Abstract
The identification of endogenous or surrogate ligands for orphan G protein-coupled receptors (GPCRs) represents one of the most important tasks in GPCR biology and pharmacology. The challenge lies in choosing an appropriate assay in the absence of ways to activate the receptor of interest. We investigated the signaling pathway for an orphan GPCR referred to here as vasopressin receptor-related receptor 1 (VRR1) by generating a chimeric receptor, V1a/VRR1. The engineered construct contained vasopressin V1a receptor with all three intracellular loops and C terminus replaced by those of VRR1. The chimera behaved like a typical GPCR when transiently and stably expressed in mammalian cell lines based on radioligand binding and receptor internalization studies. Upon arginine vasopressin stimulation, this chimeric receptor induced robust calcium mobilization and increase of adenylate cyclase activity. The observed signaling activities are through the activation of the chimera instead of endogenously expressed receptors, as single amino acid changes in the second transmembrane regions of the chimera drastically reduced receptor efficacy and potency. Our results suggest that VRR1 has dual signaling properties in coupling to both Gq and GS pathways. Analysis of native VRR1 receptor signaling pathway by using a recently identified ligand for VRR1 confirmed this conclusion and therefore validated the utility of the chimeric receptor approach for signaling pathway identification.
G protein-coupled receptors (GPCRs) can be activated by a variety of extracellular signals including neuropeptides, chemokines, biogenic amines, hormones, lipid-derived mediators, proteases, light, taste, and smell. Upon ligand binding, GPCRs transduce these signals into intracellular responses that regulate cell function via the heterotrimeric G proteins (1). A large fraction of GPCRs are orphan receptors for which the cognate ligands have not yet been identified (2, 3). The most frequent method for identifying GPCR ligands is to screen these orphan receptors against a collection of candidate ligands or tissue extracts. To be successful in this exercise, it is a prerequisite to know the appropriate assay format for a given orphan receptor. Although constitutive activity exhibited by receptor overexpression is often used as an indication of signaling pathways possessed by the receptor, many receptors do not show such activity. In this case, either multiple assays need to be performed “blindly” during ligand screening or the receptor must be forced to couple to a specific signaling pathway via promiscuous or chimeric G proteins (4).
We have identified a putative GPCR, referred to here as vasopressin receptor-related receptor 1 (VRR1), which shows 27% amino acid identity to the oxytocin and vasopressin receptors. VRR1 is selectively expressed in the hypothalamus and retina and maps to chromosome 7p14-15 between markers D7S795 and D7S526, where the retinitis pigmentosa subtype 9 locus was localized (5). VRR1 does not exhibit constitutive activity and, hence, the signaling pathway was not clear (data not shown). Here we describe the creation of a chimeric receptor from the human vasopressin V1a and VRR1 receptors to determine the signaling pathway of VRR1. Our data indicate that VRR1 couples to both Gq and Gs signal transduction pathways. We have used a recently identified endogenous ligand for VRR1 to confirm the coupling of native VRR1 to both Gq and Gs signaling pathways, validating our chimeric receptor approach.
Experimental Procedures
Cloning and Tissue Expression Analysis of VRR1. Primers for 5′- and 3′-RACEs and expression analysis were designed based on the DNA sequence that exhibits homology with vasopressin receptor V1a. VRR1 was amplified from the multiple-tissue cDNA panel (Clontech) by using PCR and the oligonucleotide primers 5′-GCCTGGAGCCTGTCTTTTCTGTTCT-3′ and 5′-GGCAGGTTCTGAATGATCACAGAGG-3′. Amplification was performed for 40 cycles using 0.5 ng of cDNA per reaction (50 μl total reaction volume) generating a 449-bp product. For RACE, hypothalamus-derived Marathon cDNA (Clontech) was used as template. The sense primer was 5′-GCCTGGAGCCTGTCTTTTCTGTTCT-3′ and antisense primer was 5′-GGCAGGTTCTGAATGATCACAGAGG-3′. Full-length cDNA was amplified from hypothalamus cDNA and cloned into pcDNA3.1 (Invitrogen).
Construction of Human Vasopressin Receptor V1a and VRR1 Chimera and Cloning. The extracellular, transmembrane, and intracellular domains for the vasopressin receptor V1a and VRR1 receptor were determined by using the tmhmm2.0 software package (6). The chimeric receptor construct consisted of the N terminus, all three extracellular loops, and all seven transmembrane domains from the V1a receptor and all three intracellular loops and the C terminus from the VRR1 receptor. V1a/VRR1 chimera was synthesized by Blue Heron Biotechnology (Bothell, WA) using the GeneMaker technology according to the sequence specified. The chimeric receptor was then amplified by using PCR and oligonucleotide primers 5′-GCGAATTCATCGATAGATCTCACCATGCGTCTCTCCGC-3′ and 5′-CAACCGGGATCCTCTAGACTAGATGAATTCTGGCTTG-3′ and subcloned into the pFLAG-CMV3 expression vector (Sigma).
Stable Expression of Chimeric Receptor V1a/VRR1 in HEK293 Cells. HEK293 cells were transfected with the expression plasmid pFLAG-CMV3 containing the chimeric receptor by using LipofectAMINE2000 (Invitrogen). After 24 h, cells were replated in selective media containing 0.4 mg/ml antibiotic Geneticin (GIBCO/BRL). Single clones were selected for their resistance to Geneticin. Further selection of clones was done by the intracellular calcium mobilization assay described below.
Measurement of Intracellular Calcium. Mobilization of intracellular calcium was measured as described (7) by using an aequorin-based luminescent assay (Euroscreen, Brussels). Control HEK293 cells and HEK293 cells stably expressing chimeric receptor were transfected with pcDNA3-aequorin expression plasmid by using LipofectAMINE 2000 (Invitrogen). Aequorin luminescence resulting from intracellular calcium mobilization upon addition of increasing concentrations of Arg-8-vasopressin (AVP, Bachem) was measured by using the microplate luminometer (EG & G Bertholt, Gaithersburg, MD). Aequorin assay was also performed on cells transiently transfected with VRR1 expression plasmid and pcDNA3-aequorin expression plasmid. Intracellular calcium mobilization in these cells was measured upon addition of increasing concentrations of the natural ligand (SFRNGVGTGMKKTSFQRAKS).
Site-Directed Mutagenesis of the Chimeric Receptor. Point mutations were introduced in the transmembrane domain II of the chimeric receptor by using the QuikChange site-directed mutagenesis kit (Stratagene) and the mutations were verified by dideoxynucleotide sequencing. One mutant chimera had a point mutation D96A, and the other had a point mutation Q107A (8).
cAMP Assay. Control HEK 293 cells, HEK293 cells stably expressing V1a/VRR1receptor, and HEK 293 cells transiently transfected with VRR1 receptor were incubated with 1 mM 3-isobutyl-1-methylxanthine for 30 min (37°C). Cells were then transferred to 96-well plates (≈1.0-1.5 × 104 cells per well) and then stimulated with increasing concentrations of either AVP or 0.1 μM of the VRR1 natural ligand for 30 min (37°C). Reactions were terminated, and stimulation of adenylyl cyclase was measured by using the Tropix cAMP Screen system (Applied Biosystems).
Membrane Preparation. Membranes were prepared from HEK293 cells (control) and HEK 293 cells stably expressing chimeric receptor V1a/VRR1. Briefly, cells were washed in cold PBS without Ca2+ and Mg2+ and then resuspended in membrane buffer (50 mM Hepes/10 mM MgCl2/2 mM EGTA and protease inhibitors; Roche Molecular Biochemicals no. 1697498). They were then homogenized at 4°C and centrifuged at 1,000 rpm in a Beckman GS-6KR centrifuge for 5 min to pellet the nuclei. The supernatants were decanted and centrifuged at 100,000 × g for 1 h at 4°C. Membrane pellets were resuspended in a small volume of membrane buffer and homogenized. Membrane protein concentration was determined by the Bradford assay (Bio-Rad) using BSA as standard. Aliquots of membranes were stored at -80°C.
Radioligand Binding Assay. Binding assay was carried out as described (7). Briefly, 10-μg membrane proteins from control HEK293 cells and HEK293 cells stably expressing V1a/VRR1 receptor were mixed with 0-10 nM 3H-labeled AVP (specific activity, 65.5 Ci/mmol; NEN; 1 Ci = 37 GBq) in the binding buffer [50 mM Hepes/10 mM MgCl2/0.15 mM Bacitracin (Sigma), Roche Molecular Biochemicals protease inhibitors/0.5% BSA, pH 7.6]. Nonspecific binding was determined in the presence of 100 μM unlabeled AVP. Incubation was carried out for 1 h at room temperature followed by filtration through a pretreated (0.5% polyethyleneimine) 96-well GF/B plate on a Packard Filtermate Cell Harvester and washed with buffer containing 50 mM Hepes and 0.1% BSA. Scintillation liquid (40 μl, Packard MicroScint) was added, and the radioactivity was counted on a Packard Microplate Topcount. All experiments were performed in triplicates, and the data were averaged. Kd and Bmax values were evaluated by the prism software package (GraphPad, San Diego).
Receptor Internalization by Immunofluorescence. Control HEK293 cells and stably transfected HEK293 cells expressing chimeric receptor were seeded on glass coverslips coated with poly(d-lysine) (Sigma). Internalization was induced by incubation of cells in the presence of 10 μM AVP for 20 min at 37°C. The cells were washed with cold PBS and then fixed with 4% paraformaldehyde for 20 min. Cells were again washed with cold PBS, then blocking agent, 3% BSA in PBS, was added on the cells for 30 min. For internal staining, cells were permeabilized with 0.2% Triton in PBS for 15 min. Permeabilized and nonpermeabilized (for surface staining) cells were incubated with 1 μg/ml primary anti-flag mouse monoclonal M2 antibody (Sigma) for 1 h followed by 1-h incubation with secondary fluorescein conjugated goat anti-mouse antibody (1:500 in 3% BSA) (Jackson ImmunoResearch). Samples were examined by standard fluorescence microscopy using a Nikon fluorescence microscope.
Results and Discussion
Molecular Cloning and Expression Analysis of VRR1. As part of an ongoing program to mine orphan GPCRs from the human genome, we identified partial sequences that display homology with vasopressin receptors (7, 9). The corresponding full-length human cDNA was then isolated by using 5′- and 3′-RACEs using Marathon cDNAs made from hypothalamus RNA. The predicted protein sequence of this gene, referred to as VRR1, is aligned and compared with other GPCRs in Fig. 1 A and B. Among the known vertebrate GPCRs, VRR1 exhibits the greatest sequence similarity with the oxytocin-vasopressin receptor family (Fig. 1 A). VRR1 and human vasopressin V1a receptor share 27% primary amino acid sequence identity as illustrated in Fig. 1B. Sequence conservation between the two receptors is higher in the transmembrane regions at 36%. Interestingly, a blast (10) search of the Drosophila melanogaster genome revealed the existence of a fly homolog, CG6111. Recently, the crustacean cardioactive peptide (CCAP) was shown to be a high-affinity ligand for CG6111 (11, 12). However, VRR1 is not activated by vasopressin, oxytocin, or CCAP, despite its sequence similarities to these receptors (data not shown).
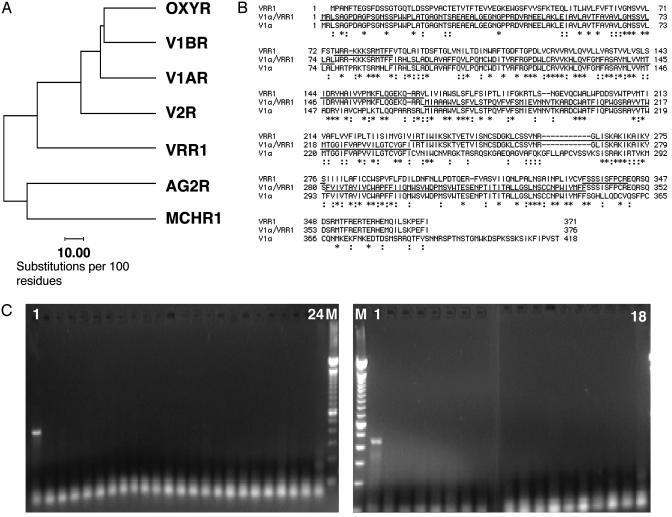
Fig. 1.
(A) Phylogenetic tree generated by the Genetics Computer Group (GCG) programs distances and growtree (GCG, Wisconsin Package Version 10.3; Accelrys, San Diego) showing VRR1 closely related to the oxytocin-vasopressin receptor family. The similarities of the protein sequences between two receptors are inversely proportional to the lengths of the horizontal lines connecting the receptors. (B) Amino acid sequence alignment of VRR1, vasopressin receptor V1a, and chimera V1a/VRR1 generated by the clustalw program (17). Asterisks indicate identical amino acid residues, and colons indicate conserved residues. The underlined V1a/VRR1 sequence matches the V1a receptor, and the overlined V1a/VRR1 sequence matches the VRR1 receptor. (C) Analysis of VRR1 expression patterns by RT-PCR. PCR primers that span two exons were used. RNA samples derived from the following human tissues were analyzed. (Left) Lane 1, hypothalamus; lane 2, whole brain; lane 3, hippocampus; lane 4, pituitary gland; lane 5, amygdala; lane 6, adipocyte; lane 7, pancreas; lane 8, adrenal gland; lane 9, stomach; lane 10, small intestine; lane 11, liver; lane 12, skeletal muscle; lane 13, heart; lane 14, kidney; lane 15, placenta; lane 16, universal; lane 17, lung; lane 18, thymus; lane 19, spleen; lane 20, skin; lane 21, prostate; lane 22, colon; lane 23, s.c. fat; lane 24, genomic DNA; lane M, marker. (Right) M, marker; lane 1, retina; lane 2, leukemia, promyelocytic; lane 3, XG prostatic adenocarcinoma; lane 4, XG ovarian carcinoma; lane 5, XG renal carcinoma; lane 6, leukemia, lymphoblastic; lane 7, colorectal adenocarcinoma; lane 8, XG melanoma; lane 9, lymphoma, Raji; lane 10, fetal brain; lane 11, HeLa; lane 12, XG lymphoma, Daudi; lane 13, XG lung carcinoma; lane 14, leukemia, myelogenous; lane 15, XG mammary carcinoma; lane 16, XG colon adenocarcinoma; lane 17, XG glioblastoma; lane 18, genomic DNA.
To help understand the potential physiological role of VRR1, we carried out studies to examine the tissue distribution of VRR1 by performing PCR on multiple human tissue cDNAs using primers that anneal to separate exons to avoid genomic amplification. As shown in Fig. 1C, VRR1 mRNA was only found in the hypothalamus and retina. No expression was detected in 25 normal tissues, including other parts of the central nervous system, and 15 cancer samples. The restricted expression pattern of VRR1 in components of the retinohypothalamic tract suggests that VRR1 may be involved in light detection and the control of circadian rhythm (12).
By searching the human genome database (www.ncbi.nlm.nih.gov/genome/seq/page.cgi?F=HsBlast.html&&ORG=Hs), the VRR1 gene was localized to the retinitis pigmentosa 9 locus. Retinitis pigmentosa is a hetergenous, retinal degenerative disease. Subtype 9 has been mapped to human chromosome 7p14-15. Together with the expression data in retina, this strongly implicates VRR1 as a candidate gene for the defect in RP9 patients. Fine mapping and the sequencing of DNA samples from the RP9 patients are necessary to confirm this hypothesis.
Construction of Chimeric Receptor V1a/VRR1 and Expression of the Chimera in HEK293 Cells. To develop a functional assay for ligand identification, we investigated the signal-transduction pathway of VRR1. This receptor does not exhibit any constitutive activity in various assays, including inositol phosphate accumulation, cAMP stimulation, and cAMP inhibition (data not shown). To allow the activation of signaling pathways downstream of VRR1, we created a chimeric receptor V1a/VRR1, which contained the predicted extracellular and transmembrane domains of vasopressin V1a receptor and the cytoplasmic regions of the VRR1 receptor (Fig. 2A). This approach has been successfully applied to the Frizzled receptor family (13-16). V1a was chosen to make the chimera because of its sequence homology with VRR1, facilitating the determination of appropriate gene fusion boundaries.
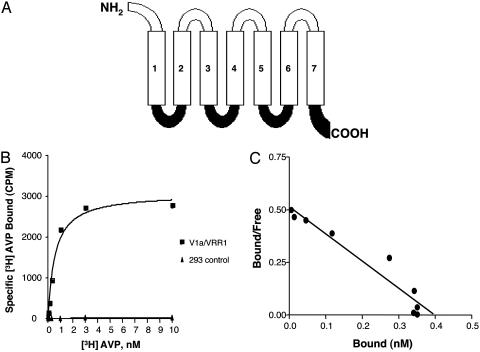
Fig. 2.
(A) Schematic representation of chimeric receptor V1a/VRR1. The chimera consists of the N terminus, extracellular loops, and transmembrane domains of the V1a receptor (open) and the intracellular loops and C terminus of the VRR1 receptor (filled). (B) Saturation binding of [3H]AVP (0 to 10 nM) to V1a/VRR1 (squares) and to control HEK293 membranes (triangles). V1a/VRR1 bound to AVP with a dissociation constant (Kd) of 0.56 ± 0.1 nM. (C) Scatchard plot. Bmax was ≈3.8 pmol/mg membrane protein, corresponding to 0.38 nM.
After transfection of flag-epitope tagged V1a/VRR1 into HEK293 cells, we identified stable cell lines by measuring the cell surface expression of V1a/VRR1 with fluorescence-activated cell sorting (FACS) analysis (data not shown). To further validate the cell surface expression of V1a/VRR1, we performed radio-ligand binding by using tritiated AVP. As shown in Fig. 2 B and C, the membranes prepared from HEK293 cells stably expressing V1a/VRR1 receptor exhibited specific and saturable binding to radiolabeled AVP with a Kd = 0.56 ± 0.1 nM and Bmax = 3.8 pmol/mg membrane protein. As controls, membranes prepared from untransfected HEK293 cells were devoid of any specific binding to the ligand.
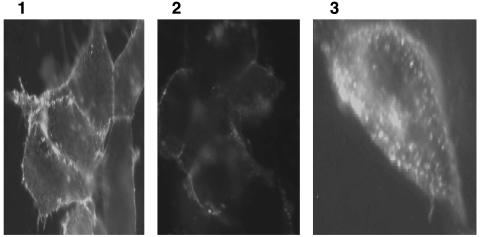
GPCRs undergo internalization after ligand binding. To further confirm that the chimeric receptor V1a/VRR1 is functional, we examined AVP-induced internalization of V1a/VRR1 receptor stably expressed in HEK293 cells by immunofluorescence. Complete internalization of the chimeric receptor was observed after 20-min incubation with 10 μM AVP (Fig. 3).
Fig. 3.
Internalization of chimeric receptor V1a/VRR1. Stably transfected HEK293 cells expressing chimeric receptor V1a/VRR1 were incubated in the absence (Left) or presence (Center and Right) of 10 μM AVP for 20 min at 37°C. The cells were fixed, permeabilized (Right) or nonpermeabilized (Left and Center), and then incubated with primary and secondary antibody (as described in Experimental Procedures). Samples were examined on a standard fluorescence microscope.
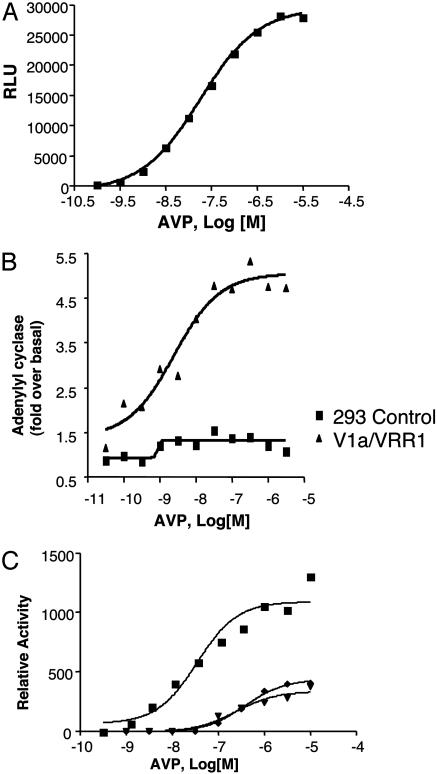
Determination of Signaling Properties of Chimeric Receptor V1a/VRR1. We next examined the signaling transduction pathways downstream of V1a/VRR1 chimera by using standard Gq, Gs, or Gi assays. Stably expressed V1a/VRR1 induced strong intracellular calcium mobilization upon stimulation with AVP. The calculated EC50 is 19 nM (Fig. 4A). The calcium increase is probably through a Gq mediated pathway, because AVP did not activate Gi/o in a cAMP inhibition assay (data not shown). To test whether V1a/VRR1 activation results in an increase in intracellular cAMP concentration, an indication of Gs pathway signaling, we carried out cAMP assay in the presence of a serial dilution of AVP. As shown in Fig. 4B, AVP stimulated cAMP accumulation in a dose-dependent manner with an EC50 of 3 nM.
Fig. 4.
Signaling of V1a/VRR1 receptor. (A) Dose-response curve of AVP-induced calcium production mediated by the HEK293 cells stably expressing chimeric receptor and transiently cotransfected with aequorin plasmid. (B) Dose-response curve of AVP for stimulation of adenylyl cyclase activity mediated by control HEK293 cells (square) and HEK293 cells stably expressing chimeric receptor (triangle). (C) Attenuation of V1a/VRR1 response to AVP by point mutations in the receptor. Shown are dose-response curves of AVP-induced calcium production mediated by HEK293 cells transiently cotransfected with aequorin and V1a/VRR1 receptor (squares) or V1a/VRR1(D96A) receptor (triangles) or with V1a/VRR1(Q107A) receptor (diamonds). The data are represented as the mean of two independent experiments, each done in duplicate or triplicate.
To exclude the remote possibility that the signaling events activated by AVP are caused by activation from endogenously expressed receptors in HEK293 cells, such as vasopressin V1a, V1b, V2, or oxytocin receptors, point mutations were made in transmembrane domain II of the chimeric receptor (as described in Experimental Procedures), which have been shown to diminish the activation of V1a receptor by AVP (8). Compared to wild-type chimeric receptor, both mutant receptors V1a/VRR1(D96A) and V1a/VRR1(Q107A) exhibited a shift in EC50 to 383 nM and the second mutant receptor showed a shift in EC50 to 337 nM (Fig. 4C), confirming that the intracellular response upon AVP stimulation is indeed via the chimeric receptor.
The aim of this study was to elucidate the signal transduction pathway of an orphan GPCR referred to here as VRR1, which did not display any constitutive activity. To circumvent the problem of not having an agonist for this receptor, we used a strategy involving the generation of a chimeric receptor between the V1a receptor and VRR1. This approach is based on the hypothesis that the intracellular loops and the C-terminal tail are responsible for the G protein coupling, whereas the ligand binding is dictated by the transmembrane and extracellular domains. Our results demonstrated that the chimeric receptor is properly targeted to the cytoplasmic membrane and is functionally responsive to AVP. AVP binding to the chimera elicited a calcium response and caused stimulation (and not inhibition) of adenylate cyclase, indicating that V1a/VRR1 couples to both Gq and Gs pathways. It is interesting to note that vasopressin V2 receptor is also known to activate these dual signaling pathways in recombinant systems (8). To expand on these results, further mutagenesis studies are required to pinpoint the regions and amino acid residues in VRR1 that are involved in G protein coupling.
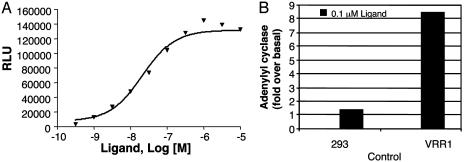
To confirm that the native VRR1 receptor indeed couples with the Gq and Gs pathways, we identified fractions from rat brain extract that specifically activated the VRR1 receptor in the Gq mode (data not shown). While the purification of this activity was in progress, Mori et al. (18) reported the identification of the endogenous ligand for VRR1 as a peptide, with a primary amino acid sequence of SFRNGVGTGMKKTSFQRAKS (PCT application, WO03/025179). This peptide is derived from a polypeptide precursor that is conserved in mammals but absent in Drosophila. Activation of VRR1 by this endogenous ligand induced intracellular calcium mobilization in a dose-dependent manner (EC50 of 22 nM) as well as adenylyl cyclase accumulation at 0.1 μM (Fig. 5), consistent with the results obtained by using V1a/VRR1 chimera.
Fig. 5.
Signaling of VRR1 receptor. (A) Dose-response curve of endogenous ligand-induced calcium production mediated by the HEK293 cells transiently cotransfected with VRR1 receptor and aequorin plasmid. (B) Response of 0.1 μM endogenous ligand for stimulation of adenylyl cyclase activity mediated by control HEK293 cells and HEK293 cells transiently transfected with VRR1 receptor.
In summary, we have identified an orphan GPCR related to vasopressin receptor subfamily. The tissue distribution of VRR1 mRNA suggests a potential function of VRR1 in circadian rhythm regulation. Additional studies, including the development of VVR1 knockout models and the generation of receptor agonists and antagonists, will be necessary to decipher the physiological functions of this receptor and its potential therapeutic utility. By using a chimeric receptor approach, we concluded that VRR1 signals through the Gq and Gs pathway. This was validated upon identification of the receptor's endogenous ligand. This finding will enable us to design specific screening assays for the identification of potential therapeutic small molecule agonists and antagonists by using high-throughput assays. In addition, our results further validate this chimera technology for the study of orphan GPCRs.
Acknowledgments
We thank Juan Du, Miki Rich, and Amy Bakleh for sequencing and technical help. We also thank Jeff Reagan for helpful discussion and comments on the manuscript.
Abbreviations: VRR1, vasopressin receptor-related receptor 1; GPCR, G protein-coupled receptor; AVP, Arg-8-vasopressin.
References
- 1.Baldwin, J. M. (1994) Curr. Opin. Cell Biol. 6, 180-190. [DOI] [PubMed] [Google Scholar]
- 2.Fredriksson, R., Lagerstrom, M. C., Lundin, L. G. & Schioth, H. B. (2003) Mol. Pharmacol. 63, 1256-1272. [DOI] [PubMed] [Google Scholar]
- 3.Vassilatis, D. K., Hohmann, J. G., Zeng, H., Li, F., Ranchalis, J. E., Mortrud, M. T., Brown, A., Rodriguez, S. S., Weller, J. R., Wright, A. C., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 4903-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostenis, E. (2001) Trends Pharmacol. Sci. 22, 560-564. [DOI] [PubMed] [Google Scholar]
- 5.Keen, T. J., Inglehearn, C. F., Green, E. D., Cunningham, A. F., Patel, R. J., Peacock, R. E., Gerken, S., White, R., Weissenbach, J. & Bhattacharya, S. S. (1995) Genomics 28, 383-388. [DOI] [PubMed] [Google Scholar]
- 6.Sonnhammer, E. L., von Heijne, G. & Krogh, A. (1998) Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175-182. [PubMed] [Google Scholar]
- 7.An, S., Cutler, G., Zhao, J. J., Huang, S. G., Tian, H., Li, W., Liang, L., Rich, M., Bakleh, A., Du, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 7576-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu, X., Gilbert, S., Birnbaumer, M. & Birnbaumer, L. (1994) Mol. Pharmacol. 46, 460-469. [PubMed] [Google Scholar]
- 9.Lin, D. C., Bullock, C. M., Ehlert, F. J., Chen, J. L., Tian, H. & Zhou, Q. Y. (2002) J. Biol. Chem. 277, 19276-19280. [DOI] [PubMed] [Google Scholar]
- 10.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 11.Park, Y., Kim, Y. J. & Adams, M. E. (2002) Proc. Natl. Acad. Sci. USA 99, 11423-11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cazzamali, G., Hauser, F., Kobberup, S., Williamson, M. & Grimmelikhuijzen, C. J. (2003) Biochem. Biophys. Res. Commun. 303, 146-152. [DOI] [PubMed] [Google Scholar]
- 13.Golombek, D. A., Ferreyra, G. A., Agostino, P. V., Murad, A. D., Rubio, M. F., Pizzio, G. A., Katz, M. E., Marpegan, L. & Bekinschtein, T. A. (2003) Front. Biosci. 8, S285-S293. [DOI] [PubMed] [Google Scholar]
- 14.Liu, T., DeCostanzo, A. J., Liu, X., Wang, H., Hallagan, S., Moon, R. T. & Malbon, C. C. (2001) Science 292, 1718-1722. [DOI] [PubMed] [Google Scholar]
- 15.Liu, X., Liu, T., Slusarski, D. C., Yang-Snyder, J., Malbon, C. C., Moon, R. T. & Wang, H. (1999) Proc. Natl. Acad. Sci. USA 96, 14383-14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeCostanzo, A. J., Huang, X. P., Wang, H. Y. & Malbon, C. C. (2002) Naunyn-Schmiedebergs Arch. Pharmacol. 365, 341-348. [DOI] [PubMed] [Google Scholar]
- 17.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori, M., Hayashi, K., Miya, H. & Sato, S. (2003) PCT Appl. WO03025179.