Abstract
To elucidate the organizational and evolutionary principles of the metabolism of living organisms, recent studies have addressed the graph-theoretic analysis of large biochemical networks responsible for the synthesis and degradation of cellular building blocks [Jeong, H., Tombor, B., Albert, R., Oltvai, Z. N. & Barabási, A. L. (2000) Nature 407, 651-654; Wagner, A. & Fell, D. A. (2001) Proc. R. Soc. London Ser. B 268, 1803-1810; and Ma, H.-W. & Zeng, A.-P. (2003) Bioinformatics 19, 270-277]. In such studies, the global properties of the network are computed by considering enzymatic reactions as links between metabolites. However, the pathways computed in this manner do not conserve their structural moieties and therefore do not correspond to biochemical pathways on the traditional metabolic map. In this work, we reassessed earlier results by digitizing carbon atomic traces in metabolic reactions annotated for Escherichia coli. Our analysis revealed that the average path length of its metabolism is much longer than previously thought and that the metabolic world of this organism is not small in terms of biosynthesis and degradation.
Keywords: bioinformatics, metabolism, small-world network
According to the formal definition, in a small-world network, (i) most nodes (metabolites in our case) have a low connection degree, and the degree distribution follows a power law also referred to as scale-freeness; (ii) high-degree nodes, called hubs, dominate the network, and most nodes are clustered around hubs; and (iii) the average path length (AL; i.e., the average of the shortest path length over all pairs of nodes in the network) remains the theoretical minimum, that of a random graph (1-3). Because of its topology with few hubs, a small-world network may be resistant to random failures: any peripheral node is likely to have a low connection degree and is therefore expendable. In biological networks, the hubs are thought to be functionally important and phylogenetically oldest (4-6).
Although several groups confirmed the small-world property of small-molecule metabolisms in multiple data sources, the details of their results differ depending on the purpose of the analysis and its data-preparation scheme (4-9). Notable differences are attributable to the reversibility of enzymatic reactions and to the treatment of metabolically ubiquitous compounds referred to as coenzymes or inorganics. Table 1 summarizes differences in the major analyses and compares the AL and hub metabolites they identified.
Table 1. Comparison of four E. coli network analyses.
| Jeong et al. (7), directed | Wagner and Fell (5), undirected | Ma and Zeng (8), directed | This study, (un)directed | |
|---|---|---|---|---|
| Top 10 hubs | H2O | l-glutamate | glycerate 3P | carbon dioxide |
| ADP | pyruvate | d-ribose 5P | pyruvate | |
| P | CoA | acetyl CoA | acetyl CoA | |
| ATP | α-keto glutarate | pyruvate | ATP | |
| l-glutamate | l-glutamine | d-xylulose 5P | d-glucose | |
| NADP+ | l-aspartate | d-fructose 6P | l-glutamate | |
| PP | acetyl CoA | 5P-d-ribose 1PP | d-galactose | |
| NAD+ | phosphoribosyl PP | l-glutamate | CoA | |
| NADPH | tetrahydrofolate | d-glyceraldehyde 3P | S-adenosyl L-methionine | |
| NADH | succinate | l-aspartate | d-5-phosphoribosyl-1P | |
| AL | 3.2 | 3.8 | 8.2 | 8.4 (8.0) |
The top 10 hub metabolites and ALs reported in each study. Wagner and Fell (5) computed several versions of the network; the one shown here is the substrate-based network where ATP, ADP, NAD, NADP, NADH, NADPH, carbon dioxide, ammonia, sulfate, thioredoxin, (ortho) phosphate (P), and pyrophosphate (PP) are removed.
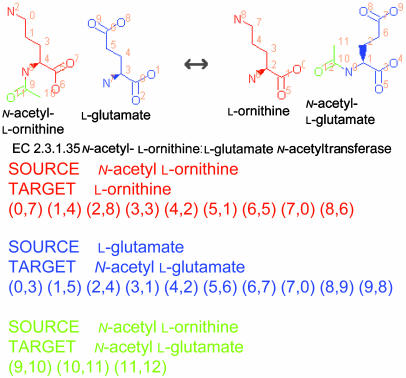
All of these studies used the same algorithmic procedure, and discrepancies are ascribable to the different aims of their network analyses. Jeong et al. (7) computed the proximity of metabolites by regarding all substrates and products in the same reaction as adjacent (Fig. 1; see also Fig. 7 and Supporting Text, which are published as supporting information on the PNAS web site). Wagner and Fell (5) computed stoichiometric relationships to estimate the transmission degree of perturbations in the metabolic network. They used the metrics with and without coenzymes such as ATP and NAD in both substrate- and reaction-based networks to compare their differences. Ma and Zeng (8) manually specified links in each reaction, aiming to delineate only physical relationships responsible for biosynthesis and degradation. To reproduce biochemical pathways in the traditional metabolic map, however, metabolites to be linked cannot be defined per se by compounds or reactions. The biochemical link between metabolites is context-sensitive; it depends on the conserved structural moieties in the adjacent reactions. To accurately compute the reaction connectivity as in the traditional metabolic map, we used digitally compiled atomic mappings, i.e., atomic position pairs between substrates and products corresponding to the substructural moieties conserved in each reaction (Figs. 1 and 2) (10). With this information, we reassessed the global properties of metabolic networks with special emphasis on the small-world hypothesis.
Fig. 1.
Two representations of the EC 2.3.1.35 reaction. In this reaction, the acetyl moiety of N-acetyl l-ornithine is transferred to l-glutamate to form N-acetyl l-glutamate. (Lower Left) In the scheme of Jeong et al. (7), its two substrates and two products are equally linked to the object representing the EC number, irrespective of their structural changes. (Lower Right) In our scheme, conserved substructural moieties, coded by color, are computationally detected, and each link is associated with the information of which atom goes where.
Fig. 2.
Three atomic mappings for the EC 2.3.1.35 reaction. Each reaction formula is decomposed to a set of substructural correspondences coded by color: each color indicates a set of atomic position pairs called an atomic mapping. Atomic positions are line numbers in the MOL file format files (see Methods for details) and are not generalizable to other metabolic databases.
Methods
Definition of Metabolic Pathways. In this work, a metabolic pathway (pathway for short) from metabolite X to Y is defined as a sequence of biochemical reactions through which at least one carbon atom in X reaches Y. Only carbon atoms are considered throughout this article. A metabolite Y is called reachable from X if there is a pathway from X to Y.
Preparation of Reaction Data. The reaction formulas annotated for Escherichia coli were originally collected from the Kyoto Encyclopedia of Genes and Genomes (www.genome.ad.jp/kegg), the Encyclopedia of Escherichia coli K12 Genes and Metabolism (http://ecocyc.org), and BRENDA (www.brenda.uni-koeln.de) databases and Enzyme Nomenclature (www.chem.qmw.ac.uk/iubmb/enzyme) (11-14). For a gene annotation with a specific EC number of the enzyme hierarchy, the corresponding reaction formulas were collected. If additional metabolites (other than those in the registered reaction formulas) were described in the comment section of the EC definition, the corresponding reaction formula was extrapolated and also included in our data set. For a gene annotation with an incomplete EC number, such as EC 1.1.1.-(the hyphen is a “do not care” symbol), the corresponding reaction formulas were collected from the pathway maps of the aforementioned databases. When the formula in the pathway maps coincided with a specific EC number (e.g., EC 1.1.1.100), the incomplete number was overwritten with the specific number. In the curating process, we attempted to match as many E. coli genes with specific reaction formulas as possible. The reactions remaining with incomplete EC numbers were labeled with our original numbers, starting from 999, to distinguish annotated genes with the same (incomplete) EC numbers from each other. Some spontaneous reactions also were included in our data set. The direction of reactions was made to conform to the direction of the arrow in the Roche Applied Science Biochemical Pathways chart (15). All reactions underwent the following process to detect the atomic correspondents between substrates and products on either side: (i) resolution of synonyms for molecule names in the data set, (ii) substitution of generic molecule names with concrete ones (e.g., ethanol or methanol for alcohol), (iii) balancing the number of atoms on either side (hydrogen atoms were not considered), and (iv) rearranging molecule orders so that their structures corresponded one-to-one on either side. The curated data set is available at www.metabolome.jp.
Definition of Atomic Correspondents. For a metabolic reaction to have an equilibrated mass balance, all atoms on the left must correspond one-to-one to the atoms on the right. In our analysis, such a structural relationship at the atomic scale is digitized as a set of atomic correspondents, i.e., atomic position pairs between substrates and products (Fig. 2). Atomic correspondents can be grouped for each substrate-product pair of molecules (see the color-coding in Fig. 2), and the set of position pairs for each color is called an atomic mapping. For example, there are three atomic mappings for the reaction of EC 2.3.1.35 (see the 9 red, 10 blue, and 3 green position pairs in Fig. 2). The atomic positions in each metabolite are labeled with the line numbers for atoms in its MOL file format structure file (16). The MOL file format is the de facto standard for describing molecular structures; it allows atoms in a molecule to be written in an arbitrary order. For this reason, information regarding atomic positions depends on our structure data and cannot be directly used in other databases. In summary, each reaction formula is decomposed to the corresponding atomic mappings, each of which represents a set of position pairs between a substrate-product pair.
Detection of Atomic Correspondents. After the reactions were manually curated, their atomic mappings were identified by a heuristic graph-matching algorithm and precompiled in our data set. Because water molecules are often involved in metabolic reactions and complicate the detection of oxygen and hydrogen traces, atomic correspondents of these atomic elements were not verified in the data set; the mapping information was archived only for carbon, nitrogen, and sulfur atoms. The computed atomic mappings were manually verified and, if necessary, corrected; ≈2% of our computed results required correction. We previously reported our method for compiling the atomic mappings for >2,500 reactions; details of the graph-matching process and the breakdown list of manual corrections are given in ref. 10.
The number of collected reactions in the data set was similar to a previous report (17) except for hydrolases catalyzing the hydrolysis of various chemical bonds (EC class 3), which were significantly underrepresented in our data. Typical enzymes in this class include peptidases and glycosylases, and many functions of these enzymes cannot be described in the form of equations; they are represented by textual descriptions like “release of an N-terminal amino acid, Xaa † Xbb-, in which Xaa is preferably Leu, but may be..., and Xbb may be Pro... “ (Enzyme Nomenclature for EC 3.4.11.1; abbreviation is the author's). Thus, the atomic correspondents of these enzymes are inherently unavailable. Because of the similar unavailability of one-to-one atomic correspondents, polymerizing or ligating reactions such as “ATP + (DNA)n + (DNA)m = AMP + diphosphate + DNA(n+m)” (Enzyme Nomenclature for EC 6.5.1.1) cannot be accurately represented in our data. In principle, we considered only reaction formulas where all atoms on the left of an equation correspond one-to-one to the atoms on the right; we did not consider generic metabolites such as “DNA” or “phosphatidyl glycerol” (with two alkyl chains of variable length). Thus, our data were restricted to the small-molecule metabolism, smaller than most sugar chains and lipids. The mapping data and the molecular structures in MOL file format are available at www.metabolome.jp.
Graph Representation and Pathway Computation. The metabolic network of E. coli is represented as a directed graph (metabolic graph, hereafter) where nodes and edges correspond to metabolites and their atomic mappings, respectively (Fig. 3). The graph representation includes atomic-level information so that each carbon, nitrogen, and sulfur atom can be traced in the network. Depending on its reversibility, each atomic mapping was converted to one (if the mapping was unidirectional) or two (if the mapping was reversible) graph edges. Candidates for pathways were obtained by applying a shortest-paths algorithm to the metabolic graph (10). For a computed sequence of reactions, the set of atomic positions conserved throughout the sequence was validated by using the atomic mappings and the symmetry information of metabolites as follows.
Fig. 3.
Graph representation of the ornithine biosynthetic pathway. In ornithine biosynthesis, l-ornithine (l-Orn) is synthesized from l-glutamate (l-Glu) through five reactions. Red arrows indicate the transfer of the carbon skeleton of l-glutamate, blue arrows indicate the transfer of acetyl moiety, and green arrows indicate the transfer of a nitrogen atom from l-glutamate. (Left) In the traditional metabolic map, multiple substrates and products are involved in each reaction, and their structural relationships are implicit. (Right) In our graph representation, physically related metabolites are linked with atomic mappings (red and blue arrows), and each reaction corresponds to a set of mappings (orange links). Note that the mapping between l-glutamate and N-acetyl l-glutamate (N-Ace l-Glu) is shared by two reactions (EC 2.3.1.1 and EC 2.3.1.35). The mapping between l-ornithine and N-acetyl l-ornithine (N-Ace l-Orn) is also shared. p, Phosphate; sa, semialdehyde.
We use an example in Fig. 4 to explain the validation method of pathways. The example sequence contains two atomic mappings, one from EC 1.1.1.38 between pyruvate and l-malate and the other from EC 4.2.1.2 between l-malate and fumarate. For a carbon position of the starting compound, e.g., position 0 in pyruvate, its transferred positions through the mappings were computed until the end of the reaction sequence. In Fig. 4, position 0 of l-malate and position 2 of fumarate originated in position 0 of pyruvate (positions 6, 7, and 8 in l-malate correspond to a carbon dioxide). In each mapping transfer, the information of metabolite symmetry was used to distribute candidate positions to their equivalents: because position 2 in fumarate is structurally equivalent to position 3 in the same compound, both positions correspond to position 0 in l-malate and, then, in pyruvate. The information on the structural symmetry of molecules is thus necessary to verify whether a given sequence of atomic mappings forms a pathway, and, in our data set, the symmetry information taking account of configurations (in D or L form) and aromaticity was precompiled for all registered metabolites. The information was used by ARM (available at www.metabolome.jp) to search for pathways between two given compounds in the metabolic graph (10).
Fig. 4.
Pathway from pyruvate to fumarate. Highlighted positions show the traces of two carbon atoms in pyruvate (positions 0 and 1). Because the two positions in fumarate (2 and 3) are equivalent, all highlighted positions become equivalent when reactions are considered reversible.
Results
The 853 total annotations for E. coli metabolic genes accounted for 1,004 reaction formulas in 614 EC enzyme subsubclasses. These reactions were converted into 1,230 atomic mappings among 906 metabolites for carbon and nitrogen metabolism. Of 1,230 mappings, 1,179 accounted for carbon-carbon relationships among 905 metabolites (the only excluded metabolite was ammonia). To obtain the distance between reachable metabolites, the shortest pathway between all pairs of carbon-containing metabolites was computed at the atomic scale. The distribution of pathway length between all pairs of metabolites is shown in Fig. 5. When no pathway was found, the length was considered 0 and excluded from the statistic. When the reversibility of the reactions was made to conform to the direction of the arrow in the Roche Applied Science Biochemical Pathways chart, 362 reactions were irreversible, and the AL became 8.4. When all edges were considered reversible, the AL decreased to 8.0. In both interpretations, the lengths of computed pathways yielded the same distribution. Thus, with our current metabolic information, the AL of the E. coli network remains ≈8, much larger than that of a random graph (18, 19). The metabolic world of E. coli is therefore not small with respect to biosynthesis/degradation pathways on the traditional metabolic map. When substrates and products in the same reaction were completely linked, and no structural information was considered in the pathway computation (Fig. 1 Lower Left), the AL became 3.2, the same value as that reported by Jeong et al. (7).
Fig. 5.
Distribution of pathway length. Filled bars indicate the population of pathways when the direction of reactions is considered (i.e., directed graph). Open bars indicate the population when all reactions are considered reversible (undirected graph).
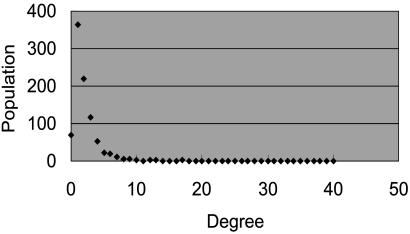
The distribution of out-degrees is shown in Fig. 6. Carbon dioxide, at 33, has the highest degree; next, in order of degree, are pyruvate, at 28; acetyl CoA, at 27; and ATP and d-glucose, both at 17. The degree was determined by the number of structural changes, which is not equal to the number of reactions where the molecule appears. For example, when a molecule X is split into two molecules Y and Z during a reaction, we counted two edges for the reaction: one from X to Y and the other from X to Z. In Fig. 3, the degree of l-glutamate is 2 (one to N-acetyl l-glutamate and the other to α-keto glutarate) although the molecule appears in three reactions. Note that edges may be shared by multiple reactions in our scheme. The edges between major cofactors, e.g., the reversible mapping between NAD and NADH or the mapping between ATP and ADP, are prevalent in the metabolic reactions, but even such a mapping is represented as only two edges (for both directions) in the metabolic graph.
Fig. 6.
Degree distribution of the graph. Degree corresponds to the number of structural changes, not frequencies.
Discussion
In the conventional graph representation, the degree of nodes corresponds to the frequency of metabolites in reaction formulas (4-9). However, their frequent appearance in reactions does not necessarily imply their biological centrality. Moreover, the translation from generic names such as l-amino acid or alcohol into specifics may bias the statistic. From a biochemical perspective, a better alternative is to focus on the pattern of structural changes of metabolites. By counting the changing patterns for each metabolite where it acts as a substrate, the most versatile metabolites (hubs) were determined to be carbon dioxide, pyruvate, acetyl CoA, ATP, d-glucose, and l-glutamate, in that order (Table 1). The biological centrality of these metabolites has been previously acknowledged (4, 6). Because the structural information is also indispensable for finding alternative metabolic routes (20, 21) or for detecting subnetworks (6, 9, 22-24), the reevaluation of previous approaches in our style may provide new perspectives.
For several reasons it is not appropriate to evaluate scalefreeness by using our graph representation. First, the proposed method represents only the number of structural changes without any quantitative aspect. The mapping between a frequently used substrate-product pair is treated as equal to the mapping between a less used pair, although they clearly have different biological roles and importance. Second, the number of edges (and thus degrees) is too small in our graph. The proposed representation is, in a way, a compressed description of a metabolic network with focus on metabolite structures only. The information on the structural changes of substrates does not suffice to fully delineate the biological importance of reactions. For the same reason, it is difficult to estimate the evolutionary importance of metabolites or the robustness of the network from our representation. Even when a certain atomic mapping is shared by multiple reactions, this does not imply that the atomic mapping (or its function) is compensated by other reactions. In this respect, our method must be used in conjunction with other informative strategies.
Although we collected known metabolic data from multiple data sources, our results identified a huge deficit in the metabolic information on E. coli (17, 25). Because d-glucose serves as the sole carbon source for E. coli, all carbon atoms in metabolites must be reachable from d-glucose (26). However, in our computation, even when all reactions are considered reversible, all carbon atoms of only half the metabolites (n = 454) are reachable from d-glucose. This finding is partly due to an artifact in our pathway-finding algorithm, because it does not allow the same compound to appear iteratively in the same pathway. (This constraint is assigned to accelerate the computation. Refer to ref. 10 for details.) In reality, however, many carbon atoms may become reachable by going through the TCA cycle and other cyclic pathways. However, inspection of the pattern of structural changes for metabolites also disclosed that half the metabolites (n = 484) have only one structural change or appear in a single reaction. Although the existence of such “dead-end” metabolites is not anomalous, more metabolites can be expected to be involved in multiple reactions.
When we computed the reachability of at least one carbon atom starting from any single metabolite, the 906 metabolites we assessed were largely separated into two groups; approximately one-third of metabolites (n = 331) could reach 10 or fewer metabolites, whereas 400 could reach >540 metabolites. Largely, the former group accounts for nonfunctional or isolated reactions, in part because the assignment system is based on the EC numbers, in part because of the unavailability of atomic mappings for certain reaction classes, and in part because of our computation artifact, generated by barring cyclic pathways. The latter, well connected group forms a core network responsible for the primary metabolism. To investigate the effect of such a deficit on network analyses, including this work, we must complete the ongoing annotation and supply appropriate reactions by considering atomic-level information.
In conclusion, metabolic pathway discussions should not be based on substrate-level network topology. Because the superficial connectivity on metabolic maps does not always correspond to pathways, structural information of metabolites is indispensable for computing biochemical pathways. Our atomic-level analysis complements virtually any metabolism-related study, from gene annotation to network evolution.
Supplementary Material
Acknowledgments
I thank Yukiko Nakanishi (Intec Web and Genome Informatics Corporation) for data curation, Ursula Petralia for editing the manuscript, and two anonymous referees for detailed comments that improved the final version.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: AL, average path length.
References
- 1.Watts, D. J. & Strogatz, S. H. (1998) Nature 393, 440-442. [DOI] [PubMed] [Google Scholar]
- 2.Strogatz, S. H. (2001) Nature 410, 268-276. [DOI] [PubMed] [Google Scholar]
- 3.Girvan, M. & Newman, M. E. (2002) Proc. Natl. Acad. Sci. USA 99, 7821-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fell, D. A. & Wagner, A. (2000) Nat. Biotechnol. 18, 1121-1122. [DOI] [PubMed] [Google Scholar]
- 5.Wagner, A. & Fell, D. A. (2001) Proc. R. Soc. London Ser. B 268, 1803-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravasz, E., Somera, A. L., Mongru, D. A., Oltvai, Z. N. & Barabási, A. L. (2002) Science 297, 1551-1555. [DOI] [PubMed] [Google Scholar]
- 7.Jeong, H., Tombor, B., Albert, R., Oltvai, Z. N. & Barabási, A. L. (2000) Nature 407, 651-654. [DOI] [PubMed] [Google Scholar]
- 8.Ma, H.-W. & Zeng, A.-P. (2003) Bioinformatics 19, 270-277. [DOI] [PubMed] [Google Scholar]
- 9.Ma, H.-W. & Zeng, A.-P. (2003) Bioinformatics 19, 1423-1430. [DOI] [PubMed] [Google Scholar]
- 10.Arita, M. (2003) Genome Res. 13, 2455-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa, M., Goto, S., Kawashima, S. & Nakaya, A. (2002) Nucleic Acids Res. 30, 42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karp, P. D., Riley, M., Saier, M., Paulsen, I. T., Collado-Vides, J., Paley, S. M., Pellegrini-Toole, A., Bonavides, C. & Gama-Castro, S. (2002) Nucleic Acids Res. 30, 56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schomburg, I., Chang, A., Hofmann, O., Ebeling, C., Ehrentreich, F. & Schomburg, D. (2002) Trends Biochem. Sci. 27, 54-56. [DOI] [PubMed] [Google Scholar]
- 14.International Union of Biochemistry and Molecular Biology (1992) Enzyme Nomenclature 1992 (Academic, San Diego).
- 15.Michal, G., ed. (1999) Biochemical Pathways: An Atlas of Biochemistry and Molecular Biology (Wiley & Spektrum, Heidelberg).
- 16.Dalby, A., Nourse, J. G., Hounshell, W. D., Gushurst, A. K. I., Grier, D. L., Leland, B. A. & Laufer, J. (1992) J. Chem. Inf. Comput. Sci. 32, 244-255. [Google Scholar]
- 17.Ouzounis, C. A. & Karp, R. D. (2000) Genome Res. 10, 568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung, F. & Lu, L. (2003) Internet Math. 1, 91-114. [Google Scholar]
- 19.Aiello, W., Chung, F. & Lu, L. (2001) Exp. Math. 10, 53-66. [Google Scholar]
- 20.Küffner, R., Zimmer, R. & Lengauer, T. (2000) Bioinformatics 16, 825-836. [DOI] [PubMed] [Google Scholar]
- 21.Kitami, T. & Nadeau, J. H. (2002) Nat. Genet. 32, 191-194. [DOI] [PubMed] [Google Scholar]
- 22.Schuster, S., Pfeiffer, T., Moldenhauer, F., Koch, I. & Dandekar, T. (2002) Bioinformatics 18, 351-361. [DOI] [PubMed] [Google Scholar]
- 23.Gagneur, J., Jackson, D. B. & Casari, G. (2003) Bioinformatics 19, 1027-1034. [DOI] [PubMed] [Google Scholar]
- 24.Stelling, J., Klamt, S., Bettenbrock, K., Schuster, S. & Gilles, E. D. (2002) Nature 420, 190-193. [DOI] [PubMed] [Google Scholar]
- 25.Saqi, M. A. S. & Sternberg, M. J. E. (2001) J. Mol. Biol. 313, 1195-1206. [DOI] [PubMed] [Google Scholar]
- 26.Neidhardt, F. C., Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E., eds. (1996) Escherichia coli and Salmonella (Amer. Soc. Microbiol., Washington, DC), pp. 189-198.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.