Abstract
Burkholderia pseudomallei is resistant to a diverse group of antimicrobials including third generation cephalosporins whilst quinolones and aminoglycosides have no reliable effect. As therapeutic options are limited, development of more effective forms of immunotherapy is vital to avoid a fatal outcome. In an earlier study, we reported on the B. pseudomallei serine MprA protease, which is relatively stable over a wide pH and temperature range and digests physiological proteins. The present study was carried out to evaluate the immunogenicity and protective efficacy of the MprA as a potential vaccine candidate. In BALB/c mice immunized with recombinant MprA protease (smBpF4), a significantly high IgG titer was detectable. Isotyping studies revealed that the smBpF4-specific antibodies produced were predominantly IgG1, proposing that immunization with smBpF4 triggered a Th2 immune response. Mice were immunized with smBpF4 and subsequently challenged with B. pseudomallei via the intraperitoneal route. Whilst control mice succumbed to the infection by day 9, smBpF4-immunized mice were protected against the lethal challenge and survived beyond 25 days post-infection. In conclusion, MprA is immunogenic in melioidosis patients whilst also eliciting a strong immune response upon bacterial challenge in mice and presents itself as a potential vaccine candidate for the treatment of melioidosis.
Keywords: melioidosis, serine protease, immunotherapy
Introduction
Burkholderia pseudomallei is the Gram negative etiological agent for melioidosis, a potentially acute fulminating disease in humans and animals in Southeast Asia and Northern Australia (Cheng and Currie, 2005; Peacock, 2006). In areas where this bacterium is endemic, infection by B. pseudomallei has been estimated to be responsible for 20–30% mortality due to septicaemia and 40% of sepsis-related mortality (Wiersinga et al., 2006). Human melioidosis presents with an array of clinical symptoms ranging from acute or chronic localized infection involving a single organ, to fulminant septicaemia in multiple organs and septic shock (Cheng and Currie, 2005). Current management of melioidosis involves the full 20-week duration of third generation cephalosporins therapy. Unfortunately, treating a B. pseudomallei infection is difficult as the bacterium is intrinsically resistant to many antibiotics and relapse following apparently successful therapy is well recognized (Cheng and Currie, 2005).
Production of an effective vaccine against B. pseudomallei is challenging due to the multi-factorial nature of this pathogen. Various vaccination strategies, including the use of attenuated stains of B. pseudomallei (Haque et al., 2006), heat-inactivated bacteria (Sarkar-Tyson et al., 2009), subunit vaccines (Harland et al., 2007), and DNA vaccines (Chen et al., 2006) have been extensively explored, but none were able to confer complete protection against the infection. Currently, the use of protection-eliciting pathogen proteins, for example virulence factors, is looked upon as an alternative and viable approach as previously demonstrated with the caseinolytic protease of Streptococcus pneumoniae (Cao et al., 2009), parasite cysteine proteases (Jorgensen and Buchmann, 2011), cytotoxic serine protease of Vibrio harveyi (Cheng et al., 2010), outer membrane proteins of Haemophilus parasuis (Yuan et al., 2011), and outer membrane secretin PilQ of Neisseria meningitidis (Haghi et al., 2012).
A common group of virulence factors shared among bacterial pathogens is protease, and protease inhibitors have proven effective therapeutic agents in treating infectious diseases in vertebrates (Ribeiro-Guimaraes et al., 2007; Serrano-Luna et al., 2007). For example, in the pathogenic bacterium Enterococcus faecalis, both gelatinase and serine protease are required for systemic infections in mammalian hosts (Engelbert et al., 2004). Similarly, the extracellular alkaline serine protease produced by the nosocomial pathogen Stenotrophomonas maltophilia is an important pathogenicity factor and has been recognized as a potential therapeutic target (Windhorst et al., 2002). The primary function of proteases in the bacterial kingdom is to provide a source of free amino acids for bacterial survival and growth, but there is increasing evidence of the role of proteases in bacterial pathogenesis in creating a niche for colonisation, suppressing host defence mechanisms and supporting host dissemination. Furthermore, the involvement of proteases in antibody degradation or modulation of cellular immune responses is also proposed (Jorgensen and Buchmann, 2011).
Many reports have alluded to the potential role of proteases in B. pseudomallei pathogenicity. The serine MprA protease has been implicated as a possible virulence factor (Lee and Liu, 2000) causing extensive damage to mammalian physiological proteins involved in circumventing the detrimental effects of bacterial secreted proteases (Chin et al., 2007). Previously, we identified an epitope of the B. pseudomallei protease by peptide biopanning against anti-B. pseudomallei antibodies, whereby selected peptides displayed a conserved motif of serine-methionine-alanine (S-M-A). This S-M-A motif mimics the catalytic serine and its flanking amino acids of the B. pseudomallei mprA gene (Chan and Nathan, 2005). As a potential antigen involved in B. pseudomallei pathogenesis, we further characterized the MprA protein and demonstrated that the expressed recombinant MprA protease maintained superior proteolytic activity over a wide pH (pH5-11) and temperature (4–68°C) range and partially digested immunoglobulins A and G, transferrin and myosin (Chin et al., 2007).
As the MprA protein carried an antibody-binding epitope and was implicated in proteolysis, we evaluated the potential of B. pseudomallei MprA protease as a vaccine candidate in a mouse model. We observed a significant delay in death of the immunized challenged animals indicating that this protease elicited a protective immune response in mice.
Materials and methods
Ethics statement
All animal experiments were performed in accordance with the Universiti Kebangsaan Malaysia animal ethics guideline and approved by the Universiti Kebangsaan Malaysia Animal Ethics Committee (UKMAEC). BALB/c mice (6 weeks old, female) were obtained from the Animal House Facility, Universiti Kebangsaan Malaysia. Mice were maintained under specific-pathogen-free conditions, subjected to a 12 h light/dark cycle and were fed on a diet of commercial pellets and distilled water ad libitum.
Bacterial strains and plasmid
B. pseudomallei human isolate (strain D286) was obtained from the Pathogen Laboratory, School of Biosciences and Biotechnology, Faculty of Science and Technology, Universiti Kebangsaan Malaysia. This bacterial strain was isolated from a patient with clinical manifestation of melioidosis at the Kuala Lumpur Hospital and previously characterized based on biochemical tests as well as by 16S rRNA sequencing (Lee et al., 2007). The recombinant MprA protease (BPSS1933) smBpF4 construct has been previously described (Chin et al., 2007).
Bacterial-cell free culture filtrate—C. elegans survival assay
B. pseudomallei bacterial cell-free culture filtrate was prepared and used to treat Caenorhabditis elegans to evaluate nematode killing. The assay and preparation of the cell-free filtrate was performed as previously described (Lee et al., 2011) with minor modifications. In brief, one B. pseudomallei colony was inoculated into 10 ml Brain Heart Infusion (BHI) broth and incubated overnight at 37°C. Phenylmethylsulfonyl fluoride (PMSF, 10 mM) and ethylenediaminetetraacetic acid (EDTA, 10 mM) were added to the overnight bacterial culture and further incubated at 37°C for 30 min. Subsequently, the mixture was centrifuged at 4000 g for 15 min at 4°C and the supernatant collected and filtered through a 0.22 μm pore size syringe membrane filter unit (Millipore, USA). The supernatant was supplemented with 1 ml S-basal medium. The assay was performed in six-well microtiter plates containing 2 ml of the supernatant and S-basal mixture. Thirty age-matched C. elegans Glp worms (Lee et al., 2011) were transferred into the conditioned liquid medium. Heat-killed Escherichia coli OP50 were supplied to prevent starvation. Mortality of the worms was monitored at 4-h intervals. E. coli OP50 was used in place of B. pseudomallei as the negative control.
Human sera samples
Culture-confirmed melioidosis patients sera (n = 5) from Malaysia presenting with acute and chronic disease manifestations and sera samples of patients presenting with Legionella infection (n = 5) but confirmed B. pseudomallei-culture negative were provided by the Institute for Medical Research, Malaysia. Healthy (confirmed HIV-negative) human sera samples (n = 5) were generously provided by Clinipath Malaysia Sdn. Bhd.
Over-expression and purification of MprA protease, smBpF4
Over-expression and purification of recombinant MprA protease smBpF4 were performed as previously described (Chin et al., 2007).
Reactivity of recombinant proteins toward melioidosis patients' sera
The 96-well microplates were coated with 0.3 μg per well of either heat-killed (80°C, 1 h) and sonicated (15 min) (Vibracell, Sonics and Materials, USA) B. pseudomallei or recombinant smBpF4 purified protein in coating buffer (0.1 M sodium bicarbonate, p.H 8.6) and incubated overnight at 4°C. The plates were blocked, washed, then incubated with human sera samples (1:200 dilution) and incubated at 37°C for 1 h as previously described (Chin et al., 2007; Hara et al., 2009). Rabbit anti-human IgG conjugated with peroxidase (1:10,000 dilution, Sigma-Aldrich, USA) was added and incubation continued for 1 h at 37°C. At the end of the incubation, the plates were washed and 50 μl ABTS:peroxidase B (1:1; KPL, USA) was added as substrate. The absorbance was read at 405 nm on an ELISA reader (Dynex MRX, USA).
Mouse immunization
BALB/c mice (n = 4, 6 weeks old, female) were immunized via three doses of 100 μg of heat-treated (80°C, 10 min) purified recombinant smBpF4 protein, via the intra-peritoneal (i.p.) route over 9 weeks as previously described (Hara et al., 2009). The first dose was administrated in Freund's complete adjuvant (Pierce Biotechnologies, USA) on day 0 and 2, boosts were administrated on days 14 and 28 with an identical dose of antigen in Freund's incomplete adjuvant (Pierce Biotechnologies, USA) (Harlow and Lane, 1988). Control mice (n = 3) were injected with Phosphate Buffered Saline (PBS) plus adjuvant as per the above schedule. The sera samples were collected and recovered prior to immunization and 1 week after the first and second booster according to the procedure of Harlow and Lane (1988) and antibody titres evaluated before the animals were challenged with B. pseudomallei. The resulting sera were subsequently used for antibody response analyses and protease neutralization assays by zymography
Analysis of antibody response
Mouse antibody titers against recombinant proteins were measured by ELISA as previously described (Chin et al., 2007; Hara et al., 2009) with some modifications. Briefly, 96-well plates were coated overnight with 1 μg of the purified recombinant protein used for immunization. Wells were blocked with 5% skimmed milk to prevent nonspecific binding. The plates were then washed thoroughly with distilled water. Serum samples were serially diluted two-fold (1:100 to 1:25,600 dilutions) across the microtiter plate in 5% skimmed milk and incubated for 1 h at 37°C. Rabbit anti-mouse IgG conjugated with peroxidase (1:4000 dilution, Pierce, USA) was added and incubation continued for 1 h at 37°C.
For the analysis of specific antibodies, Horseradish Peroxidase (HRP)-conjugated goat anti-mouse IgG (Promega, USA) (1:2500 dilution); rabbit anti-mouse IgM (μ chain) (1:2500 dilution), IgG1 (gamma 1) (1:1000 dilution), IgG2a (gamma 2a) (1:1000 dilution) or IgG2b (gamma 2b) (1:1000 dilution) (Zymed® Laboratories, USA) was added prior to the addition of ABTS:peroxidase B (1:1; KPL, USA). The absorbance was read at 405 nm on an ELISA reader (Dynex MRX, USA).
Protease neutralization by zymography
The ability of anti-smBpF4 mice sera antibodies to neutralize MprA gelatinase activity was tested by zymography as previously described (Chin et al., 2007). Briefly, 10 μg of purified smBpF4 protein was pre-mixed with 30 μg of anti-smBpF4 at 37°C for 1 h. The mixtures were equilibrated to room temperature and subsequently subjected to zymography.
Mouse protection study
For the challenge experiment, smBpF4-immunized and PBS control mice were challenged with approximately 1 × 106 CFU B. pseudomallei D286 administered intraperitoneally 2 weeks after the second booster and mice were monitored daily for signs of mortality for 35 days post-challenge as previously described (Hara et al., 2009; Su et al., 2010).
Enumeration of viable B. pseudomallei in the blood
smBpF4-immunized and challenged mice were tail-bled on days 6, 12, 18, and 25 post-infection. The blood was then plated on Ashdown agar and colonies were counted after 2 days incubation at 37°C.
Statistical analysis
Statistical analysis on human and mice sera reactivity was performed using the Student's t-test. Data are expressed as the mean ± standard deviation. Mortality of immunized vs. non-immunized groups was analyzed by Kaplan–Meier survival analysis and survival curves were compared using the Logrank test. Differences were considered statistically significant at P = 0.05. All statistical analyses were performed using GraphPad Prism® version 4.0 (GraphPad Software, USA).
Results
B. pseudomallei proteases are implicated in C. elegans killing
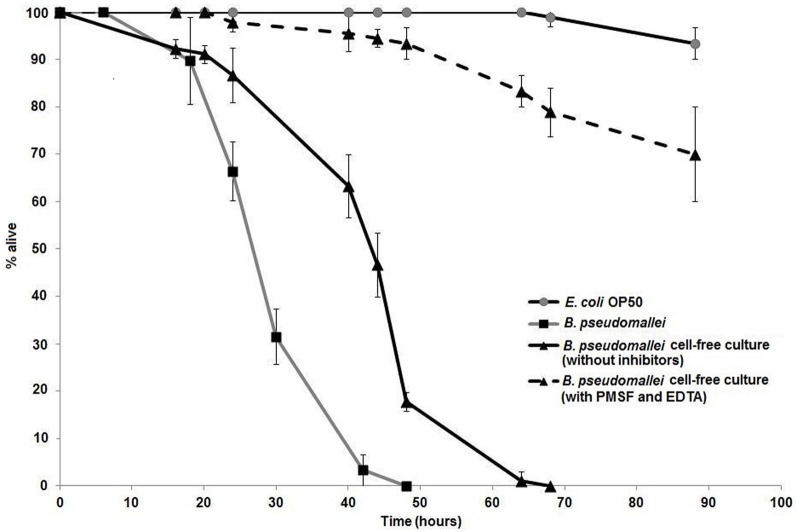
The prevalent view on the mode of action of extracellular proteases toward nematodes is that these proteases participate in cuticle penetration. To determine if B. pseudomallei proteases contribute to killing of C. elegans, a combination of serine protease inhibitors was added to the B. pseudomallei culture to inhibit the secreted proteases. Zymography was performed and the results confirmed that the B. pseudomallei cell-free culture filtrate gelatinase activity was indeed inhibited by 10 mM PMSF (serine protease inhibitor) and 10 mM EDTA (Zn2+ dependent metalloprotease inhibitor) (data not shown). Survival of the worms exposed to the laboratory food source E. coli OP50 remained at almost 100% over the course of the experiment (Figure 1). Survival of C. elegans exposed to bacterial cell-free culture filtrates treated with PMSF and EDTA was monitored over time and worms were observed to have an extended life span with a mean time to death (TDmean) of 65.63 ± 3.70 h compared to worms exposed to B. pseudomallei D286 cell-free culture filtrates in the absence of inhibitors (TDmean = 44.49 ± 1.37 h) [Logrank (Mantel–Cox) test, P < 0.0001] and B. pseudomallei D286 (TDmean = 31.35 ± 0.91 h) (Figure 1). PMSF is a common inhibitor of protease hydrolytic activity and mortality of nematodes exposed to PMSF-treated culture filtrates was significantly delayed, indicating that the proteases most likely killed nematodes through protein hydrolysis and not via the typical ligand–receptor interaction mechanism as exemplified by the Bt toxin produced by Bacillus thuringiensis (Beale et al., 2006). The survival assay validated the importance of B. pseudomallei proteases as virulence factors in pathogenesis.
Figure 1.
B. pseudomallei proteases contribute toward lethality in C. elegans. One-day old Glp worms were transferred to individual B. pseudomallei cell-free culture without (solid line, black triangles) or with protease inhibitors (PMSF and EDTA) (dashed line, black triangles), E. coli OP50 (grey line, open squares) and B. pseudomallei cells (solid line, black squares). The graph shows the mean ± SD of three replicates (30 worms/replicate) from a representative of two independent experiments.
Recombinant MprA protease (smBpF4) is a patient sero-reactive protein
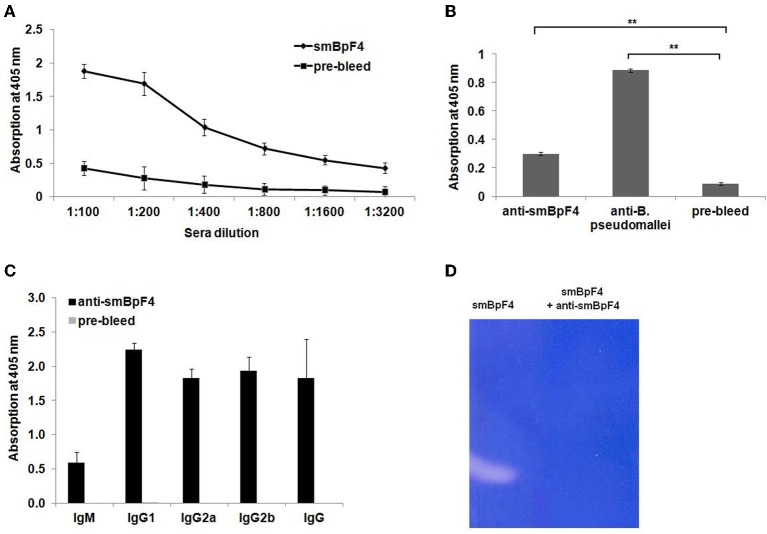
To further evaluate and study the sero-reactivity of the smBpF4 protease, the full length recombinant 55 kDa smBpF4 protein was expressed and purified as previously described (Chin et al., 2007). The expressed smBpF4 protein was subsequently tested against melioidosis patient sera, legionellosis patient sera and healthy subject (HIV-negative) sera. ELISA analyses demonstrated that smBpF4 reacted with the melioidosis sera but not toward the legionellosis and healthy sera (Figure 2). The results indicated that the presence of anti-MprA antibodies is specific to melioidosis patients.
Figure 2.
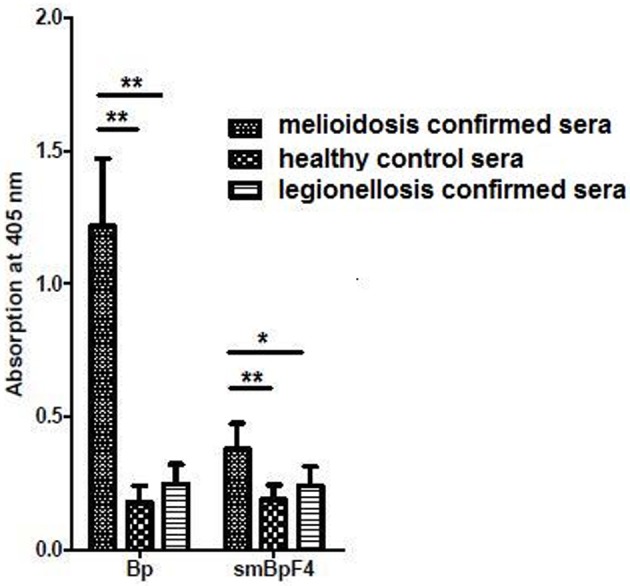
Recombinant MprA protease, smBpF4, is sero-reactive. Reactivity of purified smBpF4 and B. pseudomallei lysate (Bp) with pooled melioidosis-confirmed, healthy control and legionellosis-confirmed sera. Significance was determined by Student's t-test (*P < 0.05; **P < 0.01).
Recombinant MprA protein is immunogenic and induces strong antibody responses
To assess the immunogenicity of the recombinant smBpF4, sera were collected and pooled from smBpF4-immunized mice prior to and following B. pseudomallei challenge, and tested for reactivity with purified smBpF4 protease by ELISA. The specific antibodies against smBpF4 protease were observed in immunized groups over several dilutions screened but not in the pre-immunized sera (P < 0.001) (Figure 3A). In addition, the ELISA results (Figure 3B) demonstrated that the anti-smBpF4 also binds to proteins in the B. pseudomallei lysate. To identify the type of immune response induced, we determined the levels of specific IgG subclasses (IgG1, IgG2a, and IgG2b) and IgM by ELISA. The predominant Ig subclass elicited by smBpF4 immunization was IgG1, followed by IgG2b and to a lesser extent, IgG2a (Figure 3C). The ratio of IgG2a/IgG1 was 0.817 in smBpF4-immunized mice (Figure 3C), indicating a Th2-type immune response in mice. The ELISA results indicate that recombinant MprA protease is an immunogenic protein able to trigger specific antibodies in the immunized mice.
Figure 3.
Recombinant MprA protein is immunogenic and induces a strong antibody response. (A) Screening of smBpF4-specific IgG (anti-smBpF4) in immunized BALB/c mice. Mice (n = 4) were immunized with 100 μg of purified recombinant smBpF4. Sera collected prior to and after immunization were pooled and levels of anti-smBpF4 were analyzed by ELISA. The presence of specific antibodies to purified smBpF4 protein (black circles) was observed in sera from immunized mice but not in pre-immunized sera (black squares). (B) Interaction and recognition of anti-smBpF4 with B. pseudomallei lysate by ELISA. Significance was determined by Student's t-test (**P < 0.01). (C) Analysis of anti-smBpF4 antibody isotypes in sera obtained from immunized BALB/c mice by ELISA. Data represent the mean absorbance (OD405 nm) ± standard deviation. The IgG2a/IgG1 ratio of smBpF4-immunized mice is 0.817; (D) smBpF4 protease neutralization assay by zymography.
To evaluate the ability of the anti-SmBpF4 antibodies to neutralize the protease, the polyclonal antibodies were reacted with smBpF4 protease and analyzed by zymography. smBpF4 gelatinase activity was successfully inhibited by anti-smBpF4 (Figure 3D). Our results thus far support the proposal that when smBpF4 is utilized as an immunogen, antibodies raised against the recombinant MprA protease should be able to react with and neutralize proteases secreted during an active infection, hence preventing the proteolytic digestion of the infected individual's key physiological proteins.
Antibodies raised against smBpF4 elicit potential protective immunity in mice
To determine if the immune response induced by immunization with smBpF4 could confer protection against B. pseudomallei infection, smBpF4 immunized-animals were challenged with 10 × LD50 of B. pseudomallei D286 3 weeks after the second immunization boost and survival was observed. We had previously reported that the LD50 of B. pseudomallei D286 was 105 CFU following infection of BALB/c mice via an i.p. route (Hara et al., 2009). A challenge dose of 10 × LD50 is expected to cause 100% mortality of the infected animals within the observation period. Thus, any reduction in the number of dead animals or a significant delay in the time taken for all the animals to succumb to the challenge in immunized mice would indicate the potential protective efficacy of this recombinant smBpF4 protease.
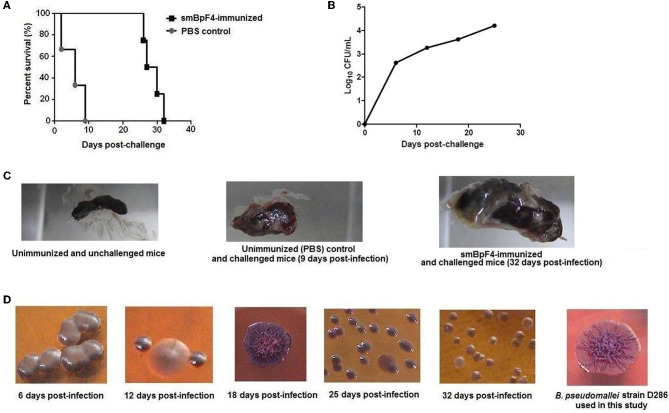
Mice immunized with smBpF4 (n = 4) demonstrated better survival rates than control immunized mice (Figure 4A). In the control group of mice (n = 3) immunized with PBS in adjuvant and subsequently challenged with B. pseudomallei, one mouse succumbed on day-2 post challenge, the second on day-6 and the last mouse died on day-9 with a median survival of 6 days (Figure 4A). All of the smBpF4-immunized mice (4/4) survived beyond 25 days post-challenge. However, viable B. pseudomallei were still detected and the CFU counts increased in the blood of smBpF4-immunized and challenged mice over the 25 days post-challenge (Figure 4B). This eventually led to bacteremia in the smBpF4-immunized and challenged mice, a hallmark of acute melioidosis. Concomitantly, one mouse died on day-26, the second on day-27 and the third on day-30. Thus, recombinant smBpF4 demonstrated the ability to confer protection against B. pseudomallei infection where the median survival was prolonged by 22.5 days (P = 0.0101) in immunized animals when compared to the control group. However, immunization with a single B. pseudomallei immunogen was unable to provide complete protection probably due to the lack of T cell stimulation (Patel et al., 2011) as well as the fact that B. pseudomallei pathogenesis is most likely multi-factorial. The challenged surviving mice also displayed splenomegaly and multiple abscess formation, demonstrating incomplete bacterial clearance from the system (Figure 4C) similar to that previously reported by Breitbach et al. (2008).
Figure 4.
Survival and assessment of bacterial burden of recombinant smBpF4-immunized mice against lethal challenge with 1×106 CFU B. pseudomallei D286 by the i.p. route of infection. (A) Mice were immunized with smBpF4 (black squares) prior to challenge with B. pseudomallei D286. All the smBpF4-immunized mice (black squares) survived up to or beyond 25 days post-infection. All the PBS-treated control mice (grey circles) died by day-9 post-challenge. (B) Bacterial loads in the blood of smBpF4-immunized and challenged mice over the 25 days post-infection before any deaths were recorded. (C) The appearance of abscesses on the spleen and splenomegaly of smBpF4-immunized B. pseudomallei challenged mice (32 days post-infection) compared to the spleen of control mice. (D) Various B. pseudomallei colony morphotypes on Ashdown's agar were observed in the smBpF4-immunized and challenged mice compared to B. pseudomallei D286 grown in vitro. These different B. pseudomallei colony morphotypes were confirmed by 16S rRNA sequencing as previously described (Lee et al., 2007).
B. pseudomallei can produce different colony morphologies in vitro and in vivo. These morphotypes are associated with phenotypes that alter the ability of B. pseudomallei to survive in adverse environmental conditions, which could have major implications for the development of therapeutics and vaccines (Chantratita et al., 2007; Tandhavanant et al., 2010). Interestingly, we also observed changes in colony morphotypes in the smBpF4-immunized and challenged mice over the infection period (Figure 4D). We identified the colony morphotypes observed in our study using the colony morphotyping scheme reported by Chantratita et al. (2007). B. pseudomallei D286 used in the challenge experiment presented a parental type 1 (the “cornflower head” morphology) colony morphology when cultured in vitro. Over time, this morphotype switched from parental type 1 to II (small and rough colonies) or type III (large and smooth colonies) (Figure 4D). The type III morphotype is reported to have a fitness disadvantage and lower intracellular replication rates in human macrophages when compared to parental type 1 morphotype (Chantratita et al., 2007; Tandhavanant et al., 2010). The switch to the type III colony morphotype predominantly seen throughout the infection period is most likely the contributing factor in the observed prolonged survival (Figure 4A) and low bacterial counts (Figure 4B) in smBpF4-immunized and challenged mice relative to the initial inoculums.
Discussion
Mortality associated with melioidosis remains high because of the lack of antimicrobial drugs, which could improve the outcome during the first 48 h after hospital admission (Peacock, 2006). This highlights the need for melioidosis-specific drugs to be administered immediately upon diagnosis to avoid a fatal outcome. Prophylactic recombinant B. pseudomallei proteins have been evaluated as potential candidate vaccines in animal models of melioidosis. The recombinant B. pseudomallei outer membrane protein A (OmpA, BPSL2522) (Hara et al., 2009) and Omp85 (BLSL2151) (Su et al., 2010) were shown to induce protective immunity against melioidosis. Pathogen surface-associated proteins have predominantly been the first pick for evaluation as potent antigens but recent findings suggest that secreted enzymes such as proteases may be effective targets as well (Jorgensen and Buchmann, 2011). Proteases from other pathogens, including parasites (Pechine et al., 2011) and bacteria (Murthy et al., 2009), are highly immunogenic and protective in animal models of infection. Thus, in this study, we extended the investigation to evaluate the prophylactic properties of recombinant B. pseudomallei serine MprA protease, a proposed virulence factor, which has been shown to cause extensive damage of mammalian physiological proteins although its role in the pathogenesis of melioidosis is still unclear.
B. pseudomallei MprA is located on Chromosome 2 of the B. pseudomallei K96243 genome at position 2,695,482–2,696,984. This protease is highly conserved among the Burkholderia spp. and subtilisins from other bacteria, particularly at the subtilisin-like catalytic triad residues (Asp 102, His 57, and Ser 195) (Lee and Liu, 2000). Our laboratory had previously utilized anti- B. pseudomallei protease polyclonal antibodies as a tool to investigate the protease-antibody interaction. We isolated phage displayed fusion peptides carrying a consensus peptide sequence, TKSMALSG, which closely resembled part of the mprA gene active site sequence, 435GTSMATPHVAG445 (Chan and Nathan, 2005). Homology modeling of the MprA protein based on subtilisin demonstrated that the MprA catalytic binding pocket inclusive of the consensus sequence TKSMALSG could be the major potential antibody binding epitope (Chan and Nathan, 2005). Furthermore, we also proposed that B. pseudomallei MprA protease is produced within the bacterial cytosol as a pre-proenzyme that is subsequently processed utilizing a mechanism similar to subtilisin to produce the secreted mature active protease (Chin et al., 2007). The mprA gene is present in all 68 B. pseudomallei isolates tested by Lee and Liu (2000) as well as 11 local Malaysia B. pseudomallei animal isolates (Chin et al., data not shown). Two studies which utilized a PCR assay targeting the mprA gene of B. pseudomallei reported 100% sensitivity and specificity in detecting the pathogen in naturally infected dromedary and clinical samples, suggesting that MprA protease could be a potential drug target upon diagnosis of melioidosis (Neubauer et al., 2007; Suppiah et al., 2010).
Our group recently reported that different isolates of B. pseudomallei have divergent ability to kill the soil nematode C. elegans (Lee et al., 2011), thus providing us with a viable model to identify and characterize virulent determinants of B. pseudomallei. In this regard, the virulence of B. pseudomallei proteases toward C. elegans was assessed by inhibiting the gelatinase activity with EDTA and PMSF. The prolonged survival of worms exposed to inhibitor-treated bacterial cell free culture (Figure 1) confirms the pathogenic role of B. pseudomallei serine proteases in melioidosis. To follow up on these findings, preliminary characterization of the recombinant MprA protease as a potential vaccine candidate in controlling B. pseudomallei infection was initiated in a mouse model of B. pseudomallei infection. We clearly demonstrated that smBpF4 protein is immunogenic in mice whereby high levels of specific neutralizing IgG capable of inhibiting gelatinase activity were induced (Figure 3D), suggesting that immunization with MprA protease can potentially induce an appropriate neutralizing immune response to protect the B. pseudomallei-infected individual from the damages inflicted by MprA protease.
A key issue in vaccine development today is the type of immune response needed to best protect against organisms for which there are currently no effective vaccines. Most of the present generation of successful vaccines depends primarily on generating high antibody titres and many are administered with the Th2-biasing adjuvant alum. However, natural protection against many organisms, particularly intracellular bacteria, is mainly Th1 in nature (Beverley, 2002). B. pseudomallei is an intracellular facultative pathogen that can successfully evade the host immune system for an extended period of time. This has made vaccine development for melioidosis difficult and challenging and has driven researchers to develop a successful anti- B. pseudomallei vaccine that can strongly induce both humoral antibodies and a cellular-mediated immune response. Animal models of B. pseudomallei infection have been used for the development of effective vaccines; however the level of protection varied depending on the different vaccine strategies (Brett and Woods, 1996; Atkins et al., 2002; Engelbert et al., 2004; Chen et al., 2006; Haque et al., 2006; Hara et al., 2009; Su et al., 2010). For instance, immunization with attenuated B. pseudomallei mutants and active immunization with B. pseudomallei lipopolysaccharide resulted in 60 and 50% survival at day-35 post-challenge of immunized mice, respectively (Nelson et al., 2004; Haque et al., 2006), whilst immunization with recombinant B. pseudomallei outer membrane protein Omp85 protected 70% of immunized mice up to 14 days (Su et al., 2010). Hence, data from our study is generally comparable to these previously reported efficacies. We demonstrated that all mice immunized with smBpF4 protease survived beyond 25 days of an active B. pseudomallei infection whereas the non-immunized mice succumbed within 9 days after infection (Figure 4A). Furthermore, candidate vaccines that achieved partial protection against the disease in mice suggests potential for human use and reduction in disease severity alone would be predicted to improve outcome of high mortality rates (Peacock et al., 2012). B. pseudomallei survival under adverse environmental conditions is likely to be facilitated by phenotypic adaptability and plasticity (Chantratita et al., 2007). The morphotype III observed in smBpF4-immunized mice was previously reported to be more resistant to the antimicrobial peptide LL-37 (Tandhavanant et al., 2010). This suggests that B. pseudomallei D286 undergoes a phenotypic change to adapt, survive, and replicate in the presence of bactericidal substances within the smBpF4-immunized host milieu.
The type of immune response induced by a vaccine antigen is very important for the success of the vaccine (Yuan et al., 2011). Different IgG subclasses have distinct effector functions. In the context of vaccination, different antigenic stimuli preferentially induce different antibodies (Findlow et al., 2006) and different domains of the same antigen can induce antibodies of different IgG subclasses (Soininen et al., 1999). In mice, immunoglobulin isotype switching from IgM to IgG1 is associated with a Th2-type response, whereas IgG2a is associated with a Th1-type response and promoted by interferon gamma (Chen et al., 2006; Yuan et al., 2011). Furthermore, IgG2a has been shown to be the most effective IgG subclass antibodies in binding to FcγR1 on phagocytic cells (Li et al., 2007). Several studies have demonstrated that mice with an induced Th1-type immune response showed greater resistance to B. pseudomallei compared to mice with a Th2-type immune response (Hoppe et al., 1999; Ulett et al., 2000; Healey et al., 2005). The antibody subclass analysis of sera from smBpF4-immunized mice revealed production of both IgG2a and IgG1, the higher levels of the latter indicate a dominant Th2 response over Th1. Furthermore, induction of a Th2-response was also noted for the prophylactic properties of the B. pseudomallei recombinant Omp85 protein (Su et al., 2010). Proteases from parasites (Jorgensen and Buchmann, 2011), Clostridium difficile (Pechine et al., 2011), V. harveyi (Cheng et al., 2010) and Vibrio parahaemolyticus (Liu et al., 2011) have also been proposed as promising vaccine candidates able to modulate Th1/Th2 responses in vaccinated animals.
In summary, our study has demonstrated that the B. pseudomallei MprA protease is a promising prophylactic candidate against B. pseudomallei although the protection provided by this single protein was incomplete. Earlier studies have also noted that administration of single B. pseudomallei antigens is insufficient to confer total protection against melioidosis, therefore, a combination of multiple immunogens should be evaluated for a more complete anti- B. pseudomallei protection.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Mohd. Nor Hasan for his technical assistance in animal handling. We are grateful to the Animal House of Universiti Kebangsaan Malayisa for the animal facilities. We thank Dr. Norazah Ahmad from the Institute for Medical Research, Malaysia, for providing the melioidosis patients' sera; Clinipath Malaysia Sdn. Bhd for providing the healthy control sera; Dr. Yuka Hara for technical assistance. This study was funded by the IRPA research grant 09-02-02-05 BTK/ER/29 awarded to Sheila Nathan by the Ministry of Science, Technology and Innovation, Malaysia.
References
- Atkins T., Prior R. G., Mack K., Russell P., Nelson M., Oyston P. C., Dougan G., Titball R. W. (2002). A mutant of Burkholderia pseudomallei, auxotrophic in the branched chain amino acid biosynthetic pathway, is attenuated and protective in a murine model of melioidosis. Infect. Immun. 70, 5290–5294 10.1128/IAI.70.9.5290-5294.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale E., Li G., Tan M. W., Rumbaugh K. P. (2006). Caenorhabditis elegans senses bacterial autoinducers. Appl. Environ. Microbiol. 72, 5135–5137 10.1128/AEM.00611-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverley P. C. (2002). Immunology of vaccination. Br. Med. Bull. 62, 15–28 10.1093/bmb/62.1.15 [DOI] [PubMed] [Google Scholar]
- Breitbach K., Kohler J., Steinmetz I. (2008). Induction of protective immunity against Burkholderia pseudomallei using attenuated mutants with defects in the intracellular life cycle. Trans. R. Soc. Trop. Med. Hyg. 102Suppl. 1, S89–S94 10.1016/S0035-9203(08)70022-1 [DOI] [PubMed] [Google Scholar]
- Brett P. J., Woods D. E. (1996). Structural and immunological characterization of Burkholderia pseudomallei O-polysaccharide-flagellin protein conjugates. Infect. Immun. 64, 2824–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Li D., Gong Y., Yin N., Chen T., Wong C. K., Xu W., Luo J., Zhang X., Lam C. W., Yin Y. (2009). Caseinolytic protease: a protein vaccine which could elicit serotype-independent protection against invasive pneumococcal infection. Clin. Exp. Immunol. 156, 52–60 10.1111/j.1365-2249.2008.03866.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. W., Nathan S. (2005). Epitope mapping of Burkholderia pseudomallei serine metalloprotease: identification of serine protease epitope mimics. FEMS Immunol. Med. Microbiol. 43, 37–44 10.1016/j.femsim.2004.06.025 [DOI] [PubMed] [Google Scholar]
- Chantratita N., Wuthiekanun V., Boonbumrung K., Tiyawisutsri R., Vesaratchavest M., Limma-thurotsakul D., Chierakul W., Wongratanacheewin S., Pukriti-yakamee S., White N. J., Day N. P., Peacock S. J. (2007). Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei. J. Bacteriol. 189, 807–817 10.1128/JB.01258-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. S., Hsiao Y. S., Lin H. H., Yen C. M., Chen S. C., Chen Y. L. (2006). Immunogenicity and anti-Burkholderia pseudomallei activity in Balb/c mice immunized with plasmid DNA encoding flagellin. Vaccine 24, 750–758 10.1016/j.vaccine.2005.08.069 [DOI] [PubMed] [Google Scholar]
- Cheng A. C., Currie B. J. (2005). Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18, 383–416 10.1128/CMR.18.2.383-416.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Zhang W. W., Zhang M., Sun L. (2010). Evaluation of the vaccine potential of a cytotoxic protease and a protective immunogen from a pathogenic Vibrio harveyi strain. Vaccine 28, 1041–1047 10.1016/j.vaccine.2009.10.122 [DOI] [PubMed] [Google Scholar]
- Chin C. Y., Othman R., Nathan S. (2007). The Burkholderia pseudomallei serine protease MprA is autoproteolytically activated to produce a highly stable enzyme. Enzyme Microb. Tech. 40, 373–377 [Google Scholar]
- Engelbert M., Mylonakis E., Ausubel F. M., Calderwood S. B., Gilmore M. S. (2004). Contribution of gelatinase, serine protease, and fsr to the pathogenesis of Enterococcus faecalis endophthalmitis. Infect. Immun. 72, 3628–3633 10.1128/IAI.72.6.3628-3633.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlow H., Southern J., Mabey L., Balmer P., Heyderman R. S., Auckland C., Morris R., Miller E., Borrow R. (2006). Immunoglobulin G subclass response to a meningococcal quadrivalent polysaccharide-diphtheria toxoid conjugate vaccine. Clin. Vaccine Immunol. 13, 507–510 10.1128/CVI.13.4.507-510.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghi F., Peerayeh S. N., Siadat S. D., Zeighami H. (2012). Recombinant outer membrane secretin PilQ (406–770) as a vaccine candidate for serogroup B Neisseria meningitidis. Vaccine 30, 1710–1714 10.1016/j.vaccine.2011.12.076 [DOI] [PubMed] [Google Scholar]
- Haque A., Chu K., Easton A., Stevens M. P., Galyov E. E., Atkins T., Titball R., Bancroft G. J. (2006). A live experimental vaccine against Burkholderia pseudomallei elicits CD4+ T cell-mediated immunity, priming T cells specific for 2 type III secretion system proteins. J. Infect. Dis. 194, 1241–1248 10.1086/508217 [DOI] [PubMed] [Google Scholar]
- Hara Y., Mohamed R., Nathan S. (2009). Immunogenic Burkholderia pseudomallei outer membrane proteins as potential candidate vaccine targets. PLoS ONE 4:e6496 10.1371/journal.pone.0006496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland D. N., Chu K., Haque A., Nelson M., Walker N. J., Sarkar-Tyson M., Atkins T. P., Moore B., Brown K. A., Bancroft G., Titball R. W., Atkins H. S. (2007). Identification of a LolC homologue in Burkholderia pseudomallei, a novel protective antigen for melioidosis. Infect. Immun. 75, 4173–4180 10.1128/IAI.00404-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. (1988). Antibodies: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory; [Google Scholar]
- Healey G. D., Elvin S. J., Morton M., Williamson E. D. (2005). Humoral and cell-mediated adaptive immune responses are required for protection against Burkholderia pseudomallei challenge and bacterial clearance postinfection. Infect. Immun. 73, 5945–5951 10.1128/IAI.73.9.5945-5951.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe I., Brenneke B., Rohde M., Kreft A., Haussler S., Reganzerowski A., Steinmetz I. (1999). Characterization of a murine model of melioidosis: comparison of different strains of mice. Infect. Immun. 67, 2891–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen L. G., Buchmann K. (2011). Cysteine proteases as potential antigens in antiparasitic DNA vaccines. Vaccine 29, 5575–5583 10.1016/j.vaccine.2011.05.091 [DOI] [PubMed] [Google Scholar]
- Lee M. A., Liu Y. (2000). Sequencing and characterization of a novel serine metalloprotease from Burkholderia pseudomallei. FEMS Microbiol. Lett. 192, 67–72 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Chong C. E., Lim B. S., Chai S. J., Sam K. K., Mohamed R., Nathan S. (2007). Burkholderia pseudomallei animal and human isolates from Malaysia exhibit different phenotypic characteristics. Diagn. Microbiol. Infect. Dis. 58, 263–270 10.1016/j.diagmicrobio.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Ooi S. K., Mahadi N. M., Tan M. W., Nathan S. (2011). Complete killing of Caenorhabditis elegans by Burkholderia pseudomallei is dependent on prolonged direct association with the viable pathogen. PLoS ONE 6:e16707 10.1371/journal.pone.0016707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Gottschalk M., Esgleas M., Lacouture S., Dubreuil J. D., Willson P., Harel J. (2007). Immunization with recombinant Sao protein confers protection against Streptococcus suis infection. Clin. Vaccine Immunol. 14, 937–943 10.1128/CVI.00046-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Chen J., Li K., Zhang X. (2011). Identification and evaluation as a DNA vaccine candidate of a virulence-associated serine protease from a pathogenic Vibrio parahaemolyticus isolate. Fish Shellfish Immunol. 30, 1241–1248 10.1016/j.fsi.2011.04.005 [DOI] [PubMed] [Google Scholar]
- Murthy A. K., Guentzel M. N., Zhong G., Arulanandam B. P. (2009). Chlamydial protease-like activity factor–insights into immunity and vaccine development. J. Reprod. Immunol. 83, 179–184 10.1016/j.jri.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., Prior J. L., Lever M. S., Jones H. E., Atkins T. P., Titball R. W. (2004). Evaluation of lipopolysaccharide and capsular polysaccharide as subunit vaccines against experimental melioidosis. J. Med. Microbiol. 53, 1177–1182 10.1099/jmm.0.45766-0 [DOI] [PubMed] [Google Scholar]
- Neubauer H., Sprague L. D., Joseph M., Tomaso H., Al Dahouk S., Witte A., Kinne J., Hensel A., Wernery R., Wernery U., Scholz H. C. (2007). Development and clinical evaluation of a PCR assay targeting the metalloprotease gene (mprA) of B. pseudomallei. Zoonoses Public Health 54, 44–50 10.1111/j.1863-2378.2007.01008.x [DOI] [PubMed] [Google Scholar]
- Patel N., Conejero L., De Reynal M., Easton A., Bancroft G. J., Titball R. W. (2011). Development of vaccines against Burkholderia pseudomallei.Front. Microbiol. 2:198 10.3389/fmicb.2011.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock S. J. (2006). Melioidosis. Curr. Opin. Infect. Dis. 19, 421–428 10.1097/01.qco.0000244046.31135.b3 [DOI] [PubMed] [Google Scholar]
- Peacock S. J., Limmathurotsakul D., Lubell Y., Koh G. C., White L. J., Day N. P., Titball R. W. (2012). Melioidosis vaccines: a systematic review and appraisal of the potential to exploit biodefense vaccines for public health purposes. PLoS Negl. Trop. Dis. 6:e1488 10.1371/journal.pntd.0001488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechine S., Deneve C., Le Monnier A., Hoys S., Janoir C., Collignon A. (2011). Immunization of hamsters against Clostridium difficile infection using the Cwp84 protease as an antigen. FEMS Immunol. Med. Microbiol. 63, 73–81 10.1111/j.1574-695X.2011.00832.x [DOI] [PubMed] [Google Scholar]
- Ribeiro-Guimaraes M. L., Tempone A. J., Amaral J. J., Nery J. A., Gomes Antunes S. L., Pessolani M. C. (2007). Expression analysis of proteases of Mycobacterium leprae in human skin lesions. Microb. Pathog. 43, 249–254 10.1016/j.micpath.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Sarkar-Tyson M., Smither S. J., Harding S. V., Atkins T. P., Titball R. W. (2009). Protective efficacy of heat-inactivated B. thailandensis, B. mallei or B. pseudomallei against experimental melioidosis and glanders. Vaccine 27, 4447–4451 10.1016/j.vaccine.2009.05.040 [DOI] [PubMed] [Google Scholar]
- Serrano-Luna J., Cervantes-Sandoval I., Tsutsumi V., Shibayama M. (2007). A biochemical comparison of proteases from pathogenic Naegleria fowleri and non-pathogenic Naegleria gruberi. J. Eukaryot. Microbiol. 54, 411–417 10.1111/j.1550-7408.2007.00280.x [DOI] [PubMed] [Google Scholar]
- Soininen A., Seppala I., Nieminen T., Eskola J., Kayhty H. (1999). IgG subclass distribution of antibodies after vaccination of adults with pneumococcal conjugate vaccines. Vaccine 17, 1889–1897 [DOI] [PubMed] [Google Scholar]
- Su Y. C., Wan K. L., Mohamed R., Nathan S. (2010). Immunization with the recombinant Burkholderia pseudomallei outer membrane protein Omp85 induces protective immunity in mice. Vaccine 28, 5005–5011 10.1016/j.vaccine.2010.05.022 [DOI] [PubMed] [Google Scholar]
- Suppiah J., Thimma J. S., Cheah S. H., Vadivelu J. (2010). Development and evaluation of polymerase chain reaction assay to detect Burkholderia genus and to differentiate the species in clinical specimens. FEMS Microbiol. Lett. 306, 9–14 10.1111/j.1574-6968.2010.01923.x [DOI] [PubMed] [Google Scholar]
- Tandhavanant S., Thanwisai A., Limmathurotsakul D., Korbsrisate S., Day N. P., Peacock S. J., Chantratita N. (2010). Effect of colony morphology variation of Burkholderia pseudomallei on intracellular survival and resistance to antimicrobial environments in human macrophages in vitro. BMC Microbiol. 10, 303 10.1186/1471-2180-10-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulett G. C., Ketheesan N., Hirst R. G. (2000). Proinflammatory cytokine mRNA responses in experimental Burkholderia pseudomallei infection in mice. Acta Trop. 74, 229–234 10.1016/S0001-706X(99)00075-3 [DOI] [PubMed] [Google Scholar]
- Valade E., Thibault F. M., Gauthier Y. P., Palencia M., Popoff M. Y., Vidal D. R. (2004). The PmlI-PmlR quorum-sensing system in Burkholderia pseudomallei plays a key role in virulence and modulates production of the MprA protease. J. Bacteriol. 186, 2288–2294 10.1128/JB.186.8.2288-2294.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W. J., Van Der Poll T., White N. J., Day N. P., Peacock S. J. (2006). Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 4, 272–282 10.1038/nrmicro1385 [DOI] [PubMed] [Google Scholar]
- Windhorst S., Frank E., Georgieva D. N., Genov N., Buck F., Borowski P., Weber W. (2002). The major extracellular protease of the nosocomial pathogen Stenotrophomonas maltophilia: characterization of the protein and molecular cloning of the gene. J. Biol. Chem. 277, 11042–11049 10.1074/jbc.M109525200 [DOI] [PubMed] [Google Scholar]
- Yuan F., Fu S., Hu J., Li J., Chang H., Hu L., Chen H., Tian Y., Bei W. (2011). Evaluation of recombinant proteins of Haemophilus parasuis strain SH0165 as vaccine candidates in a mouse model. Res. Vet. Sci. 93, 51–56 10.1016/j.rvsc.2011.04.020 [DOI] [PubMed] [Google Scholar]