Abstract
Bacteriophages are estimated to be the most abundant entities on earth and can be found in every niche where their bacterial hosts reside. The initial interaction between phages and Campylobacter jejuni, a common colonizer of poultry intestines and a major source of foodborne bacterial gastroenteritis in humans, is not well understood. Recently, we isolated and characterized a phage F336 resistant variant of C. jejuni NCTC11168 called 11168R. Comparisons of 11168R with the wildtype lead to the identification of a novel phage receptor, the phase variable O-methyl phosphoramidate (MeOPN) moiety of the C. jejuni capsular polysaccharide (CPS). In this study we demonstrate that the 11168R strain has gained cross-resistance to four other phages in our collection (F198, F287, F303, and F326). The reduced plaquing efficiencies suggested that MeOPN is recognized as a receptor by several phages infecting C. jejuni. To further explore the role of CPS modifications in C. jejuni phage recognition and infectivity, we tested the ability of F198, F287, F303, F326, and F336 to infect different CPS variants of NCTC11168, including defined CPS mutants. These strains were characterized by high-resolution magic angle spinning NMR spectroscopy. We found that in addition to MeOPN, the phase variable 3-O-Me and 6-O-Me groups of the NCTC11168 CPS structure may influence the plaquing efficiencies of the phages. Furthermore, co-infection of chickens with both C. jejuni NCTC11168 and phage F336 resulted in selection of resistant C. jejuni bacteria, which either lack MeOPN or gain 6-O-Me groups on their surface, demonstrating that resistance can be acquired in vivo. In summary, we have shown that phase variable CPS structures modulate phage infectivity in C. jejuni and suggest that the constant phage predation in the avian gut selects for changes in these structures leading to a continuing phage–host co-evolution.
Keywords: bacteriophage, Campylobacter jejuni, capsular polysaccharide, phase variation, phosphoramidate, methylation
Introduction
Bacteriophages (phages) are the most abundant group of organisms on the planet and are estimated to outnumber their bacterial hosts by 10-fold (Brüssow and Hendrix, 2002). The first step of phage infection is through the specific recognition and attachment of the phage particle to the host cell (Weinbauer, 2004). In general, phages bind to unique host-specific structures, thus allowing them to recognize a suitable host in a mixed bacterial population (Rakhuba et al., 2010). Knowledge of phage receptors is required for implementation of efficient phage therapies against human pathogens – an intervention strategy that has been promoted by the increasing occurrence of antibiotic resistance (Levy and Marshall, 2004; Kutateladze and Adamia, 2010), yet only a few bacterial phage receptors have been identified in Gram-negative bacteria (for example, Gemski and Stocker, 1967; Stirm and Freund-Molbert, 1971; Gemski et al., 1975; Yu and Mizushima, 1982; Morona et al., 1984).
Capsular polysaccharides (CPS) are highly hydrated polymeric gels that provide a thick (400 nm or more) layer protecting bacteria from hostile environments and host immune defenses, but the CPS can also be a target for phage binding and infection (Stirm and Freund-Molbert, 1971; Roberts, 1996; Comstock and Kasper, 2006). The capsule of the zoonotic foodborne pathogen, Campylobacter jejuni, is required for serum resistance, invasion of human epithelial cells, colonization of chickens, and diarrheal disease in ferrets (Bacon et al., 2001; Jones et al., 2004). Furthermore, mutation of several C. jejuni CPS genes resulted in phage resistance against a group of phages, but the actual capsular moieties involved were not identified (Coward et al., 2006). Only recently a specific component of the C. jejuni capsule was identified as the phage receptor for this species. We showed that phage F336 relies on the O-methyl phosphoramidate (MeOPN) modification of the C. jejuni NCTC11168 CPS for adsorption to the host cell surface and infection (Sørensen et al., 2011).
The CPS of C. jejuni is the major component recognized by the Penner serotyping scheme for which 47 different serotypes are known (Guerry et al., 2012). Thus, the CPS are highly diverse in structure and this is reflected by the gene content of the capsular loci in various C. jejuni strains (Karlyshev et al., 2005). In addition, a number of CPS genes possess homopolymeric tracts making them prone to phase variation which allows a rapid on/off switching of the genes and promotes variations in CPS even when the strains have identical gene contents (Linton et al., 2001; St Michael et al., 2002; Karlyshev et al., 2005; McNally et al., 2005; Chen et al., 2008). The CPS of C. jejuni 11168 consists of -2)-β-d-Ribf-(1-5)-3-MeOPN-β-d-GalfNAc-(1-4)-α-d-GlcpA6(NGro)-(1-backbone with a 3,6-di-O-methyl-4-MeOPN-d-glycero-α-l-gluco-Hepp side chain (St Michael et al., 2002; Szymanski et al., 2003; McNally et al., 2005). The CPS biosynthetic locus in strain NCTC11168 includes 28 genes and 6 of these genes (cj1420, cj1421, cj1422, cj1426, cj1429, and cj1437) contain homopolymeric G (polyG) tracts (Parkhill et al., 2000). Even though the function of only three out of the six gene products has been identified, a highly variable presence/absence of methyl, ethanolamine, aminoglycerol, and phosphoramidate modifications of the CPS have been observed (St Michael et al., 2002; Szymanski et al., 2003; McNally et al., 2005), suggesting that the remaining genes could encode enzymes involved in synthesis or transfer of these modifications to the CPS glycans. Recently, the cj1421 and cj1422 genes were shown to encode MeOPN transferases attaching MeOPN to the GalfNAc and the Hep residues, respectively (McNally et al., 2007). Similarly, cj1426 was demonstrated to encode the 6-O-Me transferase enzyme (Sternberg et al., submitted).
We previously discovered that resistance toward phage F336 evolved with a high frequency in C. jejuni NCTC11168 due to loss of the MeOPN receptor on the bacterial surface as a result of phase variation in the cj1421 gene encoding the MeOPN–GalfNAc transferase. In addition, deletion of the cj1421 gene in NCTC11168 resulted in phage resistance, proving that MeOPN attached to GalfNAc was a receptor for phage F336 (Sørensen et al., 2011). The aim of the present study was to determine if other phages rely on the MeOPN moiety for infection of C. jejuni and to explore the role of other CPS phase variable modifications for phage infection. Knowledge of phage receptors and development of phage resistance in vivo is essential for implementation of efficient phage therapy against C. jejuni in the chicken gut. We found that not only the MeOPN moiety, but also the phase variable 3-O-Me and 6-O-Me groups in the CPS of C. jejuni NCTC11168 may influence phage sensitivity. Co-infection of chickens with C. jejuni NCTC11168 and phage F336 resulted in bacterial acquisition of phage resistance through a loss of MeOPN attached to GalfNAc of the capsule or the acquisition of the 6-O-Me group. We propose that the constant exposure of C. jejuni to naturally occurring phages in the avian gut selects for different phase variable structures of the CPS resulting in a continuous phage-host co-evolution.
Materials and Methods
Bacterial strains, media, and growth conditions
The C. jejuni strains used in this study are listed in Table 1 and were routinely grown on blood agar Base II (Oxoid) supplemented with 5% calf blood (BA) or in brain heart infusion (BHI) broth under microaerobic conditions (6% CO2, 6% O2, 84% N2, and 4% H2) at 37°C. For chicken experiments, bacteria were grown on Karmali agar (Oxoid) supplemented with Campylobacter selective supplement under microaerobic conditions (10% CO2, 5% O2, 85% N2) at 37°C.
Table 1.
Campylobacter jejuni strains.
| Serotype | Description | Reference | |
|---|---|---|---|
| 1447 | HS:4c | Chicken isolate | Hansen et al. (2007) |
| NCTC12662 | HS:5j | Origin not known | National Collection of |
| Type Cultures, UK | |||
| NCTC11168 (MP21) | HS:2 | Wild type | Sørensen et al. (2011) |
| NCTC11168 MP24 | HS:2 | NCTC11168 variant passaged | This study |
| NCTC11168 MP25 | HS:2 | NCTC11168 variant passaged | This study |
| NCTC11168 MP26 | HS:2 | NCTC11168 variant passaged | This study |
| NCTC11168 V26 | HS:2 | NCTC11168 variant | Carrillo et al. (2004) |
| kpsM | Untypeable | NCTC11168 kpsM::kan does not have a capsule | Karlyshev et al. (2000) |
| 11168R | Phage F336 resistant NCTC11168 | Sørensen et al. (2011) | |
| S11168R | Phage-sensitive 11168R | Sørensen et al. (2011) | |
| 11168H | HS:2 | Hypermotile NCTC11168 | Jones et al. (2004) |
| 11168HΔ1421 | Loss of MeOPN on GalfNAc | McNally et al. (2007) | |
| 11168HΔ1421/1422 | Loss of MeOPN on GalfNAc | McNally et al. (2007) | |
| Loss of MeOPN on Hep | |||
| 11168HΔ1422 | Loss of MeOPN on Hep | McNally et al. (2007) | |
| 11168HΔ1421/1422 + 1422 | Loss of MeOPN on GalfNAc | McNally et al., 2007 | |
| Gain of MeOPN on Hep | |||
| 11168H1 | 11168H variant | This study | |
| Loss of 6-O-Me and MeOPN on Hep |
Bacteriophages
Bacteriophages used in this study are listed in Table 2. They all belong to the family of Myoviridae and have genome sizes of approximately 140 kb, but show different HhaI restriction patterns. Phages were propagated on C. jejuni NCTC12662, except for phage F336 where C. jejuni 1447 was used (Hansen et al., 2007). Stock bacteriophages were kept in SM buffer (0.05 M Tris–Cl, pH 7.5 supplemented with 5.8 g NaCl, 2.0 g MgSO4·7H2O, and 5 ml gelatin, 2% w/v solution) at 4°C.
Table 2.
Bacteriophages.
Bacteriophage propagation
Bacteriophage propagation was performed as described by Sørensen et al. (2011) using a plate lysis method developed from Frost et al. (1999), Hansen et al. (2007). The propagating strains were grown overnight on blood agar Base II plates and harvested in BHI supplemented with 1 mM CaCl2 and 10 mM MgSO4 (CBHI). Bacterial suspensions were adjusted to an optical density of OD600 = 0.35, incubated for 4 h at 37°C under microaerobic conditions, mixed with phages at a multiplicity of infection (MOI) of 0.01 and incubated for 15 min at 37°C to allow bacteriophage adsorption. Subsequently, 0.6 ml of the adsorbed suspensions were mixed with 5 ml of NZCYM overlay agar [NZCYM (Sigma) broth with 0.6% agar] at 45°C and poured onto NZCYM 1.2% agar plates. After incubation, plates with lysis were flooded with sterile SM buffer and the phages eluted overnight with gentle shaking at 4°C. Phage suspensions were harvested and filtered through a sterile 0.2 μm membrane filter.
Bacteriophage titration
Bacteriophage titration was performed as previously described (Sørensen et al., 2011). Briefly, bacterial lawns were made by mixing 5 ml NZCYM overlay agar tempered at 45°C with 0.5 ml bacteria harvested in CBHI from Base II agar plates and incubated for 4 h as described above and poured onto NZCYM plates. Phage solutions were spotted manually with three times 10 μl of 100–10−7 phage dilutions in SM buffer. Plates were incubated at 37°C for 20–24 h under microaerobic conditions and the number of plaque-forming units per milliliter (PFU/ml) of undiluted phage suspensions was calculated. Usually, phage titers of 108–109 PFU/ml could be obtained on both the propagating strains and C. jejuni MP21.
Plaque assay
Susceptibility of a C. jejuni strain to the bacteriophages was performed using the plaque assay protocol described under Bacteriophage titration and determined as the efficiency of plaquing (EOP) in percent by dividing the PFU/ml on the test strain by the PFU/ml on the control strain (NCTC11168) multiplied by 100. The values represented are the mean count standard deviations derived from two to four independent experiments.
High-resolution magic angle spinning NMR spectroscopy
High-resolution magic angle spinning (HR-MAS) NMR analysis of intact bacterial cells was performed as previously described (McNally et al., 2005).
In vivo experiments
One-day-old chickens were obtained from the Poultry Research Facility, Department of Agriculture, Food and Nutritional Science, University of Alberta. On the day of arrival, chickens were checked for C. jejuni colonization by cloacal swabs and divided in groups of four chickens. On day 2, chickens were first orally gavaged with either 107 F336 phages in SM buffer or with SM buffer alone. After approximately 30 min, the chickens were orally gavaged with a suspension containing 106 C. jejuni NCTC11168 in PBS or with PBS alone. Six days post infection, chickens were euthanized and the contents from one cecum were collected aseptically. The cecal contents were weighed and resuspended in phosphate-buffered saline at a final concentration of 0.1 g/ml. This suspension was subjected to 10-fold serial dilutions and plated onto Karmali selective agar plates to determine bacterial colonization levels. One hundred microliters of the 10−1 dilution was plated separately onto selective plates and all resulting bacterial colonies were pooled together. Half of this pool was used for sequencing and phage sensitivity and the other half was used for NMR analysis. The experiment was repeated twice with consistent results. PCR of gene cj1421 was performed as previously described (Sørensen et al., 2011) while sequencing was done using the following primer: 1421-seq2-F (5′-CTCGAGTTATAAGGATATTTTAGATGAG-3′). Gene cj1426 was PCR-amplified using the primers: 1426-PCR-F (5′-TTGAGAATTATGATAAGATGAAGG-3′) and 1426-PCR-R (5′-TTTCCTAAGAATTCTTTACTTTCG-3′) and then sequenced using the primers: 1426-seq-F (5′-AAGATCCAGATAAAAGAGATTATTTGG-3′) and 1426-seq-R (5′-ATCAGGAGAATCAAAAATGATTTTTCC-3′).
Results
The MeOPN moiety of the CPS of C. jejuni NCTC11168 is a receptor for several phages
We recently demonstrated that phage F336 requires the O-methyl phosphoramidate (MeOPN) moiety attached to the GalfNac of the C. jejuni NCTC11168 CPS for infection (Sørensen et al., 2011). In our phage collection (Hansen et al., 2007) we found that five phages, including F336, rely on capsular moieties for infection of C. jejuni NCTC11168, since they do not form plaques on a kpsM capsule mutant (Table 3). In our previous study, we isolated a phage F336 resistant NCTC11168 variant in vitro (11168R), which lost the MeOPN attached to the GalfNAc residue due to the presence of an additional G in gene cj1421 resulting in an off version of the gene (Sørensen et al., 2011). Here we found that 11168R had gained cross-resistance to all of the four phages: F198, F287, F303, and F326 (Table 3). While phages F198 and F303 did not form plaques on C. jejuni 11168R at all, plaques were observed with F287 and F326, but with a much lower EOP compared to wild type NCTC11168. Hence, the MeOPN structure is important for infection by the four other phages, although two of these phages (F287 and F326) were able to infect a strain not expressing the MeOPN modification. Similar results were obtained when screening a cj1421 MeOPN transferase deletion mutant in the NCTC11168 wild type background (data not shown). Supporting this, the EOP of four of these phages, including F336, were fully restored to wild type NCTC11168 levels, while the EOP of phage F326 was partially restored, when we tested the phages against a 11168R revertant containing an intact cj1421 gene and returned expression of the CPS MeOPN (C. jejuni S11168R; Sørensen et al., 2011; Table 3). Thus, several phages rely on the MeOPN moiety attached to the GalfNAc residue of CPS for infection of C. jejuni NCTC11168.
Table 3.
Efficiency of plaquing (EOP) of phages related to F336 as compared to NCTC11168.
| Phage | NCTC11168 | 11168R | S11168R | kpsM |
|---|---|---|---|---|
| F198 | 100a | –b | 82.5 ± 24.7 | – |
| F287 | 100 | 0.04 ± 0.03 | 2300 ± 282.8 | – |
| F303 | 100 | – | 350 ± 113.1 | – |
| F326 | 100 | 0.02 ± 0.02 | 11.6 ± 6.7 | – |
| F336 | 100 | – | 331.8 ± 64.3 | – |
aEOP is calculated in percent as the PFU/ml of the phages on the test strain divided by the PFU/ml obtained on NCTC11168 multiplied by 100.
b–No plaques formed.
The location of the MeOPN on the CPS is important for phage infection
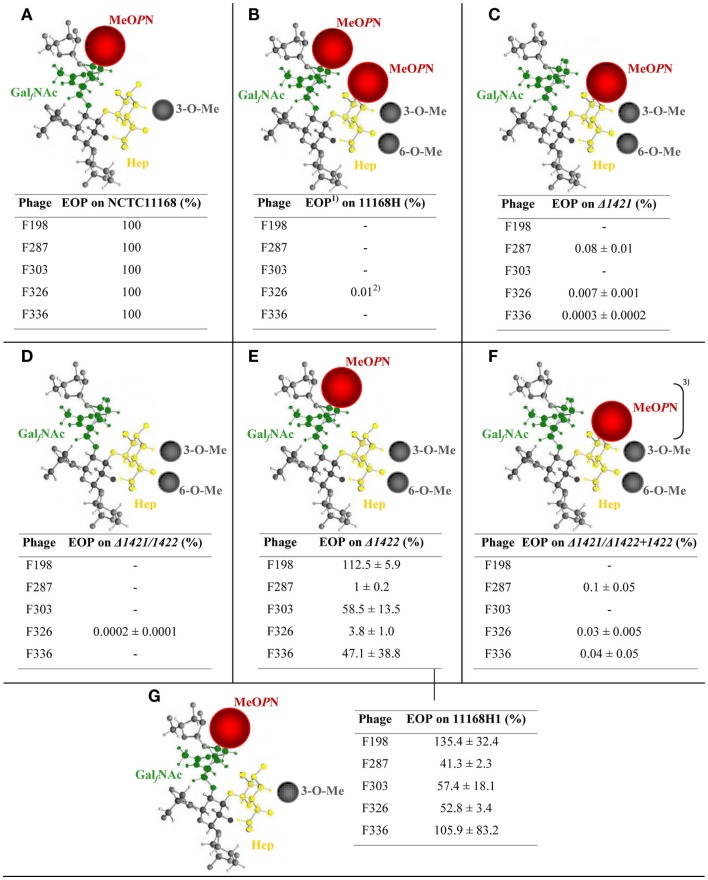
The MeOPN moiety of C. jejuni NCTC11168 can be attached to the GalfNAc and the Hep residues in the CPS (McNally et al., 2007) and we speculated that the attachment site of MeOPN may be important for phage infection. We therefore conducted plaque assays and determined EOPs of the five phages F198, F287, F303, F326, and F336 on the hypermotile NCTC11168 variant 11168H as well as on defined mutants in the cj1421 and cj1422 MeOPN transferase genes in this background and compared to the NCTC11168 wild type strain. All of these strains have previously been characterized by HR-MAS NMR and 1H–31P heteronuclear single-quantum correlation (HSQC) HR-MAS NMR for the presence/absence of MeOPN in their CPS structure (McNally et al., 2007). Our results showed that the attachment site of MeOPN in the CPS clearly influenced the efficiency of the phages to infect a particular strain (Figure 1). In general, all phages showed a higher EOP when MeOPN was situated on the GalfNAc residue (Figures 1A,E) as compared to MeOPN attached to the Hep residue in the CPS (Figure 1C). A lower EOP obtained with MeOPN attached to the Hep residue was further confirmed by screening a cj1421 + cj1422 deletion strain complemented with cj1422 (McNally et al., 2007) with our five phages (Figure 1F). Interestingly, the four phages F198, F287, F303, and F336 were not able to infect C. jejuni 11168H, which is the only strain expressing MeOPN on both GalfNAc and Hep (Figure 1B), suggesting that two MeOPN moieties may inhibit phage infection. Finally, we observed that the presence of two other phase variable CPS modifications, the 3-O-Me and 6-O-Me groups influenced the plaquing efficiency of the phages. We also isolated a 11168H single colony variant (11168H1) that by HR-MAS NMR and 1H–31P HSQC HR-MAS NMR analysis showed the same CPS profile as the NCTC11168 wild type strain (data not shown). The phage sensitivity profile observed with this strain (Figure 1G) confirmed the results obtained with NCTC11168 (Figure 1A). Thus, the presence of the 6-O-Me group on the CPS reduced the plaquing efficiencies of phages F287 and F326 (Figures 1A,E), suggesting that other phase variable CPS modifications besides MeOPN affect phage infection.
Figure 1.
Capsular polysaccharide structures expressed by defined MeOPN transferase mutants and their affect on phage sensitivity. The CPS structure is schematically illustrated (modified from Guerry and Szymanski, 2008). Phage sensitivity patterns are calculated as the 1efficiency of plaquing (EOP) as compared to plaque formation on the NCTC11168 wild type strain. (A) NCTC11168; (B) 11168H; (C) 11168HΔ1421; (D) 11168HΔ1421/Δ1422; (E) 11168HΔ1422; (F) 11168HΔ1421/Δ1422 + 1422; and (G) 11168H1. 2Plaques were only detected in one out of three experiments. 3MeOPN detected in lower amounts due to partial complementation. Abbreviations are: GalfNAc, N-acetylgalactosamine in the furanose configuration; MeOPN, O-methyl phosphoramidate; Hep, heptose; and O-Me, O-methyl.
C. jejuni phages require different combinations of phase variable CPS modifications for successful infection
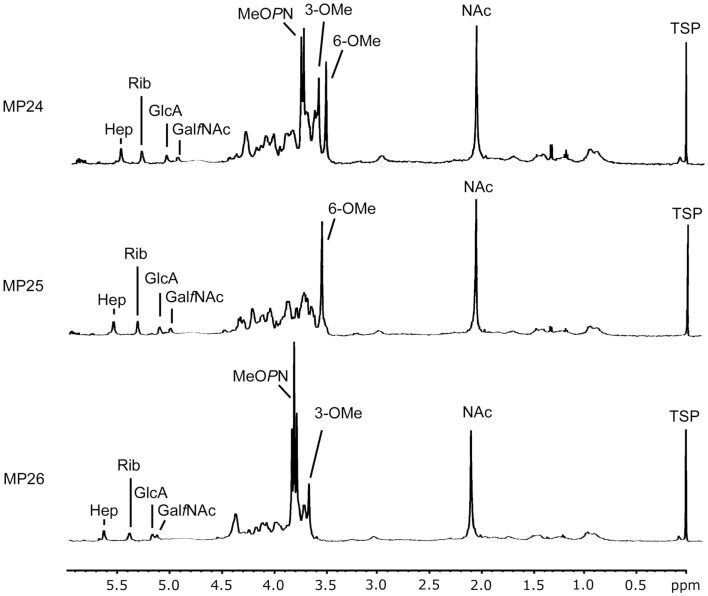
Phase variable expression of the capsular modifications in C. jejuni NCTC11168 is well documented (Parkhill et al., 2000; St Michael et al., 2002; Szymanski et al., 2003). Three frozen stocks of NCTC11168 (MP24, MP25, and MP26) that had been passaged a number of times in our laboratory were analyzed by HR-MAS NMR and 1H–31P HSQC HR-MAS NMR (Figure 2). This analysis revealed differences in the four phase variable modifications of the CPS structure, i.e., the 3-O-Me, 6-O-Me, MeOPN-GalfNAc, and MeOPN-Hep as compared to our NCTC11168 wild type (Table 4). We also included another NCTC11168 variant (V26) previously shown to have yet another different set of modifications (St Michael et al., 2002; Carrillo et al., 2004).
Figure 2.
High-resolution magic angle spinning NMR spectra showing the differences in CPS modifications between NCTC11168 variants MP24, MP25 and MP26. 1H CPMG spectra of intact C. jejuni cells. Abbreviations are: Hep, d-glycero-α-l-gluco-Hep; Rib, β-d-Ribf; GlcA, α-d-GlcpA6(NGro); GalfNAc, β-d-GalfNAc; MeOPN, O-methyl phosphoramidate; OMe, O-methyl; NAc, N-acetyl; TSP, trimethylsilyl propionic acid standard.
Table 4.
ppm Values for 1H HR-MAS NMR spectra for NCTC11168 variants and MeOPN transferase status.
| C. jejuni strain | Hepa | Rib | GlcA | GalfNAc | 3-O-Me | 6-O-Me | NAc | MeOPN | cj1421/cj1422 (GalfNAc/Hep)b |
|---|---|---|---|---|---|---|---|---|---|
| NCTC11168 MP24 | 5.582 | 5.376 | 5.103 | 5.015 | 3.629 | 3.557 | 2.071 | 3.772 | −/+ |
| NCTC11168 MP25 | 5.599 | 5.363 | 5.148 | 5.039 | – | 3.569 | 2.066 | - (3.772)c | −/− |
| NCTC11168 MP26 | 5.610 | 5.361 | 5.143 | 5.101 | 3.630 | – | 2.068 | 3.768/3.743 | +/+ |
aOnly anomeric proton resonances for carbohydrate residues are listed.
bSummary of sequencing results indicating the “on” (+) and “off” (−) status of the genes encoding the MeOPN transferases onto GalfNAc (cj1421) and Hep (cj1422).
cOnly trace amounts of MeOPN detected.
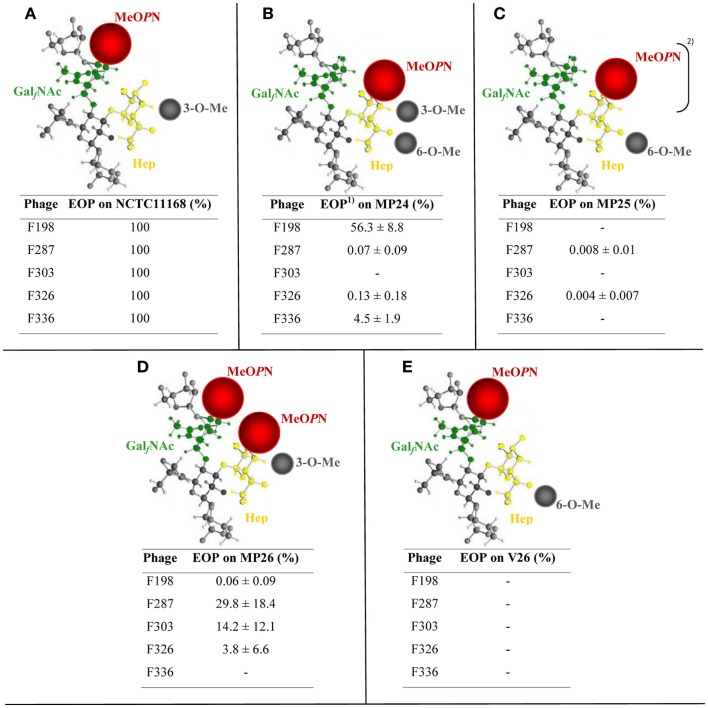
Plaque assays using our five phages showed that these C. jejuni strains had distinct phage sensitivity profiles (Figures 3B–E). Interestingly, C. jejuni MP26 that expresses both MeOPN moieties, similar to the phage-resistant C. jejuni 11168H (Figure 1B), could be infected by four of the five phages although with different efficiencies (Figure 3D). However, CPS of C. jejuni MP26 only contains the 3-O-Me group, whereas 11168H CPS contains both the 3-O-Me and 6-O-Me groups (Figures 1B and 3D). These results indicate that the presence of 6-O-Me may inhibit phage infection. This was further supported by the observation that none of the phages formed plaques when the 3-O-Me was exchanged with the 6-O-Me while retaining the MeOPN modification attached to GalfNAc (Figures 3A,E). Furthermore, the presence of the 6-O-Me group in combination with a MeOPN situated on the GalfNAc and a 3-O-Me group clearly inhibited infection by phage F287 and F326 (Figures 1E,G). Thus, our data suggest that the 6-O-Me group inhibits plaque formation in vitro and that all five phages require MeOPN attached to GalfNAc together with the 3-O-Me to obtain the most efficient infection. However, different combinations of the phase variable CPS modifications appear to influence binding of the phages differently. In particular, phage F326 showed the lowest EOP values (Table 3; Figures 1 and 3) suggesting that other combinations of receptors, not investigated in this study, might be optimal for the binding of this phage.
Figure 3.
Capsular polysaccharide structures expressed by NCTC11168 variants and the corresponding phage sensitivity profiles. The CPS structure is schematically illustrated (modified from Guerry and Szymanski, 2008). Phage sensitivity patterns are calculated as the 1efficiency of plaquing (EOP) as compared to plaque formation on the NCTC11168 wild type strain. (A) NCTC11168; (B) NCTC11168 MP24; (C) NCTC11168 MP25; (D) NCTC11168 MP26; and (E) NCTC11168 V26. 2MeOPN only detected in minor amounts. Abbreviations as above are: GalfNAc, N-acetylgalactosamine in the furanose configuration; MeOPN, O-methyl phosphoramidate; Hep, heptose; and O-Me, O-methyl.
Selection for phage-resistant variants during C. jejuni colonization of chickens
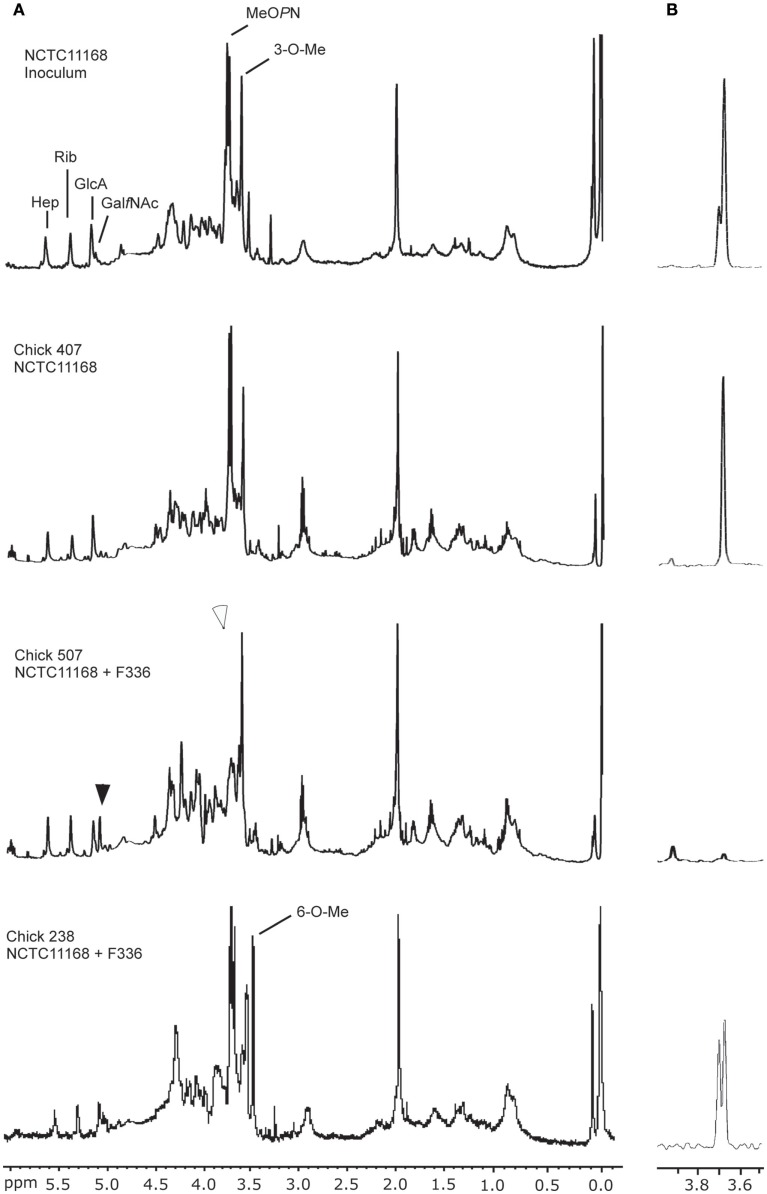
To assess whether C. jejuni is capable of becoming resistant to bacteriophage F336 in vivo during colonization of chickens, we orally gavaged 2-day-old chickens with F336 and subsequently with C. jejuni NCTC11168. After 6 days, the chickens were euthanized and the bacteria from the ceca were analyzed. We observed similar levels of colonization after 6 days of infection between the groups receiving bacteria only, as compared to those receiving bacteria and bacteriophages (i.e., 109 CFU/g cecal content, Table 5). The bacteria grown from the cecal contents were directly pooled from plates and analyzed by HR-MAS NMR. All bacteria obtained from chickens infected with C. jejuni alone showed an NMR profile that was the same as the NMR spectrum of the inoculum: the bacteria expressed one MeOPN attached to GalfNAc and the 3-O-Me group bound to Hep. However, bacteria from the group of chickens that received both bacteria and bacteriophages had lost the MeOPN in all instances, except for one (Figure 4; Table 5). In chicken #238, infected with phage F336 and C. jejuni, the bacteria showed reduced levels of MeOPN present on both GalfNAc and Hep, but the bacteria also expressed high levels of 6-O-Me on the Hep (Figure 4).
Table 5.
Presence of MeOPN and phage sensitivity of C. jejuni isolated after colonization of chickens.
| Infected with | Bacterial counts (CFU/g) | MeOPN on GalfNAc | Phage sensitivitya | |
|---|---|---|---|---|
| MP21 Inoculum | NR | NR | + | Yes |
| MP21 Karmali | NR | NR | + | Yes |
| Chick 405 | MP21 | 5.7 × 109 | + | Yes |
| Chick 406 | MP21 | 4.1 × 109 | + | Yes |
| Chick 407 | MP21 | 9.9 × 108 | + | Yes |
| Chick 408 | MP21 | 1.1 × 109 | + | Yes |
| Chick 505 | MP21 + F336 | 7.9 × 108 | − | No |
| Chick 506 | MP21 + F336 | 5.0 × 109 | − | No |
| Chick 507 | MP21 + F336 | 5.8 × 109 | − | No |
| Chick 508 | MP21 + F336 | 8.5 × 108 | − | No |
| Chick 343 | MP21 | 2.0 × 109 | + | Yes |
| Chick 344 | MP21 | 2.1 × 109 | + | Yes |
| Chick 345 | MP21 | 1.6 × 109 | + | Yes |
| Chick 346 | MP21 | 3.6 × 109 | + | Yes |
| Chick 237 | MP21 + F336 | 1.2 × 109 | − | No |
| Chick 238 | MP21 + F336 | 5.5 × 109 | +b | No |
| Chick 239 | MP21 + F336 | 4.7 × 109 | −c | No |
| Chick 240 | MP21 + F336 | 4.1 × 109 | − | No |
Chicks 405–408 and 505–508 are from the first experiment, while chicks 343–346 and 237–240 are from the second experiment.
aYes: plaques formed on isolated strain, No: no plaques formed on isolated strain.
bBacteria isolated from this chick showed reduced MeOPN levels (of both MeOPN groups) and enhanced levels of 6-O-Me.
cMeOPN levels barely above the baseline could be detected from bacteria isolated from this chick.
NR, not relevant.
Figure 4.
NMR analysis of intact C. jejuni cells recovered from chicken cecal contents. (A) 1H HR-MAS NMR CPMG spectra, displaying the capsular profile and (B) corresponding 1D 1H–31P HSQC spectra specifically demonstrating the phosphoramidate resonances. Black arrowhead: note the change in the GalfNAc resonance due to the absence of MeOPN. White arrowhead: note the absence of the MeOPN specific resonance.
To test whether the observed changes in CPS modifications resulted in phage resistance, we performed bacterial titration assays with all of the isolates recovered from the chickens. All isolates from the F336 infected birds had gained resistance to phage F336 and still showed similar levels of colonization (Table 5). Previously, we have shown that a loss of MeOPN on GalfNAc can be the result of a switch in the polyG region of the MeOPN transferase gene cj1421 (Sørensen et al., 2011). To determine whether the loss of MeOPN and subsequent phage resistance were the result of a switch in the polyG region, we sequenced gene cj1421 from the pooled colonies from both experiments. All phage-sensitive isolates showed 9 G residues in the polyG region, while all phage-resistant isolates (except from chick 238) showed either 8 or 10 G residues in this region, which in both cases results in a non-functional transferase, due to a premature stop codon 1 or 5 amino acids after the polyG region, respectively (data not shown). Examination of the pooled colonies from chick 238 showed 9 G residues in the polyG region of cj1421, consistent with the observed MeOPN expression. However, sequencing of the polyG region of cj1426 from the pooled colonies from chick 238 showed 10 G residues indicating that the 6-O-Me transferase enzyme was functional (again consistent with the acquired expression of the Me modification in Figure 4) compared to 11 G residues in the parental strain lacking this modification. Thus, C. jejuni can become resistant to phage F336 during in vivo colonization of chickens by changing the expression of its CPS phase variable modifications.
Discussion
Implementation of efficient phage therapies to target pathogenic bacteria requires detailed knowledge of the bacterial receptors recognized by the phages, as this bacteria–phage interaction is the first event in a process leading to lysis of the host organism. We previously identified the C. jejuni NCTC11168 CPS phase variable MeOPN modification as the receptor for phage F336 (Sørensen et al., 2011). The goal of the present study was to further explore the role of MeOPN and other C. jejuni phase variable CPS modifications, such as O-methylation in phage infection. We compared five different phages, including F336 that were unable to proliferate in an acapsular mutant of C. jejuni NCTC11168, and determined their ability to infect NCTC11168 variants containing the same CPS backbone sugars as NCTC11168, but with different modifications.
We found that the phages F198, F287, F303, F326, and F336 all infected C. jejuni most efficiently when the 3-O-Me modification on Hep and a MeOPN moiety was present, with higher EOPs observed when the MeOPN modification was attached to GalfNAc rather than Hep. Interestingly, when the 3-O-Me was exchanged with the 6-O-Me while retaining the MeOPN on GalfNAc, none of the phages formed plaques at all, demonstrating a clear role for the O-methyl Hep modifications for phage infection, in addition to the previously identified phage receptor, MeOPN (Sørensen et al., 2011). By looking at the predicted three dimensional (3D) structure of the C. jejuni NCTC11168 CPS (Guerry and Szymanski, 2008), it is possible that the presence or absence of one or more CPS modifications may lead to conformational changes that interfere with the phage binding process. Indeed it has been found that changes in the 3D conformation of the outer membrane protein OmpA, which serves as the receptor for many of the T-even phages infecting E. coli, leads to phage resistance (Riede and Eschbach, 1986). Furthermore, the spatial arrangement of the modifications may be important for phage recognition and some modifications may potentially block the phage binding site when they are expressed. Certainly masking the phage binding site is a well known resistance mechanism in other bacteria. One example is Staphylococcus aureus that masks its phage receptor with protein A, a cell-wall-anchored virulence factor for this bacterium (Nordstrom and Forsgren, 1974). In agreement with this hypothesis, we observed that when MeOPN was present on both the GalfNAc and Hep residues together with the 3-O-Me and 6-O-Me Hep modifications, then four of the five phages could no longer infect that particular C. jejuni 11168 strain. Thus, both the presence and absence of the capsular modifications appears to modulate phage infectivity of the NCTC1168 strain.
Our results demonstrate that the phages infecting C. jejuni are highly specific in the recognition of the CPS and that each phage has evolved to recognize a particular combination of capsular modifications. One might speculate that the phages included in this study originated from the same ancestral phage that has co-evolved with C. jejuni to recognize different capsular surface structures by minor modifications in the phage receptor binding proteins (RBP). In support of this, sequencing of viral proteins from the C. jejuni phage CP220 revealed at least two distinct tail structures in the mature virions although propagated from a single plaque (Timms et al., 2010), suggesting a possibility for the phage to recognize multiple receptors. A similar phenomena has been observed for capsular phages infecting E. coli (Scholl et al., 2001). Indeed RBPs of other phages often have hypervariable regions in the receptor recognizing domains, reflecting the ability of the phage to easily accumulate mutations thereby changing the structure of the RBP to recognize modified receptors or new types of receptors, such as switching from protein to carbohydrate recognition (Montag et al., 1990; Drexler et al., 1991; Hashemolhosseini et al., 1994). The C. jejuni phages investigated in this study have similar genome sizes and morphologies and were previously isolated from duck and broiler intestines and duck abattoirs (Hansen et al., 2007). Although these phages all are able to infect the NCTC1168 strain, they do show different host ranges for C. jejuni strains of different Penner serotypes (Hansen et al., 2007). In addition, enzymatic restriction cleavage of the phage genomes revealed differences among the phages (Table 2, Hansen et al., 2007). Thus, although a conserved head and tail architecture is maintained, it is apparent that genomic dissimilarities exist between the five phages compared in this study and this may account for the differences in host specificity.
In C. jejuni, the CPS are highly diverse, due to differences in the genetic compositions of the CPS loci, but also because many of the genes in these loci contain homopolymeric tracts making them prone to phase variation (Linton et al., 2001; St Michael et al., 2002; Szymanski et al., 2003; Karlyshev et al., 2005; McNally et al., 2005; Chen et al., 2008). Based on the number of phase variable modifications in the capsule of C. jejuni NCTC11168 alone, this may result in the expression of >700 CPS structural variants. Our results demonstrate that phase variable CPS surface structures clearly influence the phage attachments sites in C. jejuni NCTC11168. Phase variable expression also affects phage adsorption in Bordetella spp. These bacteria use phase variable expression of the BvgAS two-component regulatory system to alter their surfaces through the regulation of colonization and virulence factors and also expression of the phage BPP-1 receptor Prn (pertactin autotransporter; Liu et al., 2002). To combat these changes, the BPP-1 phage has a sophisticated mechanism of varying its RBP (∼10+13 possible sequences) that approaches the levels of diversity observed for immunoglobulins (Liu et al., 2002; Doulatov et al., 2004), and the phage is thus able to overcome the resistance by an equally efficient mechanism.
It was previously shown that CPS expression is important for C. jejuni colonization of the chicken gut as well as being important for interactions with human models of disease (Bacon et al., 2001; Jones et al., 2004). Here we show for the first time that the presence of phages selects for specific CPS variants in vivo in the chicken gut without altering the colonization levels, even though MeOPN expression influences C. jejuni invasion of human cells (van Alphen, unpublished results). Thus, the roles of phase variable CPS modifications, such as the MeOPN and the O-methyl groups, may not only be linked to adaptation to the human or chicken hosts, but may serve an important function for C. jejuni survival in the avian gut; an environment that contains a high number of bacteria as well as phages (Connerton et al., 2004; Hansen et al., 2007). Scott et al. (2007) also described that certain isolates of C. jejuni containing Mu-like prophage DNA sequences were capable of developing bacteriophage resistance in the chicken gut through a mechanism of reversible genomic rearrangement. Remarkably, these isolates inverted genomic segments up to 590 kb which also led to a decrease in chicken colonization and production of a functional Mu-like bacteriophage (Scott et al., 2007). The authors speculated that the development of phage resistance could be due to alterations in the bacterial surface receptor or through saturation of the receptor sites if the Mu phage is recognizing the same structures. Here, we have shown that phase variable modifications of the CPS influence phage infectivity, suggesting that the constant exposure of C. jejuni to phages in the avian gut selects for changes in the phase variable structures of the CPS leading to a continuing co-evolution of C. jejuni polysaccharides and phage RBPs. This should be considered when developing future strategies for phage therapy against C. jejuni in the chicken host and when determining the kinetics of resistance development and the timing for therapy administration. Since the alteration of bacterial surface receptors that reduces or eliminates phage adsorption to the host cell appears to be the most common mechanism of phage resistance (Bohannan and Lenski, 2000), continued investigations into the diversity of phage receptors in C. jejuni is required to select the most efficient combinations of phages for therapy.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to B. Wren for the gift of the C. jejuni NCTC11168 kpsM mutant. We sincerely appreciate the expert technical assistance of Lisbeth Schade Hansen, Christel Galschiøt Buerholt, Mark Miskolzie, David Simpson, Yasmin Barre, Cory Wenzel, Abofu Alemka, and Denis Arutyunov. This work was funded by a grant from the FOOD Research School at the Faculty of Life Sciences, University of Copenhagen, Denmark. Lieke B. van Alphen holds an Alberta Innovates Health Solutions (AIHS) Postdoctoral Fellowship, Shauna M. Crowley holds an AIHS Graduate Scholarship and Christine M. Szymanski holds an Alberta Innovates Scholar Award.
Abbreviations
β-D-Rib, ribose; β-D-GalfNAc, N-acetylgalactosamine in the furanose configuration; α-d-GlcpA6(NGro), glucuronic acid with 2-amino-2-deoxyglycerol at C-6; MeOPN, O-methyl phosphoramidate; Hepp, heptose. The 2-amino-2-deoxyglycerol can also be substituted with an N-ethanolamine modification at C-6.
References
- Bacon D. J., Szymanski C. M., Burr D. H., Silver R. P., Alm R. A., Guerry P. (2001). A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40, 769–777 10.1046/j.1365-2958.2001.02431.x [DOI] [PubMed] [Google Scholar]
- Bohannan B. J. M., Lenski R. E. (2000). Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3, 362–377 10.1046/j.1461-0248.2000.00161.x [DOI] [Google Scholar]
- Brüssow H., Hendrix R. W. (2002). Phage genomics: small is beautiful. Cell 108, 13–16 10.1016/S0092-8674(01)00637-7 [DOI] [PubMed] [Google Scholar]
- Carrillo C. D., Taboada E., Nash J. H., Lanthier P., Kelly J., Lau P. C., Verhulp R., Mykytczuk O., Sy J., Findlay W. A., Amoako K., Gomis S., Willson P., Austin J. W., Potter A., Babiuk L., Allan B., Szymanski C. M. (2004). Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 279, 20327–20338 10.1074/jbc.M401134200 [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Poly F., Pakulski Z., Guerry P., Monteiro M. A. (2008). The chemical structure and genetic locus of Campylobacter jejuni CG8486 (serotype HS:4) capsular polysaccharide: the identification of 6-deoxy-D-ido-heptopyranose. Carbohydr. Res. 343, 1034–1040 10.1016/j.carres.2008.09.004 [DOI] [PubMed] [Google Scholar]
- Comstock L. E., Kasper D. L. (2006). Bacterial glycans: key mediators of diverse host immune responses. Cell 126, 847–850 10.1016/j.cell.2006.08.021 [DOI] [PubMed] [Google Scholar]
- Connerton P. L., Loc Carrillo C. M., Swift C., Dillon E., Scott A., Rees C. E., Dodd C. E., Frost J., Connerton I. F. (2004). Longitudinal study of Campylobacter jejuni bacteriophages and their hosts from broiler chickens. Appl. Environ. Microbiol. 70, 3877–3883 10.1128/AEM.70.7.3877-3883.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward C., Grant A. J., Swift C., Philp J., Towler R., Heydarian M., Frost J. A., Maskell D. J. (2006). Phase-variable surface structures are required for infection of Campylobacter jejuni by bacteriophages. Appl. Environ. Microbiol. 72, 4638–4647 10.1128/AEM.00184-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S., Hodes A., Dai L., Mandhana N., Liu M., Deora R., Simons R. W., Zimmerly S., Miller J. F. (2004). Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature 431, 476–481 10.1038/nature02833 [DOI] [PubMed] [Google Scholar]
- Drexler K., Dannull J., Hindennach I., Mutschler B., Henning U. (1991). Single mutations in a gene for a tail fiber component of an Escherichia coli phage can cause an extension from a protein to a carbohydrate as a receptor. J. Mol. Biol. 219, 655–663 10.1016/0022-2836(91)90662-P [DOI] [PubMed] [Google Scholar]
- Frost J. A., Kramer J. M., Gillanders S. A. (1999). Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol. Infect. 123, 47–55 10.1017/S095026889900254X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemski P., Jr., Koeltzow D. E., Formal S. B. (1975). Phage conversion of Shigella flexneri group antigens. Infect. Immun. 11, 685–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemski P., Jr., Stocker B. A. (1967). Transduction by bacteriophage P22 in nonsmooth mutants of Salmonella typhimurium. J. Bacteriol. 93, 1588–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., Poly F., Riddle M., Maue A. C., Chen Y.-H., Monteiro M. A. (2012). Campylobacter polysaccharide capsules: virulence and vaccines. Front. Cell. Infect. Microbiol. 2:7. 10.3389/fcimb.2012.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., Szymanski C. M. (2008). Campylobacter sugars sticking out. Trends Microbiol. 16, 428–435 10.1016/j.tim.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Hansen V. M., Rosenquist H., Baggesen D. L., Brown S., Christensen B. B. (2007). Characterization of Campylobacter phages including analysis of host range by selected Campylobacter Penner serotypes. BMC Microbiol. 7, 90. 10.1186/1471-2180-7-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemolhosseini S., Montag D., Kramer L., Henning U. (1994). Determinants of receptor specificity of coli phages of the T4 family: a chaperone alters the host range. J. Mol. Biol. 241, 524–533 10.1006/jmbi.1994.1529 [DOI] [PubMed] [Google Scholar]
- Jones M. A., Marston K. L., Woodall C. A., Maskell D. J., Linton D., Karlyshev A. V., Dorrell N., Wren B. W., Barrow P. A. (2004). Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 72, 3769–3776 10.1128/IAI.72.7.3941-3950.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlyshev A. V., Champion O. L., Churcher C., Brisson J. R., Jarrell H. C., Gilbert M., Brochu D., St M. F., Li J., Wakarchuk W. W., Goodhead I., Sanders M., Stevens K., White B., Parkhill J., Wren B. W., Szymanski C. M. (2005). Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 55, 90–103 10.1111/j.1365-2958.2004.04374.x [DOI] [PubMed] [Google Scholar]
- Karlyshev A. V., Linton D., Gregson N. A., Lastovica A. J., Wren B. W. (2000). Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35, 529–541 10.1046/j.1365-2958.2000.01717.x [DOI] [PubMed] [Google Scholar]
- Kutateladze M., Adamia R. (2010). Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 12, 591–595 10.1016/j.tibtech.2010.08.001 [DOI] [PubMed] [Google Scholar]
- Levy S. B., Marshall B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–S129 10.1038/nm1145 [DOI] [PubMed] [Google Scholar]
- Linton D., Karlyshev A. V., Wren B. W. (2001). Deciphering Campylobacter jejuni cell surface interactions from the genome sequence. Curr. Opin. Microbiol. 4, 35–40 10.1016/S1369-5274(00)00161-2 [DOI] [PubMed] [Google Scholar]
- Liu M., Deora R., Doulatov S. R., Gingery M., Eiserling F. A., Preston A., Maskell D. J., Simons R. W., Cotter P. A., Parkhill J., Miller J. F. (2002). Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295, 2091–2094 10.1126/science.1067467 [DOI] [PubMed] [Google Scholar]
- McNally D. J., Jarrell H. C., Li J., Khieu N. H., Vinogradov E., Szymanski C. M., Brisson J. R. (2005). The HS:1 serostrain of Campylobacter jejuni has a complex teichoic acid-like capsular polysaccharide with nonstoichiometric fructofuranose branches and O-methyl phosphoramidate groups. FEBS J. 272, 4407–4422 10.1111/j.1742-4658.2005.04856.x [DOI] [PubMed] [Google Scholar]
- McNally D. J., Lamoureux M. P., Karlyshev A. V., Fiori L. M., Li J., Thacker G., Coleman R. A., Khieu N. H., Wren B. W., Brisson J. R., Jarrell H. C., Szymanski C. M. (2007). Commonality and biosynthesis of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. J. Biol. Chem. 282, 28566–28576 10.1074/jbc.M611027200 [DOI] [PubMed] [Google Scholar]
- Montag D., Hashemolhosseini S., Henning U. (1990). Receptor-recognizing proteins of T-even type bacteriophages. The receptor-recognizing area of proteins 37 of phages T4 TuIa and TuIb. J. Mol. Biol. 216, 327–334 10.1016/S0022-2836(05)80324-9 [DOI] [PubMed] [Google Scholar]
- Morona R., Klose M., Henning U. (1984). Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J. Bacteriol. 159, 570–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom K., Forsgren A. (1974). Effect of protein A on adsorption of bacteriophages to Staphylococcus aureus. J. Virol. 14, 198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J., Wren B. W., Mungall K., Ketley J. M., Churcher C., Basham D., Chillingworth T., Davies R. M., Feltwell T., Holroyd S., Jagels K., Karlyshev A. V., Moule S., Pallen M. J., Penn C. W., Quail M. A., Rajandream M. A., Rutherford K. M., van Vliet A. H., Whitehead S., Barrell B. G. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668 10.1038/35001088 [DOI] [PubMed] [Google Scholar]
- Rakhuba D. V., Kolomiets E. I., Szwajcer Dey E., Novik G. I. (2010). Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 59, 145–155 [PubMed] [Google Scholar]
- Riede I., Eschbach M. L. (1986). Evidence that TraT interacts with OmpA of Escherichia coli. FEBS Lett. 205, 241–245 10.1016/0014-5793(86)80905-X [DOI] [PubMed] [Google Scholar]
- Roberts I. S. (1996). The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50, 285–315 10.1146/annurev.micro.50.1.285 [DOI] [PubMed] [Google Scholar]
- Sails A. D., Wareing D. R., Bolton F. J., Fox A. J., Curry A. (1998). Characterisation of 16 Campylobacter jejuni and C. coli typing bacteriophages. J. Med. Microbiol. 47, 123–128 10.1099/00222615-47-2-123 [DOI] [PubMed] [Google Scholar]
- Scholl D., Rogers S., Adhya S., Merril C. R. (2001). Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75, 2509–2515 10.1128/JVI.75.6.2509-2515.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A. E., Timms A. R., Connerton P. L., Loc Carrillo C., Adzfa Radzum K., Connerton I. F. (2007). Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 3, e119. 10.1371/journal.ppat.0030119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M. C., van Alphen L. B., Harboe A., Li J., Christensen B. B., Szymanski C. M., Brøndsted L. (2011). The F336 bacteriophage recognizes the capsular phosphoramidate modification of Campylobacter jejuni NCTC11168. J. Bacteriol. 193, 6742–6749 10.1128/JB.05276-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Michael F. S., Szymanski C. M., Li J., Chan K. H., Khieu N. H., Larocque S., Wakarchuk W. W., Brisson J. R., Monteiro M. A. (2002). The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168. Eur. J. Biochem. 269, 5119–5136 10.1046/j.1432-1033.2002.03201.x [DOI] [PubMed] [Google Scholar]
- Stirm S., Freund-Molbert E. (1971). Escherichia coli capsule bacteriophages. II. Morphology. J. Virol. 8, 330–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski C. M., Michael F. S., Jarrell H. C., Li J., Gilbert M., Larocque S., Vinogradov E., Brisson J. R. (2003). Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from Campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J. Biol. Chem. 278, 24509–24520 10.1074/jbc.M301273200 [DOI] [PubMed] [Google Scholar]
- Timms A. R., Cambray-Young J., Scott A. E., Petty N. K., Connerton P. L., Clarke L., Seeger K., Quail M., Cummings N., Maskell D. J., Thomson N. R., Connerton I. F. (2010). Evidence for a lineage of virulent bacteriophages that target Campylobacter. BMC Genomics 11, 214. 10.1186/1471-2164-11-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer M. G. (2004). Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181 10.1016/j.femsre.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Yu F., Mizushima S. (1982). Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J. Bacteriol. 151, 718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]