonhelical linker. Hence, from these urea concentrations, a helical linker appears to be associated with greater stability of folding than a nonhelical linker, as might be expected from the greater exposure of tryptophans in the latter than in the former (Table 2).
A second obvious difference between Fig. 1 Upper and Lower is the coincidence of CD and fluorescence unfolding curves of repeats connected by a putatively helical linker (CBα15,16, HEα1,2, and HEα2,3) as opposed to the noncoincidence of CD and fluorescence unfolding curves of repeats connected by a putatively nonhelical linker (HEα4,5, HEα13,14, and HEβ8,9). Thus, for fragments with a nonhelical linker, urea-induced unfolding involves one or more intermediate states, whereas coincidence of the CD and fluorescence unfolding curves of fragments with a helical linker is consistent with the absence of intermediate states.
Heat-Induced Unfolding of Cloned Spectrin Fragments. To verify their differential responses to urea by another method, the cloned spectrin fragments were exposed to increasing temperature, and their CD222 was monitored. After an increase in temperature from 5°C to 80-85°C at a rate of 1°C per min, the temperature was decreased to 10°C at a rate of 2°C per min, and a scan was taken from 250 to 190 nm (not shown) to establish the reversibility of temperature-induced unfolding. The results of heating are presented as fraction unfolded values in Fig. 2, symbolized by triangles when obtained in 10 mM sodium phosphate, pH 8, and by circles when obtained in 0.14 M NaCl plus 10 mM sodium phosphate, pH 7.5. Each solid curve represents the best fit of related data sets to Eq. 2 (Materials and Methods), describing temperature-induced unfolding as a two-state process.
In agreement with urea-induced unfolding, fragments with putatively helical linkers (Fig. 2 Upper) unfold at higher temperatures than fragments with putatively nonhelical linkers (Fig. 2 Lower) in the absence or presence of 0.14 M NaCl. Moreover, all curves but one exhibit a single transition consistent with unfolding occurring as a single transition. The exceptional curve with two inflection points is that of HEα4,5 in 10 mM sodium phosphate, pH 8, without NaCl. This two-transition unfolding of HEα4,5 in the absence of salt was also observed by differential scanning calorimetry. HEα4,5 gave two clear endothermic peaks at 32°C and at 44°C in 10 mM sodium phosphate, pH 8, but only one peak at 41.5°C in physiological NaCl, whereas HEβ8,9 and HEα2,3 gave a single endothermic peak in both buffers (not shown).
Because thermal unfolding data for all fragments in the presence of 0.14 M NaCl were consistent with a two-state transition, unlike those for HEα4,5 in the absence of salt, we could obtain free energies of thermal unfolding, ΔGUN, by fitting the data in the presence of salt to Eq. 2 and solving for ΔGUN with Eq. 3 (Materials and Methods). The midpoints of unfolding of CBα15,16, HEα1,2 and HEα2,3 in the presence of NaCl (Fig. 2 Upper) were significantly higher (i.e., 63.0°C, 52.3°C, and 57.9°C, respectively) than the midpoints of unfolding of HEα4,5, HEα13,14, and HEβ8,9 in the presence of NaCl (Fig. 2 Lower) (i.e., 40.0°C, 36.0°C, and 33.8°C, respectively). Not only were temperatures at the midpoint of unfolding higher for CBα15,16, HEα1,2, and HEα2,3 than those for HEα4,5, HEα13,14, and HEβ8,9, but free energies for thermal unfolding in the presence of 0.14 M NaCl were higher for the former than for the latter (i.e., 10.8, 6.0, and 4.5 kcal/mol for CBα15,16, HEα1,2, and HEα2,3, respectively, vs. 2.0, 3.0, and 3.6 kcal/mol for HEα4,5, HEα13,14, and HEβ8,9, respectively). Hence, the conformation predicted for the linker of each two-repeat fragment by the DSC method is correlated with its stability of folding to heat as well as to urea; i.e., a putatively helical linker is associated with greater stability of folding and a putatively nonhelical linker is associated with lower stability of folding.
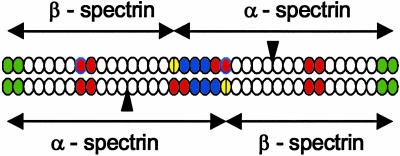
Model of Spectrin Tetramer Places Less Stably Folded Fragments Across from Each Other or Across from the Partial Repeat Association Site on Opposing Dimers and More Stably Folded Fragments Across from Each Other on Opposing Dimers. The positions on a spectrin tetramer of the two-repeat spectrin fragments characterized here are indicated on a model of the repeating unit domain of a tetramer in Fig. 3, based on the dimer model of De Matteis and Morrow (4). The less stably folded fragments, HEα4,5, HEα13,14, and HEβ8,9, are red, the more stably folded fragments, HEα1,2 and HEα2,3, are blue, the repeats formed by interaction of the two helices at the C-terminal end of β-spectrin with a single helix at the N-terminal end of α-spectrin are yellow, and the repeats, HEα20, HEα21, HEβ1, and HEβ2, which interact to form dimers, are green. The black triangles between the 9th and 10th repeating units of each HEα (13) represent Src homology 3 (SH3) domains, which are unlikely to add to the lengths of the two α-spectrin monomers because the x-ray crystal structure of the chicken brain α-spectrin SH3 domain shows that its N and C termini lie close to each other (30), in contrast with the N and C termini of a repeating unit (9, 10). The register of repeats on aligned, opposing monomers in the dimer model of De Matteis and Morrow (4) is supported by the finding that repeats HEα20 and HEα21EF bind to repeats HEβ1 and HEβ2 with high affinity (31).
Fig. 3.
Model of repeating unit domain of two antiparallel dimers of a human erythroid spectrin tetramer. Less stably folded repeats are red; more stably folded repeats are blue; repeats as yet uncharacterized with respect to their stability of folding are white; the site of interaction of two α-helices at the C terminus of β-spectrin and 1 α-helix at the N terminus of α-spectrin is represented by divided, yellow ovals; the dimer association sites are green.
Data on the folding stabilities of the repeats in this study point to the clustering of two-repeat fragments of similar stabilities of folding opposite each other in the spectrin tetramer. Thus, the less stably folded HEα4,5 on one dimer lies nearly opposite the readily dissociable (32) site of interaction of partial α and β repeats on the antiparallel, opposing dimer, and the less stably folded HEβ8,9 on one dimer lies exactly opposite the less stably folded HEα13,14 on the antiparallel, opposing dimer. It is intriguing that because HEβ8,9 and HEα13,14 are at least partially unfolded at physiological temperatures and located near each other, these fragments are well positioned to act as a hinge in the spectrin tetramer. Also clustered are the more stably folded HEα1,2 and HEα2,3, which lie nearly opposite each other in the middle of the tetramer.
Discussion
Clustering of Two-Repeat Fragments of Similar Stabilities of Folding in the Same Regions of a Spectrin Tetramer. A major finding of this investigation is the correlation between (i) the stability of folding to urea (Fig. 1) and to temperature (Fig. 2) of each of five pairs of connected repeats of human erythroid spectrin and (ii) the location of each of those pairs of repeats on a human erythroid spectrin tetramer (Fig. 3). This correlation suggests a coordinated variability in the stabilities of folding of repeats along two antiparallel spectrin dimers of a tetramer. Hence, of three two-repeat fragments found to be less stably folded than two other two-repeat fragments, HEβ8,9 on one dimer is opposite HEα13,14 on the antiparallel dimer of the same tetramer, whereas HEα4,5 on one dimer is opposite the site of interaction of partial repeats of α- and β-spectrin on the antiparallel dimer. The two two-repeat fragments, HEα1,2 and HEα2,3, found to unfold at a higher urea concentration and at a higher temperature than HEβ8,9, HEα13,14, and HEα4,5, are nearly opposite each other on antiparallel dimers of a tetramer. This “mapping” of the stabilities of folding of cloned regions of the spectrin repeating domain suggests that the varying stability of folding across the whole of intact spectrin can be assessed by similar methods.
Relationship Between Predictions of Secondary Structure and Stabilities of Folding. Predictions by the DSC algorithm of the secondary structures of their linkers enabled the selection of less stably folded fragments on one hand and more stably folded fragments on the other hand. However, the DSC predictions do not agree with (i) the prediction by a different, neural network approach, i.e., psipred, that all linkers are α-helical and (ii) our finding of an α-helical linker in HEβ8,9 by x-ray crystallography (53), although this linker was predicted to be nonhelical by DSC. It appears that DSC has predicted the relative stabilities of the linker regions more accurately in this case than their secondary structures. Nonetheless, the unfolding data presented here affirm the usefulness of secondary structure prediction methods when used with discretion.
Urea-Induced Unfolding as a Measure of Stability of Folding. Urea-induced unfolding (33) continues to be an important tool in the study of protein folding, although the mechanism of this longused method is still under investigation (34) and limitations of its application are well known (35, 36). As discussed in the latter references (35, 36) and pertinent to urea-induced unfolding in the present case, pathways of unfolding involving intermediate states are not amenable to two-state analysis. Hence, because intermediate states were indicated by the noncoincidence of CD and fluorescence urea unfolding curves of fragments with putatively nonhelical linkers (Fig. 1 Lower), evaluation of the urea data with a two-state model (29) is inappropriate. Nevertheless, the greater urea sensitivity of fragments with putatively nonhelical linkers (Fig. 1 Lower) than fragments with putatively helical linkers (Fig. 1 Lower) was large enough that visual examination of the graphs sufficed to show that the former were less stably folded than the latter. Whether intermediate states of unfolding reflect heightened susceptibility to chemical denaturation remains to be determined, although such a correlation might intuitively seem to be reasonable.
Heat-Induced Unfolding as a Measure of Stability of Folding. Data in Fig. 2 for heat-induced unfolding in the presence of salt provide quantitative corroboration of the qualitative determination by urea-induced unfolding that fragments with putatively helical linkers are more stably folded than fragments with putatively nonhelical linkers. Not only were temperatures at the midpoint of unfolding in the presence of 0.14 M NaCl higher for CBα15,16, HEα1,2, and HEα2,3, i.e., 63.0°C, 52.3°C, and 57.9°C, respectively (Fig. 2 Upper), than temperatures at the midpoint of unfolding in the presence of NaCl for HEα4,5, HEα13,14 and HEβ8,9, i.e., 40.0°C, 36.0°C, and 33.8°C, respectively (Fig. 2 Lower), free energies for thermal unfolding in the presence of 0.14 M NaCl were also higher for the former than for the latter; i.e., 10.8, and 6.0, and 4.5 kcal/mol for CBα15,16, HEα1,2, and HEα2,3, respectively, vs. 2.0, 3.0, and 3.6 kcal/mol for HEα4,5, HEα13,14, and HEβ8,9, respectively.
The data showing that certain clustered repeats within the spectrin tetramer are less structured at 37°C than other repeats confirm and expand on previous results suggesting that intact spectrin becomes more flexible at 37°C. The latter results include an increase in the tryptophan fluorescence anisotropy of intact spectrin between 10°C and 37°C (17), an increase in the areas of aliphatic peaks of [1H]NMR spectra of intact spectrin between 6°C and 37°C, indicating partial unfolding of helical, possibly repeat regions (18), and a sharp increase in the mobility of nitroxide spin labels covalently attached by maleimide to cysteines of spectrin dimers and tetramers in situ or reassociated with red cell ghosts near 37°C (19). (The fragments shown to be less stably folded here could have been targeted in the spin label study because HEα4,5 and HEβ8,9 each contain a single cysteine, whereas HEα13,14 contains three cysteines.) This collective evidence for enhanced spectrin flexibility at 37°C, in turn, is pertinent to the recent demonstration that raising the temperature to 37°C causes intact red cells to suddenly squeeze into a 1.3-μm capillary pipet (37). Such a manifestation of red cell deformability might result, at least in part, from the previously cited, enhanced spectrin flexibility at physiological temperatures. Thus, the present data on the stabilities of folding of spectrin repeats have enabled identification of specific repeats which could play a key role in the reversible deformation of red cells under physiological conditions.
Potential Hinge(s) in Intact Spectrin. Whereas the near physiological Tm values of HEα13,14 and HEβ8,9 facing each other on a tetramer are suggestive of a hinge, the role of HEα4,5 is less clear-cut because it lies opposite the dissociable tetramer formation site. This site is likely to be folded at a physiological temperature, according to temperature-dependent CD data on fragments of the partial α and β repeats and their respective adjacent complete repeats comprising this site (38), as well as a complex of these α and β fragments (39). Whether other fragments predicted to possess nonhelical linkers by DSC, but helical linkers by psipred, might serve as hinges remains to be determined. There would seem to be a limit to the number of unfolded hinge sites, however, given spectrin's highly cooperative melting temperature at 49°C (40). Finally, the unfolded states of HEα4,5, HEα13,14, and HEβ8,9 at 37°C are unlikely to affect the largely folded state of the rest of the spectrin tetramer because linker regions apparently prevent unfolding of more than one or a few repeats at a time; for example, during step-wise unfolding of repeats of intact spectrin (41) or fragments thereof (42-44) by atomic force microscopy.
Evidence for Varying Flexibility Along the Spectrin Tetramer. A number of morphological studies of isolated spectrin and spectrin networks in situ give the impression that spectrin may not be uniformly flexible along its length. The tortuous trajectory of minimally perturbed erythroid spectrin in situ imaged by electron microscopy (45), and later confirmed by atomic force microscopy of dried (46) and hydrated (47, 48) red cells, implies that spectrin can bend in a nonuniform fashion along its length. Likewise, rotary-shadowed human (49) and avian (50) spectrin dimers and tetramers exhibit different degrees of curvature, although not consistently in the same regions of the molecules. However, portions of coiled strands of spectrin in situ in higher resolution, Fourier-analyzed electron micrographs appeared to be more uniformly flexible along their lengths (51). Regionally varying flexibility might also explain differences in transient electric birefringence data between Drosophila α-spectrin fragments consisting of repeats 14, 15, and 16 on the one hand, versus fragments consisting of repeat 14, repeats 14-15, and repeats 14-17 on the other hand (52). In agreement with most of these earlier data, the present study of the stabilities of folding of cloned fragments of intact spectrin supports the possibility of varying flexibility along its length. Continued pursuit of the strategy of examining cloned repeating units of spectrin should yield a better understanding of otherwise elusive but intriguing characteristics of the intact form of this important cytoskeletal protein, such as a hinge that may facilitate the reversible deformation of red cells.
Acknowledgments
We thank Tina Byun, Lucy Gutierrez, Woo Kim, Flora Lwin, Weina Niu, Amesika Nyaku, David Nyweide, Eugene Park, Jorge Santana, Rachel Ulmer, Roseann Wu, and Kai Zhang for laboratory assistance; Robert Holmgren, Hideki Kusunoki, Robert MacDonald, George Minasov, Alfonso Mondragón, and Jonathan Widom for comments and suggestions; Leszek Kotula for the cDNA of HEα; Bernard Forget for the cDNA of HEβ; Matti Saraste for the cDNA of CBα and for support essential to the origin of this work; the staff of the P30 Reproductive Biology Center of Northwestern University for the sequencing of cDNAs; Jeongrim Lee of the Analytical Chemistry Laboratory of Northwestern University for mass spectrometry analyses; Katharina Spiegel and Joshua Hayes of the Keck Biophysics Facility of Northwestern University for instrumentation support; and Francis Neuhaus for use of his equipment. This investigation was supported by National Institute of General Medical Sciences Grant GM57692 (to R.I.M.).
Abbreviations: HEα, human erythroid α-spectrin; HEβ, human erythroid β-spectrin; CBα, chicken brain α-spectrin.
References
- 1.Speicher, D. W. & Marchesi, V. T. (1984) Nature 311, 177-180. [DOI] [PubMed] [Google Scholar]
- 2.Mohandas, N. & Evans, E. (1994) Annu. Rev. Biophys. Biomol. Struct. 23, 787-818. [DOI] [PubMed] [Google Scholar]
- 3.Lux, S. E. & Palek, J. (1995) in Blood: Principles and Practice of Hematology, eds. Handin, R. I., Lux, S. E. & Stossel, T. B. (J. B. Lippincott, Philadelphia), pp. 1701-1818.
- 4.De Matteis, M. A. & Morrow, J. S. (2000) J. Cell Sci. 113, 2331-2343. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, V. & Baines, A. J. (2001) Physiol. Rev. 81, 1353-1392. [DOI] [PubMed] [Google Scholar]
- 6.Byers, T. J., Brandin, E., Lue, R. A., Winograd, E. & Branton, D. (1992) Proc. Natl. Acad. Sci. USA 89, 6187-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muse, S. V., Clark, A. G. & Thomas, G. H. (1997) J. Mol. Evol. 44, 492-500. [DOI] [PubMed] [Google Scholar]
- 8.Djinoviæ-Carugo, K., Gautel, M., Ylänne, J. & Young, P. (2002) FEBS Lett. 513, 119-123. [DOI] [PubMed] [Google Scholar]
- 9.Grum, V. L., Li, D., MacDonald, R. I. & Mondragón, A. (1999) Cell 98, 523-535. [DOI] [PubMed] [Google Scholar]
- 10.Djinoviæ-Carugo, K., Young, P., Gautel, M. & Saraste, M. (1999) Cell 98, 537-546. [DOI] [PubMed] [Google Scholar]
- 11.Ylänne, J., Scheffzek, K., Young, P. & Saraste, M. (2001) Structure (London) 9, 597-604. [DOI] [PubMed] [Google Scholar]
- 12.MacDonald, R. I. & Pozharski, E. V. (2001) Biochemistry 40, 3974-3984. [DOI] [PubMed] [Google Scholar]
- 13.Sahr, K. E., Laurila, P., Kotula, L., Scarpa, A. L., Coupal, E., Leto, T. L., Linnenbach, A. J., Winkelmann, J. C., Speicher, D. W. & Marchesi, V. T. (1990) J. Biol. Chem. 265, 4434-4443. [PubMed] [Google Scholar]
- 14.Winkelmann, J. C., Chang, J. G., Tse, W. T., Scarpa, A. L., Marchesi, V. T. & Forget, B. G. (1990) J. Biol. Chem. 265, 11827-11832. [PubMed] [Google Scholar]
- 15.Wasenius, V. M., Saraste, M., Salven, P., Eramaa, M., Holm, L. & Lehto, V. P. (1989) J. Cell Biol. 108, 79-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King, R. D. & Sternberg, M. J. (1996) Protein Sci. 5, 2298-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshino, H. & Marchesi, V. T. (1984) J. Biol. Chem. 259, 4496-4500. [PubMed] [Google Scholar]
- 18.Begg, G. E., Ralston, G. B. & Morris, M. B. (1994) Biophys. Chem. 52, 63-73. [DOI] [PubMed] [Google Scholar]
- 19.Cassoly, R., Daveloose, D. & Leterrier, F. (1980) Biochim. Biophys. Acta 601, 478-489. [DOI] [PubMed] [Google Scholar]
- 20.Gerstein, M. & Krebs, W. (1998) Nucleic Acids Res. 26, 4280-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giorgi, M., Cianci, C. D., Gallagher, P. G. & Morrow, J. S. (2001) Exp. Mol. Pathol. 70, 215-230. [DOI] [PubMed] [Google Scholar]
- 22.Parry, D. A. D., Dixon, T. W. & Cohen, C. (1992) Biophys. J. 61, 858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, D. T. (1999) J. Mol. Biol. 292, 195-202. [DOI] [PubMed] [Google Scholar]
- 24.McGuffin, L. J., Bryson, K. & Jones, D. T. (2000) Bioinformatics 16, 404-405. [DOI] [PubMed] [Google Scholar]
- 25.Pantazatos, D. P. & MacDonald, R. I. (1997) J. Biol. Chem. 272, 21052-21059. [DOI] [PubMed] [Google Scholar]
- 26.Winograd, E., Hume, D. & Branton, D. (1991) Proc. Natl. Acad. Sci. USA 88, 10788-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, S., Liao, X., Johnson, M. E. & Fung, L. W.-M. (1999) J. Biomol. NMR 15, 345-346. [DOI] [PubMed] [Google Scholar]
- 28.Greenfield, N. & Fasman, G. D. (1969) Biochemistry 8, 4108-4116. [DOI] [PubMed] [Google Scholar]
- 29.Jackson, S. E., Moracci, M., elMasry, N., Johnson, C. M. & Fersht, A. R. (1993) Biochemistry 32, 11259-11269. [DOI] [PubMed] [Google Scholar]
- 30.Musacchio, A., Noble, M., Pauptit, R., Wierenga, R. & Saraste, M. (1992) Nature 359, 851-855. [DOI] [PubMed] [Google Scholar]
- 31.Harper, S. L., Begg, G. E. & Speicher, D. W. (2001) Biochemistry 40, 9935-9943. [DOI] [PubMed] [Google Scholar]
- 32.An, X., Lecomte, M. C., Chasis, J. A., Mohandas, N. & Gratzer, W. (2002) J. Biol. Chem. 277, 31796-31800. [DOI] [PubMed] [Google Scholar]
- 33.Pace, C. N. (1986) Methods Enzymol. 131, 266-280. [DOI] [PubMed] [Google Scholar]
- 34.Bennion, B. J. & Daggett, V. (2003) Proc. Natl. Acad. Sci. USA 100, 5142-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soulages, J. L. (1998) Biophys. J. 75, 484-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eftink, M. R. (2000) Methods Enzymol. 323, 459-473. [DOI] [PubMed] [Google Scholar]
- 37.Artmann, G. M., Kelemen, C., Porst, D., Büldt, G. & Chien, S. (1998) Biophys. J. 75, 3179-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lecomte, M. C., Nicolas, G., Dhermy, D., Pinder, J. C. & Gratzer, W. B. (1999) Eur. Biophys. J. 28, 208-215. [DOI] [PubMed] [Google Scholar]
- 39.Mehboob, S., Luo, B.-H., Patel, B. M. & Fung, L. W.-M. (2001) Biochemistry 40, 12457-12464. [DOI] [PubMed] [Google Scholar]
- 40.Brandts, J. F., Erickson, L., Lysko, K., Schwartz, A. T. & Taverna, R. D. (1977) Biochemistry 16, 3450-3454. [DOI] [PubMed] [Google Scholar]
- 41.Rief, M., Pascual, J., Saraste, M. & Gaub, H. E. (1999) J. Mol. Biol. 286, 553-561. [DOI] [PubMed] [Google Scholar]
- 42.Lenne, P. F., Raae, A. J., Altmann, S. M., Saraste, M. & Hörber, J. K. (2000) FEBS Lett. 476, 124-128. [DOI] [PubMed] [Google Scholar]
- 43.Altmann, S. M., Grünberg, R. G., Lenne, P. F., Ylänne, J., Raae, A., Herbert, K., Saraste, M., Nilges, M. & Hörber, J. K. (2002) Structure (London) 10, 1085-1096. [DOI] [PubMed] [Google Scholar]
- 44.Law, R., Carl, P., Harper, S., Dalhaimer, P., Speicher, D. W. & Discher, D. E. (2003) Biophys. J. 84, 533-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ursitti, J. A., Pumplin, D. W., Wade, J. B. & Bloch, R. J. (1991) Cell Motil. Cytoskeleton 19, 227-243. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi, M., Miyamoto, H., Sako, Y., Komizu, H. & Kusumi, A. (1998) Biophys. J. 74, 2171-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowakowski, R., Luckham, P. & Winlove, P. (2001) Biochim. Biophys. Acta 1514, 170-176. [DOI] [PubMed] [Google Scholar]
- 48.Swihart, A. H., Mikrut, J. M., Ketterson, J. B. & MacDonald, R. C. (2001) J. Microsc. 204, 212-225. [DOI] [PubMed] [Google Scholar]
- 49.Shotton, D. M., Burke, B. E. & Branton, D. (1979) J. Mol. Biol. 131, 303-329. [DOI] [PubMed] [Google Scholar]
- 50.Coleman, T. R., Fishkind, D. J., Mooseker, M. S. & Morrow, J. S. (1989) Cell Motil. Cytoskeleton 12, 248-263. [DOI] [PubMed] [Google Scholar]
- 51.McGough, A. M. & Josephs, R. (1990) Proc. Natl. Acad. Sci. USA 87, 5208-5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bjørkøy, A., Mikkelsen, A. & Elgsaeter, A. (1999) Biochim. Biophys. Acta 1430, 323-340. [PubMed] [Google Scholar]
- 53.Kusunoki, H., MacDonald, R. I. & Mondragón, A. (2004) Structure (London), in press. [DOI] [PubMed]