Abstract
The yeast gene YDR533C encodes a protein belonging to the DJ-1/ThiJ/PfpI superfamily. This family includes the human protein DJ-1, which is mutated in autosomal recessive early-onset Parkinson's disease. The function of DJ-1 and its yeast homologue YDR533Cp is unknown. We report here the crystal structure of YDR533Cp at 1.8-Å resolution. The structure indicates that the closest relative to YDR533Cp is the Escherichia coli heat shock protein Hsp31 (YedU), which has both chaperone and protease activity. As expected, the overall fold of the core domain of YDR533Cp is also similar to that of DJ-1 and the bacterial protease PfpI. YDR533Cp contains a possible catalytic triad analogous to that of Hsp31 and an additional domain that is present in Hsp31 but is not seen in DJ-1 and other members of the family. The cysteine in this triad (Cys-138) is oxidized in this crystal structure, similar to modifications seen in the corresponding cysteine in the crystal structure of DJ-1. YDR533Cp appears to be a dimer both in solution and the crystal, but this dimer is formed by a different interface than that found in Hsp31 or other members of the superfamily.
Cells of the budding yeast Saccharomyces cerevisiae respond to environmental stress by mounting both general and specific programs of changes in gene expression (1). One such stress is the chemically induced presence of large amounts of misfolded proteins in the cytosol, which mimics some, but not all, features of heat shock (2). This accumulation of misfolded proteins can be induced with the amino acid analogue azetidine-2′-carboxylic acid (AZC), which resembles proline but has one fewer carbon atom in the ring (3). In contrast with what has been observed in mammalian cells, treatment of yeast with AZC does not evoke the unfolded protein response of the endoplasmic reticulum, but instead induces a reversible cell-cycle arrest with some of the characteristics of the quiescent state known as stationary phase (2). AZC-induced arrest is accompanied by significant changes in the expression of >500 genes in the yeast genome (4), including the 20-fold or more down-regulation of essentially all of the ribosomal protein genes plus a number of other genes linked to protein synthesis, although the mechanism of this down-regulation is not known. Genes whose expression is upregulated by 20-fold or more include the heat shock chaperones Hsp12, Hsp104, and other members of the heat shock protein family. The biochemical function of these proteins is to help unfolded or misfolded proteins assume their correct structures or to protect the cell from the potentially toxic affects of their aggregation.
YDR533C, which encodes a “hypothetical” protein of unknown function, is transcriptionally up-regulated nearly as much as Hsp12 when yeast cells are treated with AZC (4). This gene was chosen for further study because its function is unknown and its small size (predicted to encode a soluble protein of 237 aa) makes it an attractive candidate for possible functional characterization by structure determination. Knockouts of the gene in both haploid and diploid yeast cells are viable (J.L.C., unpublished work), making YDR533C amenable to functional characterization by genetic techniques.
Analysis of genomewide expression data from our laboratory and publicly available microarray databases indicate that YDR533C is not a component of the general environmental stress response in yeast. The transcription of YDR533C is upregulated from a basal level of ≈500 protein molecules per cell in exponential growth (5) to 5,000-10,000 protein molecules per cell in response to a subset of stress conditions. Specifically, production of YDR533Cp (yeast community convention uses italicized uppercase for the gene name, Roman case followed by the letter p to denote the protein product, and lowercase italics to specify a mutation in, or deletion of, the gene) increases markedly during heat shock, production of misfolded proteins, oxidative stress by hydrogen peroxide or the glutathione depletion agent diamide, or entry into stationary phase caused by carbon starvation.
YDR533C has three homologues in the S. cerevisiae genome: YMR322C, YOR391C, and YPL280W. Each of these three genes is predicted to encode a protein product that is ≈70% identical to YDR533Cp and 99.5% identical to each other. This striking conservation is the result of the location of these homologues in the yeast genome: in contrast to YDR533C, an interstitial gene on chromosome 4, YMR322C, YOR391C, and YPL280W all are subtelomeric on their respective chromosomes. Subtelomeric regions are well-known hotspots for recombination (6, 7), and it appears that this mini-gene family arose from an initial duplication of the parental gene (presumably YDR533C) into a subtelomeric location, followed by recombination to produce the other copies. The flanking regions around YMR322C, YOR391C, and YPL280W are identical for thousands of base pairs as well, reflecting the homogenizing of sequence that occurs through repeated recombination. Some divergence of regulation has already taken place in this mini-gene family, however, as the expression of YMR322C, YOR391C, and YPL280W is not upregulated by AZC treatment or carbon starvation, but is increased dramatically upon starvation for nitrogen (J. Gray and J.L.C., unpublished work).
Comparison of the predicted amino acid sequence of YDR533Cp with proteins from other organisms shows that YDR533Cp is a member of the DJ-1/ThiJ/PfpI superfamily, which can be divided into a number of subfamilies with distinct (albeit poorly characterized) functions (S. Bandyopadhyay and M. R. Cookson, personal communication). YDR533Cp orthologs in a variety of prokaryotic and eukaryotic species cluster together, and most are of unknown function. The most closely related protein of known function is Hsp31 (YedU), a heat shock chaperone from Escherichia coli. The 3D structure of Hsp31 has been solved and reveals a dimeric molecule with a cysteine protease-like catalytic triad and a Zn2+ binding site (8-10). The importance of both the catalytic triad and the Zn2+ site are unclear, but their presence suggests that Hsp31 may have a number of distinct and still uncharacterized functions (9).
Two other major subfamilies of the DJ-1/ThiJ/PfpI superfamily as indicated by sequence comparison are represented by the bacterial proteins ThiJ and PfpI. ThiJ and its close orthologs are kinases involved in the biosynthesis of thiamine. PfpI and its close relatives are annotated as cysteine proteases; a crystal structure of one of them (PH1704) shows the presence of a cysteine protease-like catalytic triad, but the glutamate in the triad is contributed by a neighboring subunit in the hexameric protein structure (11). Still other, more distantly related subfamilies contain glutamine amidotransferases (12) and AraC-like transcriptional regulators (13, 14).
Despite differences in function, proteins in the DJ-1/ThiJ/PfpI superfamily appear to share some important common features. In particular, all DJ-1/ThiJ/PfpI superfamily members contain an absolutely conserved cysteine residue in a “nucleophile elbow” strand-loop-helix motif (15). This cysteine is the catalytic nucleophile in both the cysteine proteases and the GATA enzymes. An adjacent histidine residue is conserved in some, but not all, of the subfamilies; in the proteases this histidine is a member of the catalytic triad. In addition, all structurally characterized members of the superfamily appear to be oligomers, and the oligomeric state of these proteins appears to be crucial either to their stability or biochemical activity. The oligomeric state of members of the superfamily varies widely, ranging from dimers to trimers and hexamers, and the oligomerization interfaces differ from protein to protein. This highly variable oligomerization suggests that different modes of selfassociation may be correlated with functional diversity in the DJ-1/ThiJ/PfpI superfamily.
The most medically important member of the superfamily is DJ-1, mutations in which have been directly implicated in autosomal-recessive, early-onset Parkinson's disease (16, 17). All of the well-characterized mutations associated with disease appear to be loss-of-function mutations. The biochemical function of DJ-1 is unknown, but there is speculation that it may be involved in the cellular response to oxidative stress (18, 19). Several groups have determined the crystal structure of DJ-1, and these studies show that DJ-1 is a dimer and lacks the putative catalytic triad seen in the members of the protease subfamily, although it does have a permuted version that includes a conserved cysteine residue (Cys-106) (10, 20-23). Furthermore, structural and biochemical studies indicate that the Parkinson's-associated mutation L166P destabilizes the dimer and leads to rapid degradation, resulting in a loss of function (24, 25). As a consequence of its medical importance, much recent effort has been expended in the search for the biological function(s) of DJ-1, particularly the function that is disrupted in DJ-1-associated early-onset Parkinson's disease.
To better understand the possible functions and evolutionary relationships among proteins in the DJ-1/ThiJ/PfpI superfamily, the crystal structure of YDR533Cp has been determined to a resolution of 1.8 Å. The structure shows that YDR533Cp has the core flavodoxin fold that is common to all members of the DJ-1/ThiJ/PfpI superfamily and is most similar to the bacterial chaperone Hsp31. Based on both the structural and expression data for YDR533Cp, we propose that its biochemical function is likely to be as a chaperone for proteins that are misfolded or have been damaged by oxidative stress. The possibility that it may degrade or nick these proteins is also consistent with the presence of a putative catalytic triad in YDR533Cp. In contrast with the structure of Hsp31, there is no Zn2+ binding site in YDR533Cp. YDR533Cp and DJ-1 are found to be significantly different in structure; however, they share one striking similarity: Cys-138 in YDR533Cp, like its counterpart Cys-106 in DJ-1, is highly reactive and readily oxidized, most probably to sulfinic acid. Lastly, YDR533Cp has a high degree of sequence identity (45-60%) with two proteins from pathogenic fungi that have been implicated in immunological protein binding, although the implications of this similarity are unclear.
Materials and Methods
Protein Expression and Purification. The gene for YDR533Cp was subcloned into the NdeI and XhoI restriction sites of the expression construct pET21a (Novagen) such that the expressed protein contained a C-terminal 6×His tag. BL21(DE3) E. coli (Novagen) transformed with the expression construct was grown in LB broth supplemented with 100 μg/ml ampicillin, and overexpression of YDR533Cp was induced with 0.4 mM isopropyl β-d-thiogalactoside for 1 h. Cells were harvested by centrifugation, lysed on ice in 50 mM sodium phosphate (pH 7.0), 300 mM NaCl, and 10 mM imidazole supplemented with 1 mg/ml lysozyme, and sonicated, and cell debris was removed by centrifugation. The cleared lysate was purified by Ni2+-NTA agarose column chromatography followed by DEAE anion exchange chromatography. The protein was dialyzed into crystallization buffer (50 mM Mes, pH 6.0/1 mM EDTA/1 mM DTT) and stored at -80°C.
Protein Crystallization and Data Collection. Crystals of YDR533Cp with the symmetry of space group C2221 were obtained by the hanging drop vapor diffusion method by mixing 2 μl of YDR533Cp at 10 mg/ml with 2 μl of reservoir solution (35-40% PEG 2000/200 mM NH4CH3COO/100 mM NaCH3COO, pH 4.0). Plate-shaped crystals appeared after 5-10 days of incubation at room temperature and typically measured 0.5 × 0.5 × 0.15 mm. The crystals were intolerant of the addition of standard cryoprotection agents; however, the concentration of PEG 2000 in the mother liquor (35-40%) was sufficient to partially cryoprotect the crystals. Acceptable reflection profiles and mosaicities could be obtained only after annealing the crycooled crystals by returning them to reservoir solution for 3 min at room temperature, followed by direct reimmersion into liquid N2 (26).
Structure Determination and Refinement. Initial attempts at molecular replacement using a variety of search models from other members of the superfamily failed to produce an outstanding solution, and experimental-phase information was required to determine the structure of YDR533Cp. Phase information was obtained by multiple isomorphous replacement using a native data set and two heavy-atom derivatives [K2HgI4 and Pb(NO3)2]. All data sets (see Table 1) used in the phasing were collected at room temperature from capillary-mounted crystals on a Rigaku (Tokyo) RU-300 rotating Cu anode source operating at 38 kV × 24 mA. All data were integrated and scaled with denzo and scalepack, respectively (27). Because crystals of YDR533Cp were nonisomorphous even before derivitization, the native and derivative data sets were collected from three fragments of a single large native crystal that was broken into pieces before derivitization. Derivative crystals were obtained by soaking for 10 min in reservoir solution saturated with K2HgI4 or in a 10-mM solution of Pb(NO3)2 in the reservoir solution. This “quick-soak” method resulted in better isomorphism and less crystal damage than longer soaks at lower concentrations of the heavy atom compounds (28).
Table 1. Data and refinement statistics.
| High resolution | Native | Pb(NO3)2 | K2Hgl4 | |
|---|---|---|---|---|
| Data set | ||||
| Resolution range, Å | 69-1.8 | 23-2.5 | 23-2.5 | 35-2.5 |
| Space group | C2221 | C2221 | C2221 | C2221 |
| a, Å | 46.282 | 46.414 | 46.771 | 46.402 |
| b, Å | 68.776 | 69.817 | 69.631 | 69.894 |
| c, Å | 137.197 | 140.084 | 140.030 | 140.273 |
| Unique reflections | 18,620 | 7,390 | 7,203 | 7,617 |
| Completeness, % | 99.5 (99.0) | 97.7 (87.7) | 95.4 (78.1) | 98.1 (95.1) |
| Multiplicity | 6.3 (6.2) | 4.4 (2.5) | 4.24 (1.9) | 4.2 (2.9) |
| Rmerge, %* | 7.9 (50.9) | 9.8 (37.3) | 11.2 (38.1) | 8.0 (25.4) |
| I/(I) | 24.7 (3.95) | 14.0 (3.3) | 13.6 (2.8) | 17.1 (5.0) |
| Phasing statistics | ||||
| No. of sites | 1 | 1 | ||
| FOMsolve | 0.42 | |||
| FOMresolve | 0.78 | |||
| Refinement statistics | ||||
| No. of residues | 235 | |||
| No. of waters | 229 | |||
| Rwork, %† | 20.0 (26.7) | |||
| Rfree, %‡ | 23.8 (35.1) | |||
| rmsd bond, Å | 0.019 | |||
| rmsd angle, ° | 1.57 |
Values in parentheses refer to statistics in the highest-resolution shell. FOM, figure of merit.


Rfree was calculated as Rworks where the F(hkl)o were taken from a set of 2,020 reflections (10% of the data) that were not included in the refinement
Heavy atom sites were located, and phases were calculated to 2.5-Å resolution by using solve (29). Statistical density modification and subsequent automated model building were performed with resolve (30), which placed 33% of the residues. The resolve-built partial model was used as a guide to manually build the remainder of the protein into the density-modified electron density maps with the program O (31). The model was subsequently refined against a 1.8-Å resolution native data set collected at 100 K at beamline 8.3.1 of the Advanced Light Source at the Lawrence Berkeley National Laboratory (Berkeley, CA). Initial torsion angle molecular dynamics-simulated annealing (32), conjugate-direction coordinate minimization, and restrained atomic isotropic B-factor refinement were performed with the CNS refinement package (33). The final cycles of refinement included translation-libration-screw (TLS) refinement of anisotropic displacement parameters (34, 35) and were performed with refmac5 in the CCP4 suite of programs (36). It is worth noting that TLS refinement with a single rigid group reduced the R and Rfree (37) values by 4%. All refinements were performed against all of the measured data (excluding the test set for calculation of Rfree) by using a maximum-likelihood target function based on amplitudes and including both an anisotropic scale correction and a bulk solvent correction.
Results and Discussion
Structure Description. YDR533Cp is a 237-aa residue protein containing 10 α-helices and 10 β-strands that form two α/β-domains (Fig. 1). The major domain of YDR533Cp (domain A) is an α/β sandwich that is characteristic of all members of the DJ-1/ThiJ/PfpI superfamily and contains β-strands β1 and β4-β10 plus α-helices αA, αC-αG, and αJ. Domain A resembles a flavodoxin-like fold but contains seven, rather than five, β-strands in a parallel β-sheet with strand order: β4, β1, β5, β6, β10, β7, and β8. Strand β9 is the only antiparallel strand in domain A and packs against a face of the central parallel β-sheet.
Fig. 1.
A ribbon diagram of the monomer of YDR533Cp. Domain A, common to other members of the DJ-1/ThiJ/PfpI superfamily, is colored blue, and domain P, specific to YDR533Cp and its close relatives, is colored yellow. β-Strands are numbered 1-10, and α-helices are lettered A-J. The diagram was made with molscript (45).
A smaller second domain (domain P), formed by antiparallel strands β2 and β3, α-helices αB, αH, and αI, and extended loop regions, caps domain A and is found in the sequences of only a subset of the proteins in the DJ-1/ThiJ/PfpI superfamily, including one of known structure, the E. coli chaperone Hsp31 (8-10). Domain P partially occludes a cleft in domain A that contains an absolutely conserved cysteine (Cys-138 in YDR533Cp) located at the nucleophile elbow region of domain A (see below). Overall, the structure of YDR533Cp is similar to Hsp31, with some notable differences (see below).
Oligomerization State. Gel filtration chromatography and dynamic light scattering indicate that YDR533Cp is a dimer with an apparent molecular mass of ≈45 kDa in solution, in reasonable agreement with the expected dimeric mass of 51 kDa (data not shown). The dimer in the crystal is generated by a crystallographic twofold axis, buries 1,080 Å2 of surface area, and involves contacts between strands β7, β8, and β9 on each monomer. The β-strands form a quasi-six-stranded β-barrel at the dimer interface (Fig. 2), which has not been observed in the structures of other members of the superfamily, including Hsp31. The differing dimerization interfaces in YDR533Cp and Hsp31 is caused, in part, by a 45-residue N-terminal extension in Hsp31 that contains most of the residues involved in the dimer interface for this protein. This stretch of residues is absent in YDR533Cp, eliminating this potential interface.
Fig. 2.
Ribbon and electrostatic surface representation of the YDR533Cp dimer. (A and B) Views of the YDR533Cp dimer that is generated by crystallographic symmetry. The dimer twofold axis is represented as an arrow and is in the plane of the page in A, and it is perpendicular to the plane of the page and is represented as an ellipse in B. In A and B, monomer A is blue (domain A) and yellow (domain B), and monomer B is green (domain A) and orange (domain B). (C and D) The electrostatic surface for the dimer calculated by grasp in the same orientation as A and B. The saddle-shaped depression in the dimer is highly negatively charged. The figure was made with grasp (46) and molscript (45).
Dimerization of YDR553Cp forms a large and highly negatively charged saddle-shaped depression on one face of the dimer and another smaller depression on the opposite side (Fig. 2). The concentration of negative charge in the “saddle” region of the dimer is striking and is reminiscent of the gross surface features of Hsp31, despite the different dimerization interface. In addition, the dimerization interface in YDR533Cp is modest and buries only half of the surface area that is buried in the Hsp31 dimer (8-10), suggesting that the YDR533Cp dimer may not be as robust as the Hsp31 dimer.
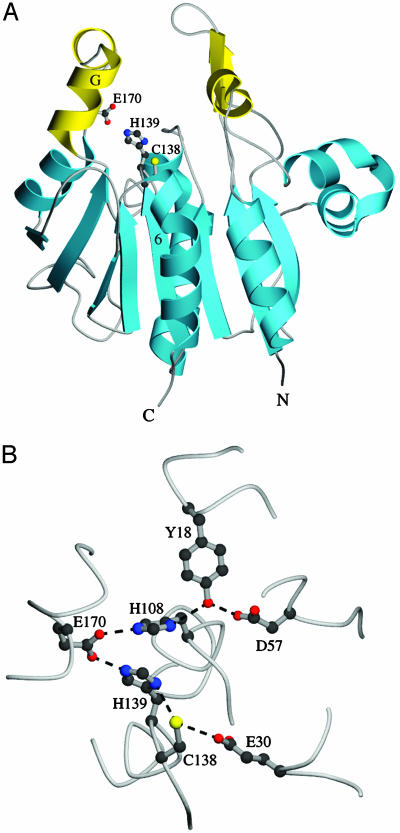
A Triad at the Nucleophile Elbow Region and Oxidative Modification at Cys-138. The nucleophile elbow (15) is a motif common to all members of known structures in the DJ-1/ThiJ/PfpI superfamily and contains an absolutely conserved cysteine residue in an unusually strained backbone conformation. The conserved cysteine at this position is catalytically essential in the cysteine protease and glutamine amidotransferase members of the superfamily and may be involved in possible enzymatic activities of the other members of the superfamily with unknown function. In YDR533Cp, Cys-138 is at the nucleophile elbow between β6 and an extended region that is in a highly distorted quasi-helical conformation. This cysteine is a member of a putative Cys-138-His-139-Glu-170 catalytic triad (Fig. 3A), where Glu-170 is contributed by αG in domain P, very similar to the arrangement of residues seen in Hsp31 (8-10) but notably different from the intermolecular catalytic triad seen in the hexameric structure of PhpI (11), where the glutamate is donated by another monomer. Importantly, because the activity of YDR533Cp is unknown, it cannot be said with certainty whether the observed triad is indeed catalytic, although the presence of the triad strongly suggests a possible hydrolase/protease activity for YDR533Cp. Such activity has been observed for Hsp31, particularly against small peptide substrates (10).
Fig. 3.
Two views of the catalytic triad in YDR533Cp. (A) The location of triad residues C138, H139, and E170 on the monomer. (B) The hydrogen-bonding network that encompasses the triad. E170 hydrogen-bonds with both the triad residue H139 and a second histidine, H108. This second histidine is also found in the structurally homologous E. coli chaperone Hsp31. The figure was made with molscript (45).
The triad in YDR533Cp is at the bottom of a predominantly hydrophobic cleft, but the triad is surrounded by polar residues that form an extensive hydrogen-bonding network (Fig. 3B). In addition, Glu-30, a highly conserved residue whose importance remains obscure, makes a 3.2-Å hydrogen bond with the Sγ of Cys-138. Glu-30 is also found near the nucleophile elbow cysteine in the structures of Hsp31 (8-10), PhpI (11), and DJ-1 (10, 20-23), but not in the more distantly related glutamine amidotransferases (38-41), suggesting that the presence of a glutamate at this position may be correlated with a functional distinction between different clades of the superfamily.
Electron density maps indicate that Cys-138 is modified, probably by oxidation, in the crystal (Fig. 4). The electron density around Cys-138 is not completely consistent with any single oxidative modification of cysteine, but is consistent with a mixture of species that likely includes sulfinic acid. A carboxylate oxygen of Glu-30 makes close contact with the modified Cys-138, which places two groups with acidic pKas in close proximity and thus requires that either Glu-30 or oxidized Cys-138 be protonated. The importance of the modification at Cys-138 and the close contact with Glu-30 is uncertain, although the high degree of conservation of both residues suggests that the modified Cys-138 may be important. Oxidation would decrease the nucleophilicity of Sγ in the modified cysteine residue, possibly providing a means of altering or regulating the activity of YDR533Cp in response to oxidative stress. This idea is speculative, as both the in vivo oxidation status of Cys-138 and the function of YDR533Cp are currently unknown. It is particularly noteworthy that the homologous cysteine residue in DJ-1 (Cys-106) was found to be very sensitive to modification by x-ray radiation in a previous structure (21), suggesting that unusual reactivity and susceptibility to oxidation are common features of this absolutely conserved cysteine in members of the superfamily.
Fig. 4.
A view of the electron density around Cys-138 in the triad. Sigma-A weighted 2 Fo - Fc electron density contoured at 1.0 σ is shown in blue, and sigma-A weighted Fo - Fc electron density contoured at 3.0 σ is shown in green. The Fo - Fc difference electron density around the Sγ of C138 suggests oxidative modification to a mixture of species, including sulfinic acid. E30 is close to the Cys at the nucleophile elbow in several structures of the DJ-1/ThiJ/PfpI superfamily. The figure was made with povscript+ (47).
Comparison with Other Members of the DJ-1/ThiJ/PfpI Superfamily. A dali search (42) reveals that YDR533Cp is most structurally similar to the E. coli chaperone Hsp31 [Cα rms deviation (rmsd) = 2.5 Å, 29% sequence identity, Z score = 23.2], and both proteins share a global similarity in fold, including the presence of domain P (Fig. 5A). However, YDR533Cp lacks a region that corresponds to the first 45 aa of Hsp31, resulting in a smaller domain P and a more solvent-accessible catalytic triad in YDR533Cp. In addition, although YDR533Cp is dimeric, like Hsp31, the dimer interface in YDR533Cp is different from that observed in Hsp31 (see above). Lastly, YDR533Cp lacks the two His residues that contribute to the Zn2+ binding site in Hsp31 (9, 10), again suggesting either a possible functional distinction between these two structurally similar proteins or that Zn2+ binding to Hsp31 may not be functionally important. We favor the former hypothesis, as two independent groups have observed Zn2+ bound to crystalline Hsp31 in two different space groups, even when no Zn2+ was added to the protein (9, 10). This finding strongly suggests that the Zn2+ binding site in Hsp31 is specific and of probable functional importance.
Fig. 5.
Superposition of YDR533Cp, Hsp31, and DJ-1 monomers. (A) YDR533Cp (blue) and Hsp31 (red) are superimposed (Cα rmsd = 2.5 Å), showing the high degree of structural similarity between these two proteins. Most of the structural differences are in the P domains. (B) YDR533Cp (blue) and DJ-1 (yellow) are superimposed in the same orientation as in A (Cα rmsd = 2.1 Å). Despite the better agreement in the core domain A, DJ-1 lacks a P domain, and YDR533Cp and DJ-1 are likely members of different branches of the DJ-1/ThiJ/PfpI superfamily. The figure was made with molscript (45).
Of the proteins in the DJ-1/ThiJ/PfpI superfamily, DJ-1 has attracted the most attention because of its involvement in early-onset Parkinson's disease (16, 17, 43). YDR533Cp and DJ-1 are structurally similar in the core flavodoxin-like fold that is shared by all members of the superfamily, but the presence of an additional domain (domain P) of unknown function, a possible catalytic triad, and a different dimerization interface in YDR533Cp distinguishes it from DJ-1 and indicates that these two proteins are members of different branches of the DJ-1/ThiJ/PfpI superfamily (Fig. 5B). Consequently, YDR533Cp and DJ-1 are not likely to share the same function. The possibility that they may exhibit some overlap in function is not ruled out by our data, however, and will require further biochemical and genetic studies.
Recently, a 29-kDa IgE-binding protein from Candida albicans that has 62% sequence identity with YDR533p was identified (44). In addition, a 36-kDa macrophage-binding protein from Coccidioides immitis with 45% sequence identity to YDR533Cp was identified previously (G.T. Cole, personal communication). The high degree of sequence identity between and YDR533Cp and these binding proteins from two fungal pathogens, which is significantly greater than the sequence identity between YDR533Cp and Hsp31 (29%), suggests a possible functional connection, although this idea remains speculative pending further experiments.
Based on the structural and likely functional similarities between YDR533Cp and the heat shock protein Hsp31, we propose that the yeast gene YDR533C be renamed HSP31. This name does not imply a specific biochemical function for the gene product, which remains to be established, but emphasizes the evolutionary relationship between the two genes and is consistent with the up-regulation of YDR533C in response to heat shock and other environmental stresses. A new name can be assigned later if needed once the biochemical and cellular functions are established. Following the established convention for gene naming in S. cerevisiae, we also propose that the YDR533C homologues YPL280W and YOR391C be named HSP32 and HSP33, respectively. The fourth member of the yeast mini-gene family, YMR322C, already has the standard name SNO4, based on its interaction with members of the SNZ gene family in a yeast two-hybrid screen. We believe that this name may be misleading, as YMR322C does not appear to be related in sequence or likely biochemical function to any of the three SNO genes, which have been annotated as being involved in the synthesis of pyridoxine derivatives. Parsimony suggests that HSP34 would be a preferable name for YMR322C, emphasizing its relationship with the other members of its gene family in yeast and its structural relative in bacteria.
Abbreviations: AZC, azetidine-2′-carboxylic acid; rmsd, rms deviation.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1RW7).
References
- 1.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trotter, E. W., Berenfeld, L., Krause, S. A., Petsko, G. A. & Gray, J. V. (2001) Proc. Natl. Acad. Sci. USA 98, 7313-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckmann, R. P., Mizzen, L. A. & Welch, W. J. (1990) Science 248, 850-854. [DOI] [PubMed] [Google Scholar]
- 4.Trotter, E. W., Kao, C. M. F., Berenfeld, L., Botstein, D., Petsko, G. A. & Gray, J. V. (2002) J. Biol. Chem. 277, 44817-44825. [DOI] [PubMed] [Google Scholar]
- 5.Ghaemmaghami, S., Huh, W., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K. & Weissman, J. S. (2003) Nature 425, 737-741. [DOI] [PubMed] [Google Scholar]
- 6.Louis, E. J. & Haber, J. E. (1990) Genetics 124, 533-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mefford, H. C. & Trask, B. J. (2002) Nat. Rev. Genet. 3, 91-102. [DOI] [PubMed] [Google Scholar]
- 8.Quigley, P. M., Korotkov, K., Baneyx, F. & Hol, W. G. J. (2003) Proc. Natl. Acad. Sci. USA 100, 3137-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao, Y. H., Liu, D. Q., Kaluarachchi, W. D., Bellamy, H. D., White, M. A. & Fox, R. O. (2003) Protein Sci. 12, 2303-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, S. J., Kim, S. J., Kim, I. K., Ko, J., Jeong, C. S., Kim, G. H., Park, C., Kang, S. O., Suh, P. G., Lee, H. S. & Cha, S. S. (2003) J. Biol. Chem. 278, 44552-44559. [DOI] [PubMed] [Google Scholar]
- 11.Du, X. L., Choi, I. G., Kim, R., Wang, W. R., Jancarik, J., Yokota, H. & Kim, S. H. (2000) Proc. Natl. Acad. Sci. USA 97, 14079-14084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massiere, F. & Badet-Denisot, M. A. (1998) Cell. Mol. Life Sci. 54, 205-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soisson, S. M., MacDougall-Shackleton, B., Schleif, R. & Wolberger, C. (1997) J. Mol. Biol. 273, 226-237. [DOI] [PubMed] [Google Scholar]
- 14.Gallegos, M. T., Schleif, R., Bairoch, A., Hofmann, K. & Ramos, J. L. (1997) Microbiol. Mol. Biol. Rev. 61, 393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., Harel, M., Remington, S. J., Silman, I., Schrag, J., et al. (1992) Protein Eng. 5, 197-211. [DOI] [PubMed] [Google Scholar]
- 16.Bonifati, V., Rizzu, P., Squitieri, F., Krieger, E., Vanacore, N., van Swieten, J. C., Brice, A., van Duijn, C. M., Oostra, B., Meco, G. & Heutink, P. (2003) Neurol. Sci. 24, 159-160. [DOI] [PubMed] [Google Scholar]
- 17.Bonifati, V., Rizzu, P., van Baren, M. J., Schaap, O., Breedveld, G. J., Krieger, E., Dekker, M. C., Squitieri, F., Ibanez, P., Joosse, M., et al. (2003) Science 299, 256-259. [DOI] [PubMed] [Google Scholar]
- 18.Cookson, M. R. (2003) Neuron 37, 7-10. [DOI] [PubMed] [Google Scholar]
- 19.Cookson, M. R. (2003) Proc. Natl. Acad. Sci. USA 100, 9111-9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao, X. & Tong, L. (2003) J. Biol. Chem. 278, 31372-31379. [DOI] [PubMed] [Google Scholar]
- 21.Wilson, M. A., Collins, J. L., Hod, Y., Ringe, D. & Petsko, G. A. (2003) Proc. Natl. Acad. Sci. USA 100, 9256-9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honbou, K., Suzuki, N. N., Horiuchi, M., Niki, T., Taira, T., Ariga, H. & Inagaki, F. (2003) J. Biol. Chem. 278, 31380-31384. [DOI] [PubMed] [Google Scholar]
- 23.Huai, Q., Sun, Y. J., Wang, H. C., Chin, L. S., Li, L., Robinson, H. & Ke, H. M. (2003) FEBS Lett. 549, 171-175. [DOI] [PubMed] [Google Scholar]
- 24.Macedo, M. G., Anar, B., Bronner, I. F., Cannella, M., Squitieri, F., Bonifati, V., Hoogeveen, A., Heutink, P. & Rizzu, P. (2003) Hum. Mol. Genet. 12, 2807-2816. [DOI] [PubMed] [Google Scholar]
- 25.Miller, D. W., Ahmad, R., Hague, S., Baptista, M. J., Canet-Aviles, R., McLendon, C., Carter, D. M., Zhu, P. P., Stadler, J., Chandran, J., et al. (2003) J. Biol. Chem. 278, 36588-36595. [DOI] [PubMed] [Google Scholar]
- 26.Harp, J. M., Timm, D. E. & Bunick, G. J. (1998) Acta Crystallogr. D 54, 622-628. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski, Z. & Minor, W. (1997) Macromol. Crystallogr. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 28.Sun, P. D., Radaev, S. & Kattah, M. (2002) Acta Crystallogr. D 58, 1092-1098. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger, T. C. (2000) Acta Crystallogr. D 56, 965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 32.Rice, L. M. & Brunger, A. T. (1994) Proteins Struct. Funct. Genet. 19, 277-290. [DOI] [PubMed] [Google Scholar]
- 33.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 34.Schomaker, V. & Trueblood, K. N. (1968) Acta Crystallogr. B 24, 63-76. [Google Scholar]
- 35.Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001) Acta Crystallogr. D 57, 122-133. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 37.Brunger, A. T. (1992) Nature 355, 472-475. [DOI] [PubMed] [Google Scholar]
- 38.Tesmer, J. G., Klem, T. J., Deras, M. L., Davisson, V. J. & Smith, J. L. (1996) Nat. Struct. Biol. 3, 74-86. [DOI] [PubMed] [Google Scholar]
- 39.Knochel, T., Ivens, A., Hester, G., Gonzalez, A., Bauerle, R., Wilmanns, M., Kirschner, K. & Jansonius, J. N. (1999) Proc. Natl. Acad. Sci. USA 96, 9479-9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, H. M., Ryan, T. J., Chave, K. J. & Van Roey, P. (2002) J. Biol. Chem. 277, 24522-24529. [DOI] [PubMed] [Google Scholar]
- 41.Korolev, S., Skarina, T., Evdokimova, E., Beasley, S., Edwards, A., Joachimiak, A. & Savchenko, A. (2002) Proteins Struct. Funct. Genet. 49, 420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holm, L. & Sander, C. (1995) Trends Biochem. Sci. 20, 478-480. [DOI] [PubMed] [Google Scholar]
- 43.Dawson, T. M. & Dawson, V. L. (2003) Science 302, 819-822. [DOI] [PubMed] [Google Scholar]
- 44.Chou, H., Tam, M. F., Chang, C. Y., Lai, H. Y., Huang, M. H., Chou, C. T., Lee, S. S. & Shen, H. D. (2003) Allergy 58, 1157-1164. [DOI] [PubMed] [Google Scholar]
- 45.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 46.Nicholls, A., Bharadwaj, R. & Honig, B. (1993) Biophys. J. 64, A166-A166. [Google Scholar]
- 47.Fenn, T. D., Ringe, D. & Petsko, G. A. (2003) J. Appl. Crystallogr. 36, 944-947. [Google Scholar]